Abstract
Background
TGFβ is critical to control hepatocyte proliferation by inducing G1-growth arrest through multiple pathways leading to inhibition of E2F transcription activity. The retinoblastoma protein pRb is a key controller of E2F activity and G1/S transition which can be inhibited in viral hepatitis. It is not known whether the impairment of pRb would alter the growth inhibitory potential of TGFβ in disease. We asked how Rb-deficiency would affect responses to TGFβ-induced cell cycle arrest.
Results
Primary hepatocytes isolated from Rb-floxed mice were infected with an adenovirus expressing CRE-recombinase to delete the Rb gene. In control cells treatment with TGFβ prevented cells to enter S phase via decreased cMYC activity, activation of P16INK4A and P21Cip and reduction of E2F activity. In Rb-null hepatocytes, cMYC activity decreased slightly but P16INK4A was not activated and the great majority of cells continued cycling. Rb is therefore central to TGFβ-induced cell cycle arrest in hepatocytes. However some Rb-null hepatocytes remained sensitive to TGFβ-induced cell cycle arrest. As these hepatocytes expressed very high levels of P21Cip1 and P53 we investigated whether these proteins regulate pRb-independent signaling to cell cycle arrest by evaluating the consequences of disruption of p53 and p21Cip1. Hepatocytes deficient in p53 or p21Cip1 showed diminished growth inhibition by TGFβ. Double deficiency had a similar impact showing that in cells containing functional pRb; P21Cip and P53 work through the same pathway to regulate G1/S in response to TGFβ. In Rb-deficient cells however, p53 but not p21Cip deficiency had an additive effect highlighting a pRb-independent-P53-dependent effector pathway of inhibition of E2F activity.
Conclusion
The present results show that otherwise genetically normal hepatocytes with disabled p53, p21Cip1 or Rb genes respond less well to the antiproliferative effects of TGFβ. As the function of these critical cellular proteins can be impaired by common causes of chronic liver disease and HCC, including viral hepatitis B and C proteins, we suggest that disruption of pRb function, and to a lesser extend P21Cip1 and P53 in hepatocytes may represent an additional new mechanism of escape from TGFβ-growth-inhibition in the inflammatory milieu of chronic liver disease and contribute to cancer development.
Background
Transforming growth factor β (TGFβ) has characteristically diverse biological effects. Depending on the cell type and state of differentiation, TGFβ can either stimulate or inhibit proliferation, affect differentiation, promote extracellular matrix (ECM) formation and epithelial-to-mesenchymal transition (EMT), regulate cell adhesion, promote or inhibit cell migration and induce apoptosis (reviewed in reference [1]). In the liver, TGFβ is a critical mediator of multiple responses to injury [2]. Liver cell death in acute and chronic liver diseases is accompanied by inflammatory cell infiltration of the parenchyma and cytokine release including TGFβ. In such settings there develops an autocrine release of TGFβ from activated stellate cells, stimulating synthesis of ECM resulting in fibrosis [3]. At the same time, while hepatocytes and other intrinsic liver cells are stimulated to proliferate to compensate for cell loss, TGFβ is one of the signals that limit the proliferation of regenerating hepatocytes [4]. In advanced human liver cancer (hepatocellular carcinoma, HCC), there is commonly ectopic TGFβ production by the malignant hepatocytes in addition to that released by the non-parenchymal cells ([5,6]), giving rise to the idea that HCC cells have acquired a resistance to TGFβ-mediated growth inhibition. Current evidence suggests there is heterogeneity of resistance mechanisms that include decreased TGFβ-receptor II expression in early and late stage HCC [6] or induction of the inhibitory SMAD7 in advanced HCC [6-8]. These two adaptations have not been found in premalignant hepatocytes (dysplastic foci and regenerative nodules) in the chronically diseased liver, which are nevertheless similarly exposed to local TGFβ. It is reasonable to suppose that any resistance of pre-malignant hepatocytes to the anti-proliferative effects of TGFβ is likely to provide selective growth advantage in chronic liver disease favouring expansion into dysplastic nodules that are the precursor of HCC.
Studies in a variety of epithelial cells, including hepatocytes, have suggested at least two interconnected mechanisms by which TGFβ normally inhibits proliferation: downregulation of c-myc in early G1 and inhibition of cyclin-dependent kinase (CDK) activities by regulation of cyclin-dependent kinase inhibitors (CDKI) ([9] and therein) leading to the maintenance of pRb in the active hypophosphorylated form ([10]) and inhibition of S phase entry. These pathways point at critical proteins whose function is often altered during hepatocarcinogenesis, specifically the tumor suppressor pRb and P53 and the CDKI P21Cip1 [10-13]. pRb, P53 and P21Cip1are critical regulators of the cell cycle that have all been involved in the antiproliferative effect of TGFβ in various systems. However, the fact that pRb phosphorylation is the main target of the regulation G1/S progression by P53 and P21Cip1 makes it difficult to identify other possible pathways, independent of pRb.
Interestingly, both hepatitis B and hepatitis C viruses (HBV and HCV) express proteins that decrease expression or inhibit the function of pRb [14-16], P53 [17-19] and P21Cip1 [20-22]. We hypothesised that such dysfunctional pRb, P53 and P21Cip1 in chronic liver disease reduce the growth inhibitory response of affected hepatocytes to the TGFβ-rich environment of cirrhosis [5,6]. Using primary murine hepatocytes deficient in these genes singly or in combination we sought to determine firstly whether there was a loss of sensitivity to TGFβ-mediated cell cycle arrest and apoptosis and also to determine the relative contribution from each of p53, p21Cip1 and Rb-deficiency.
