Abstract
Adhesins play a central role in the cellular response of eukaryotic microorganisms to their host environment. In pathogens such as Candida spp. and other fungi, adhesins are responsible for adherence to mammalian tissues, and in Saccharomyces spp. yeasts also confer adherence to solid surfaces and to other yeast cells. The analysis of FLO11, the main adhesin identified in Saccharomyces cerevisiae, has revealed complex mechanisms, involving both genetic and epigenetic regulation, governing the expression of this critical gene. We designed a genomewide screen to identify new regulators of this pivotal adhesin in budding yeasts. We took advantage of a specific FLO11 allele that confers very high levels of FLO11 expression to wild “flor” strains of S. cerevisiae. We screened for mutants that abrogated the increased FLO11 expression of this allele using the loss of the characteristic fluffy-colony phenotype and a reporter plasmid containing GFP controlled by the same FLO11 promoter. Using this approach, we isolated several genes whose function was essential to maintain the expression of FLO11. In addition to previously characterized activators, we identified a number of novel FLO11 activators, which reveal the pH response pathway and chromatin-remodeling complexes as central elements involved in FLO11 activation.
UNICELLULAR eukaryotes are generally able to adhere to different surfaces or cells in response to environmental conditions. This capability is essential for developmental processes such as the dimorphic switch, flocculation, and biofilm formation (Gimeno et al. 1992; Guo et al. 2000; for review see Verstrepen and Klis 2006). Pathogenic yeast such as Candida albicans and Candida glabrata require this adhesive property for infectivity (reviewed in Verstrepen et al. 2004). Proteins involved in this adhesion phenotype are grouped into families called adhesins, represented, for example, by the FLO family in Saccharomyces cerevisiae (Guo et al. 2000) and by the ALS (reviewed in Hoyer 2001) and EPA (De Las Penas et al. 2003) proteins in C. albicans.
Expression of adhesins is regulated by environmental stress such as nitrogen or carbon source depletion, growth on alcohol as the sole carbon source, or pH changes. Moreover, upregulation of adhesins is critical prior to entry into the pathogenic program of some microorganisms (for review see Hoyer 2001; Verstrepen and Klis 2006).
In S. cerevisiae, Flo11p/Muc1p (Lambrechts et al. 1996) is the main cell-surface protein involved in adhesion-related phenotypes (Lo and Dranginis 1998; Reynolds and Fink 2001). The ability to study cell adhesion in a genetically tractable system such as S. cerevisiae and the analysis of Flo11p as a model system for adhesins has significantly contributed to our understanding of mechanisms leading to cell adhesion and the regulatory pathways governing adhesin expression.
Control of FLO11 expression is relatively complex. Its promoter covers a region of ∼3 kb, one of the largest promoters to be found in the whole S. cerevisiae genome, containing at least four activation sequences and nine repression domains. Most of these regulatory regions are targets for the MAPK pathway, the cAMP cascade, and the Gnc4p-controlled signaling pathway (Pan and Heitman 1999; Rupp et al. 1999; Braus et al. 2003). The MAPK pathway converges on Ste12p activation (Madhani and Fink 1997). Ste12p then activates Tec1p, which can bind specific FLO11 promoter sequences independently or in combination with Ste12p to induce FLO11 expression (Madhani and Fink 1997; Rupp et al. 1999; Kohler et al. 2002; Zeitlinger et al. 2003). Activation of the cAMP pathway induces phosphorylation of Flo8p and Sfl1p transcription factors, promoting the release of Sfl1p-P and the binding of Flo8p-P to the FLO11 promoter, which in turn leads to the transcriptional activation of the FLO11 gene (Pan and Heitman 2002). FLO11 expression is also activated via Phd1p and Ash1p, two transcription factors that function independently of the MAPK and cAMP pathways (Pan and Heitman 2000). On the other hand, FLO11 expression is repressed through two other transcription factors, Nrg1p and Nrg2p, both negatively regulated by Snf1p (Kuchin et al. 2002).