Methods
Hepatocyte culture
Mouse primary hepatocytes (male, 6–12 weeks old), were isolated by a standard two-steps retrograde perfusion procedure [23] and purified using percoll gradient [24]. The obtained hepatocytes were cultured in supplemented serum-free medium selecting against survival of non-parenchimal cells [25,26]. Where appropriate, hepatocytes cultured for 24 hours were treated daily with 160 pM of TGF-β1 (TGFβ) for the indicated time.
Mice were produced by crossing p53-/- [27] with Rb-floxed mice (homozygous for exon 19 of Rb flanked by LoxP sequences) [28] and p21Cip1-null mice (p21-/-) [29] as previously described [30]. Rb-deficient isogenic cells were obtained by deletion of the Rb-floxed alleles in vitro by infection with an adenovirus expressing Cre-recombinase (Ad-Cre) using a multiplicity of infection of 10 [30]. Control cells, infected with a replication-deficient adenovirus (Ad-Dl70) are phenotypically wild-type. All animals used in this study received humane care. The study protocols are in compliance with the UK Home Office regulation and the local institutional policies.
Proliferation
In the present isolation and culture conditions, isolated hepatocytes are more or less synchronous, with the majority of cycling wild type cells entering S phase 72 hours after plating and going into mitosis around 96 hours after plating [25,31]. As TGFβ inhibits proliferation via G1 block, we assessed changes in proliferation by quantifying the number of hepatocytes undergoing replicative DNA synthesis (S phase) by immunodetection of BrdU incorporation. Briefly, hepatocytes were incubated with 40 μM BrdU for 6 hours and fixed in 80% ethanol. Immunodetection was performed using Rat anti-BrdU IgG (SeraLabs, Sussex) 1/100 dilution and rabbit anti-rat IgG HRP-conjugate 1/100 dilution. Slides were counter-stained with haematoxylin and light-green. Negative controls omitted BrdU.
To compare the effect of TGFβ in hepatocytes of different genotypes, we calculated the percentage of inhibition of proliferation by TGFβ for 2 (for p53p21-/- and TRPL) to 6 independent experiments: proliferation was integrated between 48 and 96 hours after plating (i.e. cells treated or not with TGFβ for 24 to 72 hrs) using Kaleidagraph (SynergySoftware) giving the "mean proliferation" between these time points. The inhibition of proliferation was calculated as 100-(100*(mean proliferation of TGFβ-treated cells/mean proliferation of untreated cells)).
Immunofluorescence
Hepatocytes were fixed in acetone/methanol (1:1 v/v). Immunodetection was performed using anti-P53 mouse monoclonal antibody (1:1000) (AB-1, OncogeneScience UK), rat anti-P16INK4A polyclonal (AB3004, Chemicon), monoclonal mouse anti-human-P21Cip1 (SX118, Dako), mouse monoclonal anti-P27KIP1 (BD-Pharmingen), the appropriate Alexafluor (Molecular Probes) secondary antibody (1/200), followed by Topro-3 nuclear counterstain. Quantification was performed by manual counting on 25 representative fields (×40) photographed with a Zeiss confocal microscope. Scanning was performed using multitracking, and settings constant throughout the experiments.
E2F and MYC transcriptional activity
Hepatocytes in culture for 48 hours were transfected using TFx-50 (Promega) reagent (ratio 1/5 w/w DNA/lipid) [32]) with p-TA-Luc (control reporter), p-E2F-TA-Luc (E2F reporter), or p-myc-TA-Luc (c-myc reporter) (all from Pathway Profiling System4, Clontech). All drive the firefly luciferase gene (Luc) (for detailed map see [33]). Thirty hours after transfection, luciferase activity was quantified using Luciferase Assay reagents according to the manufacturer's instructions (Promega) and corrected for the quantity of protein (Biorad assay). The data are given relative to the expression in untreated control cells.
Gene expression
Total RNA was isolated at indicated times using QIAGEN RNeasy mini Kit according to the manufacturer's instructions. RNA quality was determined with a Bioanalyzer (RNA6000 NanoLabChip kit, Agilent 2100 Bioanalyzer, USA). Expression analysis was performed using the GEArrayQ Series Mouse Cell Cycle Kit, (Superarray, USA) where each gene is represented by 4 independent spots. The cDNA was prepared from total RNA using Superarray AmpoLabeling-LPR Kit, USA and labelled with Biotin-16-dUTP (Rocha).
Images of the arrays were obtained using a Versadoc detector (BioRad, UK) and converted into raw data using Scanlyzer (Michael Eisen, Lawrence Berkely Bation Lab, USA). The data was analysed using the GEArrayAnalzer Software (Version 1.0) with background subtraction (using plasmid DNA PUC18 as negative control) and normalisation with the housekeeping gene Ppia (cyclophlinA). The normalization removes differing intensity scales from the experimental readings, allowing comparison between experiments.
Statistical analyses
Data and statistical analyses were done with Minitab 13.0 and Spotfire Decision site softwares. The proportion of affected cells was arcsine transformed to normalise the distribution, and differences between means were evaluated with Analysis of Variance (ANOVA). Differences were taken to be significant when p < 0.05. Satisfactory homogeneity of variances was determined with Bartlett's test. Where a significant difference between means was identified with ANOVA, the differences between individual means were analysed further with Bonferroni simultaneous tests for multiple comparisons.