Mss11p has been described as the pivotal element underlying all of these regulatory networks controlling FLO11 expression. It is essential for FLO11 activation via the MAPK cascade, the cAMP pathways, and Phd1p/Ash1p, as well as for repression through the Nrg1p and Nrg2p proteins (van Dyk et al. 2005). In addition to the Mss11p-related networks, FLO11 is also regulated by amino acid starvation via the Gcn4p-controlled signaling pathway, which is required for FLO11 activation (Braus et al. 2003).
Apart from these well-known signaling pathways, FLO11 expression is also subjected to epigenetic silencing, in both a positional and a promoter-specific way, probably through the Sfl1p transcription factor (Halme et al. 2004). Hda1p is the histone deacetylase responsible for this silencing effect. In addition to this silencing mechanism, a role for Rme1p and Msn1p in FLO11 activation has been suggested, hypothetically acting through a chromatin-dependent mechanism (Sidorova and Breeden 1999; van Dyk et al. 2005).
In budding yeasts, Flo11p is involved in a wide repertoire of phenotypic variations involved in adapting to adverse environmental conditions including filamentation, invasive growth, flocculation, and adherence to solid surfaces. The central role that this adhesin plays in response to environmental changes probably explains the complexity of its regulation. We have recently described a FLO11 allele (named FLO11F), found in certain wild “flor” strains of S. cerevisiae, which is highly expressed and confers a number of additional properties to these yeasts, such as the formation of compact fluffy colonies and the ability to form a buoyant biofilm in liquid media required during sherry wine production (Fidalgo et al. 2006).
To better analyze the characteristics of this particular FLO11F gene, we generated a haploid flor-laboratory hybrid strain (133d) containing FLO11F instead of the laboratory FLO11 allele (FLO11L). The 133d strain behaves as a conventional laboratory strain, but manifests all the FLO11F-associated phenotypes found in wild flor yeasts (Fidalgo et al. 2006). These phenotypes include a very high level of FLO11F expression, even in media containing a high glucose concentration, where the FLO11L allele is in a repressed state, and fluffy colonies, an easily distinguishable phenotype associated with FLO11F expression (Fidalgo et al. 2006). On the basis of these distinctive properties, we have developed a genomewide screen, utilizing insertional mutagenesis to isolate positive regulators required for FLO11 expression. Using this powerful approach, we have identified several novel activators of FLO11. Further investigation allowed us to establish that the pH response pathway is a new pathway controlling FLO11 expression and that chromatin-remodeling complexes are central elements involved in FLO11 activation.
MATERIALS AND METHODS
Strains, plasmids, media, and genetic methods:
The yeast strains used in this study are listed in Table 1. Most of directed deletions were carried out by amplifying the alleles containing the target gene replaced with KanMX4 in the strain BY4741. These alleles were amplified by PCR with oligonucleotide primers flanking the target open reading frame. The PCR products were then used to transform the 133d strain by using the lithium acetate/single-strand DNA/PEG procedure (Gietz et al. 1995). PCR-mediated disruption (Lorenz et al. 1995) was used for other gene deletions. Double deletions using the same marker were performed as described in Guldener et al. (1996). Standard YPED and synthetic complete dextrose medium (SCD) lacking the appropriate amino acids for plasmid or transposon selection were used. The YPED medium was supplemented with 200 mg/liter geneticin for selection of geneticin-resistant transformants. Solid media contained 2% agar.
TABLE 1.