Results
pRb and other pocket proteins are central to the regulation of G1/S transition by inhibition of E2F activity and transcription of multiple target genes involved in DNA synthesis and cell cycle regulation. In Rb-deficient hepatocytes, although E2F activity is elevated [30], we found using gene expression array (see methods) that expression of p107 and p130 were increased (2.1, 2 and 3.1 fold for p107; 1.3, 1.1 and 1.4 fold for p130 at 48, 72 and 96 hours after plating). This may help maintain some regulation of G1/S transition including inhibition of proliferation by TGFβ. Studies in a variety of epithelial cells, including hepatocytes, have shown at least two interconnected mechanisms by which TGFβ normally inhibits proliferation: downregulation of c-myc in early G1 and inhibition of cyclin-dependent kinase (CDK) activities by regulation of cyclin-dependent kinase inhibitors (CDKI) ([9] and therein) leading to the maintenance of pRb, and other pocket proteins in the active hypophosphorylated form [10] and inhibition of E2F responsive promoters. We asked what would be the consequences of Rb deletion on TGFβ regulation of hepatocytes proliferation.
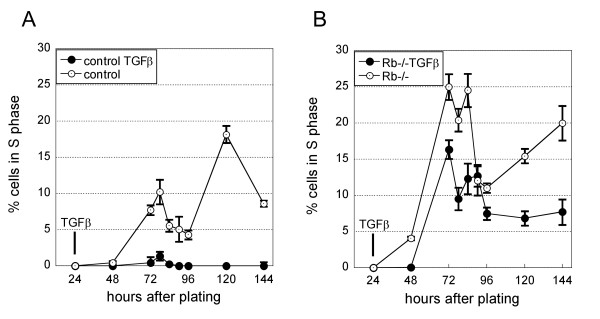
In our culture conditions, primary hepatocytes enter S phase in a more or less synchronous manner. The first control cells reach S phase around 72 hours after plating (Figure 1A) and M phase is observed around 96 hours [25,30,31]. Hepatocytes can enter a second cell cycle but often in a less synchronous manner. As we have previously reported [30] following Rb deletion within the first 24 hours after plating a higher number of cells enter S phase and there is an earlier onset of DNA synthesis which is detected as soon as 48 hours after plating (Figure 1A & B compare curves with open symbols).
Figure 1.
Rb deficiency reduces TGFβ inhibition of proliferation. The figure shows the percentage of cells in S phase at the indicated times after plating of one representative experiment. All hepatocytes for the experiment were isolated from the same Rb-floxed mouse. Control and Rb-/- cells are Rb-floxed cells treated at the time of plating with either adenovirus control (wild-type phenotype) (A); or adenovirus expressing Cre (Rb-/-) (B) respectively. Each point is the average proliferation in 2 independent cultures where 500 hepatocytes were counted +/- SEM. The experiment was repeated multiple times with similar results. Where appropriate (close symbols), TGFβ was added daily from 24 hours after plating.
Rb is central to TGFβ-induced inhibition of proliferation
In these conditions, TGFβ-treatment of control cells was highly effective to cause growth arrest almost completely blocking proliferation (Figure 1A). By contrast, many of the sister cells subjected to inducible Rb-deletion escaped the inhibition by TGFβ and entered S phase (Figure 1B).
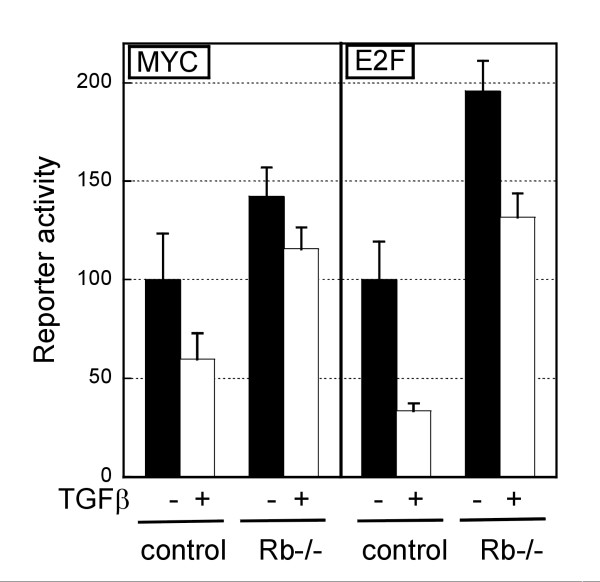
E2F and MYC activities were found to be significantly higher in Rb-null cells compared with wild type (Figure 2, compare black bars). This was detectable from 48 hours for E2F (data not shown) and 72 hours after plating for MYC (Figure 2). TGFβ-treatment decreased MYC activity in both control and Rb-null hepatocytes, although this was less efficient in Rb-null cells (40.3% and 18.5% decrease for control and Rb-null respectively) and the activity remained higher than in untreated control hepatocytes (Figure 2). E2F activity also decreased after TGFβ-treatment in cells of either genotype (32.6 in Rb-/- and 66.4% in control).
Figure 2.
TGFβ affects c-MYC and E2F transcriptional activity. The graph represents the transcriptional activities of c-MYC and E2F quantified using a reporter assay. Control and Rb-/- hepatocytes were in culture for 78 hours, treated or not with TGFβ for 30 hours at the time of the assay. The bars represent the average +/- SEM of duplicate transfections.
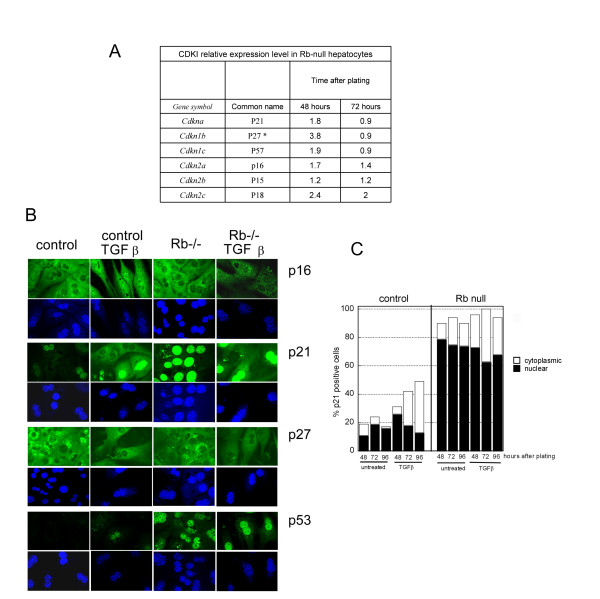
MYC is a negative regulator of CDKI expression [34-37]. Accordingly, we found that the high levels of CDKI expression initially observed in response to Rb deletion (48 after plating), returned to wild type levels at 72 hours, together with the increased MYC activity (Figure 2, Figure 3A). We can only speculate about the mechanism behind the initial increase of CDKI expression, but the suggestion that E2F activity can regulate CDKI expression [38] suggests that this may be an early response to the rapid increase of E2F activity resulting from the induced Rb deletion, before the system reaches an equilibrium.
Figure 3.
Effect of TGFβ on P53, P16INK4A, P21Cip1 and P27KIP1 expression A: Expression of CDKI in Rb-null hepatocytes. The table gives the level of expression of the various CDKI in Rb-null hepatocytes relative to control hepatocytes. The values were obtained using a gene expression array as described in methods. * The level of p27KIP1 expression in control cells was low, and the ratio may therefore be overestimated. B & C. Hepatocytes were treated or not with TGFβ 24 hours after plating. B. Immunofluorescence for P16INK4A, P21Cip1, P27KIP1 and P53. Photos were taken 48 hours after treatment. Green: specific immunofluorescence, blue: Topro-3 nuclear counterstain. C: Quantification of P21Cip1 immunopositivity. Black bars: percentage of cells exhibiting nuclear staining. White bars, cytoplasmic staining.
Decreased MYC expression, as observed here in response to TGFβ is known to alleviate inhibition of transcription of various CDKI. We therefore investigated CDKI expression in response to TGFβ. We were unable to detect P15INK4B in hepatocytes of either genotype; P27KIP1 expression was similar in control and Rb-null cells and was not affected by TGFβ (Figure 3B). P16INK4A appeared to localise in the nuclei after TGFβ-treatment of control but not Rb-null cells (Figure 3B). P16INK4A inhibits CDK4 and CDK6 that are known to phosphorylate pockets proteins. P16INK4A may therefore contribute to TGFβ-induced cell cycle arrest by inhibition of pocket protein phosphorylation and reduction of E2F activity in control but not in Rb-null cells. In control cells TGFβ-treatment was also accompanied by early activation of P21Cip1 (Figure 3B & C) which relocalised in the cytoplasm: the percentage of nuclear positivity increased initially to rapidly drop back to the level of untreated cells but with a concomitant increase of cytoplasmic P21Cip1-staining (Figure 3B, 3C) from 72 hours after plating. In Rb-null cells, which show a constitutive high level of P21Cip1 due to P53 activation [30], there was no change in nuclear positivity but an increased number of cells exhibited cytoplasmic staining (Figure 3B, 3C). Finally, TGFβ marginally affected P53, as few strongly positive cells were observed in TGFβ-treated control cells (<2%) and the small proportion of "less positive"Rb-null hepatocytes became intensively fluorescent with TGFβ-treatment (Figure 3B).
Some Rb-deficient hepatocytes remain sensitive to TGFβ-induced inhibition of proliferation through a P53-dependent pathway
Interestingly, a certain proportion of Rb-/- hepatocytes were nevertheless prevented to by TGFβ from entering S phase (Figure 1B) as the number of cells incorporating BrdU is reduced (Figure 1B compare curves with open and black symbols).
As Rb-null greatly differed from control hepatocytes in respect of the high level of P21Cip1and P53, we investigated whether these proteins could contribute to TGFβ-induced cell cycle arrest independently of pRb. To address this question, we compared the inhibition of proliferation in hepatocytes knocked-out for these genes, undergoing or not a further inducible deletion of the Rb gene.