Yeast strains used in this study
| Strains | Genotype | Source/reference |
|---|---|---|
| 133d | MATaura3-52 | Fidalgo et al. (2006) |
| 133dLa | MATaura3-52 leu2Δ | This study |
| 133d flo8Δ | MATaura3-52 flo8Δ∷KanMX4 | This study |
| 133d msn1Δ | MATaura3-52 msn1Δ∷KanMX4 | This study |
| 133d mss11Δ | MATaura3-52 mss11Δ∷KanMX4 | This study |
| 133d ash1Δ | MATaura3-52 ash1Δ∷KanMX4 | This study |
| 133d gal11Δ | MATaura3-52 gal11Δ∷KanMX4 | This study |
| 133d tup1Δ | MATaura3-52 tup1Δ∷KanMX4 | This study |
| 133d sap30Δ | MATaura3-52 sap30Δ∷KanMX4 | This study |
| 133d pho23Δ | MATaura3-52 pho23Δ∷KanMX4 | This study |
| 133d rxt2Δ | MATaura3-52 rxt2Δ∷KanMX4 | This study |
| 133d sds3Δ | MATaura3-52 sds3Δ∷KanMX4 | This study |
| 133d snf5Δ | MATaura3-52 snf5Δ∷KanMX4 | This study |
| 133d snf2Δ | MATaura3-52 snf2Δ∷KanMX4 | This study |
| 133d yta7Δ | MATaura3-52 yta7Δ∷KanMX4 | This study |
| 133d rim20Δ | MATaura3-52 rim20Δ∷KanMX4 | This study |
| 133d rga2Δ | MATaura3-52 rga2Δ∷KanMX4 | This study |
| 133d rdr1Δ | MATaura3-52 rdr1Δ∷KanMX4 | This study |
| 133d rri2Δ | MATaura3-52 rri2Δ∷KanMX4 | This study |
| 133d bud4Δ | MATaura3-52 bud4Δ∷KanMX4 | This study |
| 133d ena1Δ | MATaura3-52 ena1Δ∷KanMX4 | This study |
| 133d atp10Δ | MATaura3-52 atp10Δ∷KanMX4 | This study |
| 133d gph1Δ | MATaura3-52 gph1Δ∷KanMX4 | This study |
| 133d kre11Δ | MATaura3-52 kre11Δ∷KanMX4 | This study |
| 133d yhr177wΔ | MATaura3-52 yhr177wΔ∷KanMX4 | This study |
| 133d snf6Δ | MATaura3-52 snf6Δ∷KanMX4 | This study |
| 133d swi3Δ | MATaura3-52 swi3Δ∷KanMX4 | This study |
| 133d rim101Δ | MATaura3-52 rim101Δ∷KanMX6 | This study |
| 133d rim20Δrim101Δ | MATaura3-52 rim101Δ rim20Δ∷KanMX4 | This study |
| L5684 | MATaura3-52 leu2Δ | G. R. Fink |
| L5684 mss11Δ | MATaura3-52 leu2Δ mss11Δ∷KanMX6 | This study |
| L5684 snf5Δ | MATaura3-52 leu2Δ snf5Δ∷KanMX6 | This study |
| L5684 pho23Δ | MATaura3-52 leu2Δ pho23Δ∷KanMX6 | This study |
| L5684 tup1Δ | MATaura3-52 leu2Δ tup1Δ∷KanMX6 | This study |
| L5684 rim101Δ | MATaura3-52 leu2Δ rim101Δ∷KanMX6 | This study |
| BY4741b | MATaura3Δ leu2Δ his3Δ met15Δ | Euroscarf |
Strain used for the mutagenesis.
For deletions of genes using the KanMX4 marker, mutants for target genes in this background were used.
To obtain the plasmid pFLT133dGFP, the 133d FLO11 promoter was amplified by PCR and cloned into the EcoRI site of pRS316. The GFP was cloned downstream from the FLO11F promoter into the SmaI–KspI sites, and the ADH1 terminator was cloned downstream from the GFP into the SacII site. Bacterial transformations and plasmid isolation were performed as described (Sambrook and Russell 2001).
Yeast mutagenesis:
The 133d strain was mutagenized by transformation with NotI-cleaved DNA carrying random Tn3∷lacZ∷LEU2 insertions (Burns et al. 1994). Yeast cells carrying the transposon as a recombinational replacement of the genomic copy with the transposon-mutagenized version were selected on SCD with auxotrophic supplements lacking leucine.
The site of the insertion in selected mutants was determined by plasmid rescue and DNA sequence analysis as described (Burns et al. 1994). Briefly, mutant yeast cells were transformed with linearized pRSQ1 plasmid. Transformants were selected on SCD plates lacking both leucine and uracil. Yeast genomic DNA from each mutant was recovered and digested with EcoRI or EcoRV. The fragments were circularized and recovered in bacteria. Plasmids were sequenced using a primer complementary to the 5′-end of the transposon. DNA homology searches were performed using the Saccharomyces Genome Database.
Northern blot analysis:
To analyze FLO11 gene expression, cells were incubated in YPED liquid medium overnight at 30° and then transferred to fresh YPED medium and incubated to an optical density at 600 nm (OD600) of ∼0.8. If the analysis was in glucose-rich medium, then cells were collected and RNA extraction was performed as described below. If the analysis was in low-glucose medium, then cells were washed and transferred to YPED with 0.2% of glucose for 2 hr. Cells were washed with cool water, and total RNA was isolated with the QIAGEN (Valencia, CA) RNeasy mini kit, separated by formaldehyde denaturing agarose gel electrophoresis, and transferred overnight by capillary action to nylon membranes. The 400-bp regions at the 5′-end of FLO11 and ACT1 genes were then used to probe the membranes. The radioactive bands were visualized and quantified using a Molecular Dynamics PhosphoImager.
Flow cytometry:
To quantify the GFP levels in the obtained mutants, cells were grown in YPED overnight at 30° and then replaced to a fresh YPED medium and incubated to an OD600 of 0.8. Just prior to analysis, cells were pelleted, washed, and resuspended in 50 mm sodium citrate. The fluorescence of 10,000 cells was measured using a FACSCalibur flow cytometer (Becton Dickinson) with a 530/30 band-pass filter.
Light and fluorescence microscopy:
To study colony morphology, single-colony photographs were taken directly from petri plates using a Leica DMRE microscope with a ×10 objective. To analyze colony fluorescence, a Leica MZFLIII stereomicroscope was used.
Invasive growth assay:
The plate washing assay was performed as described (Roberts and Fink 1994) with several modifications. Cells were grown in YPED overnight at 30° and then replaced to a fresh YPED medium and incubated to an OD600 of ∼0.8 and then cells were spotted onto YPED, incubated for 4 days at 28°, and photographed. Plates were then washed under a stream of water by rubbing with Digralsky spreader and then photographed again.
Assay for adherence to plastic and hydrophobicity:
Assays for adherence to the wells of a polystyrene 96-well microtiter plate and hydrophobicity were carried out as described (Reynolds and Fink 2001) with minor modification. For adherence to plastic assays, cells were grown in YPD to an OD600 of ∼0.8, collected, washed, and resuspended in YPD to an OD600 of 1. Cells (0.1 ml) were transferred to the wells of a microtiter plate and incubated for 1 hr at 28°. The cells were then stained with 1% crystal violet, and the wells washed repeatedly with water and photographed. For quantification, the crystal violet was solubilized by adding 100 μl of SDS 10%, plates were incubated for 15 min, and then wells were mixed with 100 μl of water and the absorbance at 530 nm (A530) was measured using a microplate reader. For hydrophobicity assay, cells were grown in SCD to an OD600 of ∼0.8 and then 1.2 ml of the culture was overlaid with 600 μl of octane and vortexed for 3 min. The OD600 of the aqueous layer was taken and the relative difference with the initial OD600 was used to determine the percentage of hydrophobicity.
RESULTS
High levels of FLO11F expression are necessary to confer the fluffy-colony morphology shown by wild flor yeast:
Naturally occurring S. cerevisiae flor yeast show a fluffy-colony morphology in contrast to the smooth morphology of S. cerevisiae laboratory strains. 133d is a haploid flor-laboratory hybrid strain harboring the FLO11F allele from wild flor yeasts (Fidalgo et al. 2006) and produces fluffy colonies (Figure 1A). This phenotype is FLO11F dependent, since a FLO11F loss-of-function mutant forms smooth colonies, which are easily distinguishable from fluffy colonies by visual inspection (Figure 1A). As the main difference between the laboratory FLO11L and flor FLO11F alleles is the higher level of expression conferred by the FLO11F promoter (Fidalgo et al. 2006), it strongly suggests that the fluffy-colony phenotype is directly related to elevated FLO11 expression. To confirm this relationship, MSS11, one of the main FLO11 activators (van Dyk et al. 2005), was deleted in the 133d strain. Deletion of MSS11 yielded smooth colonies (Figure 1B) similar to those observed for FLO11F-deleted cells. This relationship between colony morphology and FLO11 expression allowed us to develop a screening strategy to detect novel FLO11 activators. To ensure that changes in colony morphology are a direct result of decreased FLO11 expression, we generated a plasmid containing a GFP reporter under the control of the FLO11F promoter from the 133d strain (pFLT133dGFP). Deletion of MSS11 in 133d pFLT133dGFP yielded smooth nonfluorescent colonies, in contrast to the fluffy fluorescent colonies of the 133d pFLT133dGFP strain (Figure 1B). Therefore, we can use 133d pFLT133dGFP as a novel method for rapidly identifying new positive regulators involved in FLO11 expression on the basis of colony morphology and fluorescence changes.