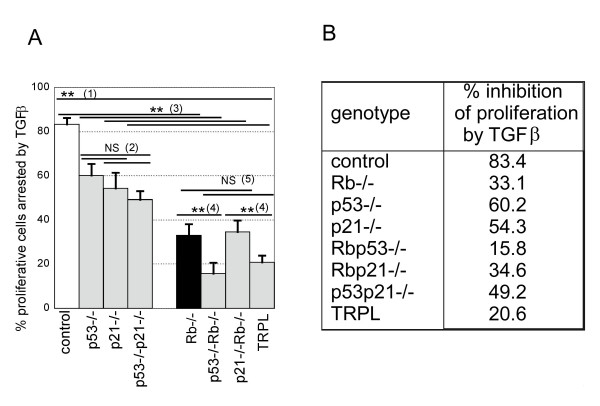
p21Cip1 and p53 deficiencies similarly affected cell responses to TGFβ with about 55–60% of the proliferating cells being inhibited by TGFβ (Figure 4 legend (2)). Simultaneous deficiencies in both p21Cip1 and p53 did not significantly differ from either alone (Figure 4 legend (2)). As expected Rb-deficiency had the strongest effect, sharply decreasing the susceptibility to TGFβ-induced cell cycle arrest regardless of p53 and p21Cip1 genotypes (Figure 4 legend (3) all p < 0.0001). This confirms the central role played by pRb in the response to TGFβ.
Figure 4.
Rb is central to TGFβ inhibition of proliferation via multiple pathways. A. The figure shows the effect of p53, p21Cip1 or Rb deficiency on TGFβ induced cell cycle arrest +/- SDV. The percentage inhibition of proliferation by TGFβ was calculated for 2 (for p53p21-/- and TRPL) to 6 independent experiments and differences analysed by ANOVA (** p < 0.0001; NS non significant). As for all experiments the Rb-null hepatocytes were obtained by infection at the time of plating of the Rb-floxed hepatocytes of corresponding genotypes with adenovirus expressing Cre recombinase. (1) all deficient hepatocytes respond less well to inhibition of proliferation by TGFβ than control cells and Rb deficient has the strongest effect. (2) p53 and p21Cip1deficiency, singly or together have a similar effect on inhibition of proliferation by TGFβ. (3) Rb deletion significantly reduces TGFβ-induced cell cycle arrest regardless of p53 and p21Cip1status (compare control with Rb-/-; p53-/- with p53-/-Rb-/-; p21-/- with p21-/-Rb-/- and p53-/-p21-/- with TRPL). (4) double deficiency in Rb and p53 further decreases hepatocytes responses to TGFβ in term of regulation of proliferation, independently of p21Cip1status. (5) by contrast the effect of TGFβ on hepatocytes deficient in both p21Cip1 and Rb is not significantly different to that of Rb null cells, and this is independent of p53 status. B. corresponding percentages of inhibition of proliferation for each genotype. TRPL: triple null hepatocytes.
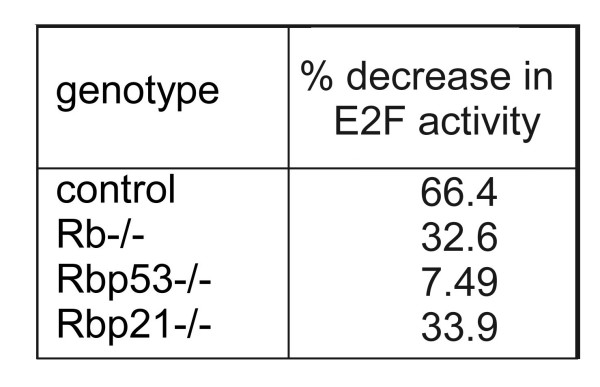
In Rb-null cells additional loss of p53 (Figure 4 legend (4)) but not p21Cip1 caused a further reduction in sensitivity to TGFβ (Rb-/- versus Rb-/-p53-/-; p = 0.0001 and Rb-/- versus p21-/-Rb-/- p = 0.20). This was accompanied by a reduced inhibition of E2F activity by TGFβ in Rb-/- p53-/- but not in Rb-/-p21-/- (7.49% reduction in Rb-/- p53-/- and 33.9 in Rb-/-p21-/- versus 32.6 in Rb-/- hepatocytes) (Figure 5).
Figure 5.
Deficiency in p53 but not p21Cip1 further decreases E2F activity in Rb-deficient hepatocytes. The values are the percentage decrease in E2F activity in TGFβ-treated hepatocytes of different genotypes compared with untreated cells. The E2F activity was quantified using a reporter assay as described in figure 2.
Discussion
We have investigated here the consequences of Rb-deficiency in hepatocytes – as happens in viral liver diseases – with respect to TGFβ regulation of proliferation. Our data show that pRb is central to the anti-proliferative action of TGFβ with the majority of Rb-null cells escaping from this effect. This may have consequences for the development of cancer. We found however that a proportion of Rb-null hepatocytes remain sensitive to TGFβ and undergo cell cycle arrest and investigated whether P53 and P21Cip1could be involved.
TGFβ is a potent inhibitor of cell proliferation by activation of a cell cycle arrest in G1 through potentially multiple pathways: First, protein complexes containing SMAD3 [39-43] and P107 [44,45] downregulate MYC activity leading to CDKI upregulation ([1] and therein). The active CDKs that can feedback on SMAD3 to reduce its activity [46,47], are therefore kept in check by the CDKIs. Consistent with the involvement of P107 rather than pRb, we observed decreased MYC activity in both wild-type and Rb-null hepatocytes, albeit with somewhat reduced efficiency in the later. In the absence of P16INK4A activation, SMAD3 may be inhibited by CDKs thus preventing optimum inhibition of MYC [39,40] in Rb-deficient cells.