Figure 1.—
Colony morphology and fluorescence level. (A) Deletion of FLO11F in 133d generates smooth colonies. (B) Deletion of MSS11 in 133d pFLT133dGFP generates smooth nonfluorescent colonies in contrast to the fluffy fluorescent colonies for 133d pFLT133dGFP. The smooth colonies are semitransparent in contrast to the opaque, dark, fluffy colonies (133d).
Isolation of FLO11F low-expression-level mutants:
To generate low FLO11F expression mutants, the 133d pFLT133dGFP strain was mutagenized by integrative transformation with a yeast genomic library carrying random Tn3∷LEU2∷lacZ gene insertions (Burns et al. 1994). First, we screened for reduced colony fluorescence in 182,000 Leu+ transformants. Colonies exhibiting reduced fluorescence were selected and streaked onto selective media to analyze colony morphology, and only smooth colonies were chosen. Following these two rounds of selection, we were left with 63 candidate mutant strains.
To identify the position of transposon insertion in each of the 63 mutants, the mutants were subjected to plasmid rescue and DNA sequencing (Burns et al. 1994). We found that they represented 45 independent insertions affecting 25 different genes (Table 2). To ensure that single insertions in the identified genes were responsible for the FLO11F low-expression phenotype, we performed directed deletion of each of the 25 identified genes in the 133d pFLT133dGFP strain and determined colony morphology and fluorescence of the deleted mutants. Sixteen of these genes showed a smooth morphology and reduced fluorescence when deleted (Table 2). As expected, a number of the identified genes correspond to known FLO11 regulators (Table 2). Of these, MSS11, FLO8, MSN1, and ASH1 have been previously described to encode FLO11 activators (Gagiano et al. 1999b; Pan and Heitman 1999; Rupp et al. 1999; Pan and Heitman 2000). TUP1 encodes a protein that has been previously described as a component of the general corepressor Tup1-Cyc8, a complex involved in the repression of FLO11 expression (Conlan and Tzamarias 2001). Here, in contrast, we have identified Tup1p as a FLO11F activator, which, when deleted, reduces FLO11 expression as judged by colony morphology and reporter expression. Thus, our findings suggest that Tup1p can act as an activating factor, at least in the 133d pFLT133dGFP genetic background.
TABLE 2.
Genes found in the screen
| Gene | No. of independent insertions in the gene | Colony morphology in directed deletions | Short description |
|---|---|---|---|
| FLO8 | 1 | Smooth | Transcription factor |
| MSN1 | 6 | Smooth | Transcriptional activator |
| MSS11 | 2 | Smooth | Transcription factor |
| ASH1 | 2 | Smooth | Transcription factor |
| GAL11 | 3 | Smooth | Component of the Mediator complex |
| TUP1 | 1 | Smooth | General repressor of transcription |
| SAP30 | 4 | Smooth | Subunit of a histone deacetylase complex |
| PHO23 | 4 | Smooth | Component of the Rpd3 histone deacetylase complex |
| RXT2 | 3 | Smooth | Subunit of the histone deacetylase Rpd3L complex |
| SDS3 | 1 | Smooth | Component of the Rpd3p/Sin3p deacetylase complex |
| SWI1 | 1 | Lethal | Subunit of the SWI/SNF chromatin-remodeling complex |
| ARP7 | 1 | Lethal | Component of both the SWI/SNF and RSC chromatin-remodeling complexes |
| SNF5 | 3 | Smooth | Subunit of the SWI/SNF chromatin-remodeling complex |
| SNF2 | 1 | Smooth | Catalytic subunit of the SWI/SNF chromatin-remodeling complex |
| YTA7 | 2 | Smooth | Protein of unknown function |
| RIM20 | 1 | Smooth | Protein involved in proteolytic activation of Rim101p in response to alkaline pH |
| RGA2 | 1 | Fluffy | GTPase-activating protein |
| RDR1 | 1 | Fluffy | Transcriptional repressor |
| RRI2 | 1 | Fluffy | Subunit of the COP9 signalosome (CSN) complex |
| BUD4 | 1 | Fluffy | Protein involved in bud-site selection and required for axial budding pattern |
| ENA1 | 1 | Fluffy | P-type ATPase sodium pump |
| ATP10 | 1 | Fluffy | Mitochondrial inner membrane protein |
| GPH1 | 1 | Fluffy | Nonessential glycogen phosphorylase |
| KRE11 | 1 | Fluffy | Protein involved in biosynthesis of cell-wall β-glucans |
| YHR177W | 1 | Fluffy | Putative protein of unknown function |
The number of independent transposon insertions in each gene is shown in the second column. The fourth column is a short description, obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/), of the proteins encoded by the identified genes.
We also identified Gal11p as an activator of FLO11F. GAL11 encodes an element of the mediator complex, which acts by transferring information from enhancers and other regulatory elements to the RNApolII. This mediator complex is required for the transcriptional control of all RNApolII-dependent genes (Bjorklund and Gustafsson 2005).
Interestingly, our screen allowed us to identify a number of novel FLO11F transcriptional activators. These include Rim20p, which is involved in the alkaline pH response pathway (Xu and Mitchell 2001; Boysen and Mitchell 2006), and a large number of genes involved in chromatin remodeling, such as components of the histone deacetylase Rpd3L complex (SAP30, PHO23, RXT2, and SDS3) (Carrozza et al. 2005) and the SWI/SNF complex (SNF2, SNF5) (Cairns et al. 1994; Peterson et al. 1994, 1998).
Deacetylases remove the acetate groups from the acetylated lysine residues in the histones amino-terminal domains (Rundlett et al. 1996; Kadosh and Struhl 1998). Usually, Rpd3p acts by repressing gene transcription, although in some genes an activation activity has been described. (Vidal and Gaber 1991; Kadosh and Struhl 1997, 1998; De Nadal et al. 2004; Puig et al. 2004).
The SWI/SNF ATP-dependent remodeling complex regulates gene expression by remodeling chromatin structure and altering histone acetylation patterns (Peterson and Workman 2000). This multi-subunit complex is able to delocalize histone octamers to generate nucleosome-free regions available for transcription factors (Whitehouse et al. 1999). In addition to SNF2 and SNF5, we also isolated ARP7 and SWI1 as FLO11 activators. Directed deletions against the latter genes showed lethality, and no further analysis was possible. However, because these two genes also belong to the SWI/SNF complex (Cairns et al. 1994; Peterson et al. 1994), their isolation further supports a role for the SWI/SNF complex in FLO11 activation.
Another candidate FLO11 activator, related to chromatin remodeling, is YTA7, encoding an ATPase with a bromo-like domain (Jambunathan et al. 2005). Yta7p is involved in binding to acetylated histones and other chromatin-associated proteins (Zeng and Zhou 2002; Yang 2004; De La Cruz et al. 2005), similar to those present in components of the SWI/SNF complex. YTA7 has also been identified in the DPB4 chromatin-remodeling complex, which binds to DNA regions near the FLO11 gene (Tackett et al. 2005). Thus, the discovery of several genes encoding components of pH response and chromatin-remodeling complexes in our screen suggests that these pathways may play an important role in controlling FLO11 activation.
The role of the remaining nine genes identified in FLO11 expression was not confirmed by directed deletion. In these cases, the phenotype observed in the original mutant could have been caused by additional insertions or by a dominant-negative effect. Further experiments are required to discern if these genes play any role in regulating FLO11 expression.