A reported effect of c-myc downregulation is to reduce P15INK4B repression and inhibit cyclinD/CDK4 and cyclinE/CDK2 by both direct binding and relocalisation of P27KIP1 from cyclinD/CDK4 to cyclinE/CDK2 [1,48]. Although in untreated Rb-null hepatocytes, all CDKI are overexpressed in G1, we were unable to detect expression of P15INK4B and did not observe any changes in P27KIP1 after TGFβ-treatment. P16INK4A which localised in the nuclei of control but not Rb-null cells after TGFβ-treatment can inhibit CDK4 & 6 and may therefore contribute to TGFβ-induced cell cycle arrest by inhibition of pocket protein phosphorylation and reduction of E2F activity in control but not in Rb-null cells.
Downregulation of c-myc also allows activation of p21Cip1[49], and our finding that TGFβ-treatment greatly increased nuclear P21Cip1 could certainly contribute to cell cycle arrest via inhibition of pRb phosphorylation in cells containing Rb.
By contrast, it is interesting that p21Cip1 deficiency in Rb-null hepatocytes did not reduce the sensitivity of hepatocytes to TGFβ (correlated with a similar decrease in level of E2F activity; 32.6 and 33.9% in Rb-/- and Rbp21-/- respectively). We have indeed previously shown that P21Cip1 provides pRb-independent control of hepatocytes proliferation: in standard culture conditions p21-/-Rb-/- hepatocytes proliferate more than hepatocytes bearing only one knock-out gene [30]. Various potential mechanisms have been discussed and include the inhibition of PCNA [50], or cyclin E/cdk2 [51,52] or reduction of MYC activity [53] by P21Cip1. The present results therefore suggest that whatever the mechanism involved, it is not enhanced by TGFβ-treatment and that proliferation rate and inhibition of proliferation by TGFβ are unrelated.
Thus activation of cyclin kinase inhibitors P21Cip1 and P16INK4A can therefore inhibit pRb and other pocket proteins phosphorylation and prevent E2F transcriptional activity leading to very efficient cells cycle arrest in control cells.
In Rb-null hepatocytes where neither CDKI seem involved after TGFβ-treatment, E2F activity was nevertheless reduced suggesting that a different mechanism may affect other pocket protein(s) and contribute to cell cycle arrest of Rb-null hepatocytes. P21Cip1 and P53 were both strongly increased in Rb-null hepatocytes and shown to change with TGFβ treatment; we therefore investigated if they could contribute to TGFβ-induced inhibition of proliferation through an Rb-independent pathway.
By comparing proliferative responses of hepatocytes of various genotypes, a P53-dependent, P21Cip1-independent pathway was highlighted. This was correlated with a greater reduction of E2F activity in Rb-/-p53-/- cells suggesting that high P53 prevents inhibition of E2F activity by TGFβ. This may involve P53-dependent inhibition of CDK4 expression – indeed we find that in p53-/- hepatocytes CDK4 expression in G1 and S phase is reduced to 76% and 72% of the level in control cells – or repression of CDK4 synthesis [54] and prevent E2F release from P107 which we have shown to be twofold increased in Rb-null cells.
Conclusion
Loss of responsiveness to TGFβ antiproliferative effects is believed to be important in carcinogenesis, yet the known mechanisms of TGFβ resistance happen late in the progression of established liver cancer [55,8]. The present results show that otherwise genetically normal hepatocytes with disabled p53, p21Cip1 or Rb genes respond less well (by differing degrees) to the antiproliferative effects of TGFβ. As the function of these critical cellular proteins can be impaired by common causes of chronic liver disease and HCC, including viral hepatitis B and C proteins [14-22], we suggest that disruption of pRb function, and to a lesser extend P21Cip1 and P53 in hepatocytes may represent an additional new mechanism of escape from TGFβ growth inhibition in the inflammatory milieu of chronic liver disease.
Abbreviations
Transforming growth factor β: TGFβ; 5-bromo-2-deoxyuridine: BrdU; Standard error of the mean: SEM; hepatocellular carcinoma: HCC; HCV: hepatitis C virus; HBV hepatitis B virus; CDK: cyclin-dependent kinase; CDKI: cyclin-dependent kinase inhibitor.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
SS carried initiated the study and carried out some of the experiments, COB contributed to the experimental design, interpretation of data, supervision and gave critical review of the manuscript, DRD helped with the interpretation of data using Spotfire Decision site software, DJH contributed to the experimental design and gave general supervision and funding support, SP made substantial contribution to the conception & design, acquisition and interpretation of data and wrote the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This work was supported by grants from the Melville Trust for the Care and Cure of Cancer to SP and SS. The p21Cip1-/- mice were kind gift from Philip Leder (Harvard Medical School, Boston, MA) and the Rb-floxed (Rblox/lox) mice from Anton Berns (Netherlands Cancer Institute, Amsterdam), to whom we are very grateful. We wish to thanks Helen Caldwell for technical assistance. The microarray data presented here were obtained by Zhenjie Xu under the supervision of Dr Pin Lu and SB. SS is currently at BioSciences Institute, University College Cork, Ireland. DRD is funded by The Wellcome Trust's Functional Genomics Development Initiative and Cardiovascular Research Initiative.
Contributor Information
Sharon Sheahan, Email: sheahan@hotmail.com.
Christopher O Bellamy, Email: c.bellamy@ed.ac.uk.
Donald R Dunbar, Email: donald.dunbar@ed.ac.uk.
David J Harrison, Email: david.Harrison@ed.ac.uk.
Sandrine Prost, Email: s.prost@ed.ac.uk.
References
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Breitkopf K, Godoy P, Ciuclan L, Singer MV, Dooley S. TGF-beta/Smad signaling in the injured liver. Z Gastroenterol. 2006;44:57–66. doi: 10.1055/s-2005-858989. [DOI] [PubMed] [Google Scholar]
- Gressner AM, Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-beta as major players and therapeutic targets. J Cell Mol Med. 2006;10:76–99. doi: 10.1111/j.1582-4934.2006.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oe S, Lemmer ER, Conner EA, Factor VM, Leveen P, Larsson J, Karlsson S, Thorgeirsson SS. Intact signaling by transforming growth factor beta is not required for termination of liver regeneration in mice. Hepatology. 2004;40:1098–1105. doi: 10.1002/hep.20426. [DOI] [PubMed] [Google Scholar]
- Bedossa P, Peltier E, Terris B, Franco D, Poynard T. Transforming growth factor-beta 1 (TGF-beta 1) and TGF-beta 1 receptors in normal, cirrhotic, and neoplastic human livers. Hepatology. 1995;21:760–766. [PubMed] [Google Scholar]
- Paik SY, Park YN, Kim H, Park C. Expression of transforming growth factor-beta1 and transforming growth factor-beta receptors in hepatocellular carcinoma and dysplastic nodules. Mod Pathol. 2003;16:86–96. doi: 10.1097/01.MP.0000047308.03300.9C. [DOI] [PubMed] [Google Scholar]
- Halder SK, Beauchamp RD, Datta PK. Smad7 induces tumorigenicity by blocking TGF-beta-induced growth inhibition and apoptosis. Exp Cell Res. 2005;307:231–246. doi: 10.1016/j.yexcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Park YN, Chae KJ, Oh BK, Choi J, Choi KS, Park C. Expression of Smad7 in hepatocellular carcinoma and dysplastic nodules: resistance mechanism to transforming growth factor-beta. Hepatogastroenterology. 2004;51:396–400. [PubMed] [Google Scholar]
- Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Laiho M, DeCaprio JA, Ludlow JW, Livingston DM, Massague J. Growth inhibition by TGF-beta linked to suppression of retinoblastoma protein phosphorylation. Cell. 1990;62:175–185. doi: 10.1016/0092-8674(90)90251-9. [DOI] [PubMed] [Google Scholar]
- Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25:3778–3786. doi: 10.1038/sj.onc.1209547. [DOI] [PubMed] [Google Scholar]
- Lee TK, Man K, Poon RT, Lo CM, Ng IO, Fan ST. Disruption of p53-p21/WAF1 cell cycle pathway contributes to progression and worse clinical outcome of hepatocellular carcinoma. Oncol Rep. 2004;12:25–31. [PubMed] [Google Scholar]
- Suriawinata A, Xu R. An update on the molecular genetics of hepatocellular carcinoma. Semin Liver Dis. 2004;24:77–88. doi: 10.1055/s-2004-860865. [DOI] [PubMed] [Google Scholar]
- Cho J, Baek W, Yang S, Chang J, Sung YC, Suh M. HCV core protein modulates Rb pathway through pRb down-regulation and E2F-1 up-regulation. Biochim Biophys Acta. 2001;1538:59–66. doi: 10.1016/S0167-4889(00)00137-3. [DOI] [PubMed] [Google Scholar]
- Choi BH, Choi M, Jeon HY, Rho HM. Hepatitis B viral X protein overcomes inhibition of E2F1 activity by pRb on the human Rb gene promoter. DNA Cell Biol. 2001;20:75–80. doi: 10.1089/104454901750070274. [DOI] [PubMed] [Google Scholar]
- Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2005;102:18159–18164. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Nagano-Fujii M, Tanaka M, Nomura-Takigawa Y, Ikeda M, Kato N, Sada K, Hotta H. NS3 protein of Hepatitis C virus associates with the tumour suppressor p53 and inhibits its function in an NS3 sequence-dependent manner. J Gen Virol. 2006;87:1703–1713. doi: 10.1099/vir.0.81735-0. [DOI] [PubMed] [Google Scholar]
- Gong GZ, Jiang YF, He Y, Lai LY, Zhu YH, Su XS. HCV NS5A abrogates p53 protein function by interfering with p53-DNA binding. World J Gastroenterol. 2004;10:2223–2227. doi: 10.3748/wjg.v10.i15.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Chen SY, Chen JY, Wu Lee YH. Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene. 2004;23:2472–2483. doi: 10.1038/sj.onc.1207368. [DOI] [PubMed] [Google Scholar]
- Ahn JY, Chung EY, Kwun HJ, Jang KL. Transcriptional repression of p21(waf1) promoter by hepatitis B virus X protein via a p53-independent pathway. Gene. 2001;275:163–168. doi: 10.1016/S0378-1119(01)00604-7. [DOI] [PubMed] [Google Scholar]
- Han HJ, Jung EY, Lee WJ, Jang KL. Cooperative repression of cyclin-dependent kinase inhibitor p21 gene expression by hepatitis B virus X protein and hepatitis C virus core protein. FEBS Lett. 2002;518:169–172. doi: 10.1016/S0014-5793(02)02694-7. [DOI] [PubMed] [Google Scholar]
- Kwun HJ, Jang KL. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Res. 2004;32:2202–2213. doi: 10.1093/nar/gkh553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton KW, Deloria LB, Mannering GJ. Effects of Polyriboinosinic Acid, Polyribocytidylic Acid and A Mouse Interferon Preparation on Cytochrome P-450-Dependent Mono-Oxygenase Systems in Cultures of Primary Mouse Hepatocytes. Molecular Pharmacology. 1978;14:672–681. [PubMed] [Google Scholar]
- Kreamer BL, Staecker JL, Sawada N, Sattler GL, Hsia MTS, Pitot HC. Use of A Low-Speed, Iso-Density Percoll Centrifugation Method to Increase the Viability of Isolated Rat Hepatocyte Preparations. In Vitro Cellular & Developmental Biology. 1986;22:201–211. doi: 10.1007/BF02623304. [DOI] [PubMed] [Google Scholar]
- Bellamy C. PhD Thesis. Edinburgh University; 1997. The roles of P53 in hepatocytes. [Google Scholar]
- Bellamy CO, Prost S, Wyllie AH, Harrison DJ. UV but not gamma-irradiation induces specific transcriptional activity of p53 in primary hepatocytes. J Pathol. 1997;183:177–181. doi: 10.1002/(SICI)1096-9896(199710)183:2<177::AID-PATH909>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Purdie CA, Harrison DJ, Peter A, Dobbie L, White S, Howie SE, Salter DM, Bird CC, Wyllie AH, Hooper ML. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene. 1994;9:603–609. [PubMed] [Google Scholar]
- Vooijs M, te RH, van d V, Berns A. Tumor formation in mice with somatic inactivation of the retinoblastoma gene in interphotoreceptor retinol binding protein-expressing cells. Oncogene. 2002;21:4635–4645. doi: 10.1038/sj.onc.1205575. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-X. [DOI] [PubMed] [Google Scholar]
- Sheahan S, Bellamy CO, Treanor L, Harrison DJ, Prost S. Additive effect of p53, p21 and Rb deletion in triple knockout primary hepatocytes. Oncogene. 2004;23:1489–1497. doi: 10.1038/sj.onc.1207280. [DOI] [PubMed] [Google Scholar]
- Sheahan S. PhD Thesis. Edinburgh University; 2002. The roles of p53, p21 and Rb in regulation of proliferation and apoptosis in hepatocytes. [Google Scholar]
- Prost S, Ford JM, Taylor C, Doig J, Harrison DJ. Hepatitis B x protein inhibits p53-dependent DNA repair in primary mouse hepatocytes. J Biol Chem. 1998;273:33327–33332. doi: 10.1074/jbc.273.50.33327. [DOI] [PubMed] [Google Scholar]
- 2006. http://www.clontech.com/products/detail.asp?tabno=2&catalog_id=631914&page=all [accessed 3 April 2007]
- Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL. Myc represses the p21((WAF1/CIP1)) promoter and interacts with Sp1/Sp3. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Shen J, Wu M, Arsura M, FitzGerald M, Suldan Z, Kim DW, Hofmann CS, Pianetti S, Romieu-Mourez R, Freedman LP, Sonenshein GE. Repression of transcription of the p27(Kip1) cyclin-dependent kinase inhibitor gene by c-Myc. Oncogene. 2001;20:1688–1702. doi: 10.1038/sj.onc.1204245. [DOI] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, Eilers M. Repression of p15(INK4b) expression by Myc through association with Miz-1. Nature Cell Biology. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, Kel-Margoulis OV, Farnham PJ, Bartley SM, Wingender E, Zhang MQ. Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J Mol Biol. 2001;309:99–120. doi: 10.1006/jmbi.2001.4650. [DOI] [PubMed] [Google Scholar]
- Feng XH, Liang YY, Liang M, Zhai W, Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B) Mol Cell. 2002;9:133–143. doi: 10.1016/S1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- Frederick JP, Liberati NT, Waddell DS, Shi Y, Wang XF. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol Cell Biol. 2004;24:2546–2559. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/S0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Kowalik TF. Smad about E2F. TGFbeta repression of c-Myc via a Smad3/E2F/p107 complex. Mol Cell. 2002;10:7–8. doi: 10.1016/S1097-2765(02)00584-1. [DOI] [PubMed] [Google Scholar]
- Liu F, Matsuura I. Inhibition of Smad antiproliferative function by CDK phosphorylation. Cell Cycle. 2005;4:63–66. doi: 10.4161/cc.4.1.1366. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta – induced cell-cycle arrest. Proc Natl Acad Sci USA. 2000;97:9498–9503. doi: 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunesku T, Mittal S, Protic M, Oryhon J, Korolev SV, Joachimiak A, Woloschak GE. Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int J Radiat Biol. 2001;77:1007–1021. doi: 10.1080/09553000110069335. [DOI] [PubMed] [Google Scholar]
- Lukas J, Herzinger T, Hansen K, Moroni MC, Resnitzky D, Helin K, Reed SI, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Bronson RT, Jacks T. p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J Cell Biol. 1998;141:503–514. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta. 2000;1471:M43–M56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- Ewen ME, Oliver CJ, Sluss HK, Miller SJ, Peeper DS. p53-dependent repression of CDK4 translation in TGF-beta-induced G1 cell-cycle arrest. Genes Dev. 1995;9:204–217. doi: 10.1101/gad.9.2.204. [DOI] [PubMed] [Google Scholar]
- Musch A, Rabe C, Paik MD, Berna MJ, Schmitz V, Hoffmann P, Nischalke HD, Sauerbruch T, Caselmann WH. Altered expression of TGF-beta receptors in hepatocellular carcinoma – effects of a constitutively active TGF-beta type I receptor mutant. Digestion. 2005;71:78–91. doi: 10.1159/000084523. [DOI] [PubMed] [Google Scholar]