FLO11F expression is reduced in all selected mutants:
To analyze and quantify FLO11F expression in the 14 selected deletion mutants, Northern blot analysis (Figure 2) and flow-cytometry-mediated GFP quantification on mutant yeasts transformed with pFLT133dGFP (data not shown) were performed. Consistent with our previous observation, all 14 mutants expressed greatly reduced levels of FLO11F, as determined by both Northern blot and flow cytometry. The deletion of TUP1 resulted in low levels of FLO11F expression, confirming the role of this protein as an FLO11F activator. As expected for RIM20, given its mild effect on colony morphology, the effect of the deletion in FLO11F expression was the lowest observed. When genes encoding components of the two chromatin-remodeling complexes, Rpd3L and SWI/SNF, were deleted, FLO11F expression was almost completely abolished. In contrast, YTA7 deletion did not result in such a dramatic drop in FLO11F expression, suggesting that FLO11F regulation via YTA7 is distinct from that achieved through the other chromatin-remodeling complexes identified in this screen. Surprisingly, deletion of previously described FLO11L activators (MSN1, MSS11, FLO8, and ASH1) also resulted in a more moderate decrease in FLO11F expression. Thus, FLO11F activation is more dependent on chromatin-remodeling complexes than previously described activators.
Figure 2.—
FLO11F expression levels in the deleted mutants. Northern blot analysis was performed to determine FLO11F expression level. ACT1 was used as probe for loading control.
FLO11-related phenotypes are altered in the isolated mutants:
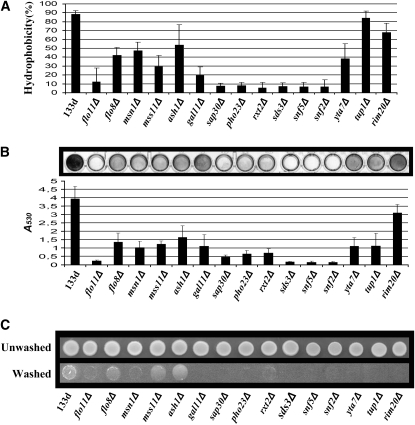
To study the role of the identified genes on a number of FLO11-dependent phenotypes, we analyzed the effect of the deletion for the selected genes on cellular hydrophobicity, invasive growth, and solid surface biofilm (Lo and Dranginis 1998; Gagiano et al. 1999b; Reynolds and Fink 2001).
In the majority of cases, effects on FLO11F expression and cellular hydrophobicity were closely related (Figure 3A). The largest effect on hydrophobicity was shown by mutants for genes involved in chromatin remodeling, where the decrease in hydrophobicity was similar to that shown by flo11FΔ. The only exception for this class of genes was yta7, which exhibited a weaker reduction in hydrophobicity, although this correlated with the smaller reduction of FLO11F expression. The only example in which hydrophobicity did not correlate with FLO11F expression was observed for TUP1. Remarkably, the hydrophobicity level for the tup1Δ mutant was similar to that observed for the wild-type strain, despite the fact that the FLO11F expression level was dramatically reduced to levels similar to mss11Δ or gal11Δ mutants.
Figure 3.—
FLO11F-related phenotypes in the deleted mutants. (A) Hydrophobicity. Cells cultures were overlaid with octane and mixed. The OD600 of the aqueous layer was taken and the relative difference with the initial OD600 was used to determine the percentage of hydrophobicity. (B) Biofilm on solid surface. Exponentially growing cells were placed in microtiter plate wells and incubated for 1 hr at 28°. The cells were then stained with 1% crystal violet, and the wells were washed repeatedly with water and photographed. For biofilm quantification, the crystal violet was solubilized using SDS (10%), and the absorbance at 530 nm (A530) was measured. Data presented represent averages of three independent assays. Error bars correspond to standard deviation. (C) Invasive growth. Dots of exponentially growing cells were spotted on YPED solid medium and photographed before (unwashed) and after (washed) washing under a stream of water.
Next we examined the ability of each of the selected mutants to form solid surface biofilms. Quantification of biofilm formation determined that the quality of the biofilm formed by each mutant was closely related to the level of FLO11F expression (Figure 3B). It is significant that, in contrast to hydrophobicity, biofilm formation in tup1Δ mutants corresponded to its reduced expression of FLO11F.
Similar to their effect on hydrophobicity and solid biofilm formation, the very low levels of FLO11F expression in mutants for genes involved in chromatin remodeling did not allow invasive growth (Figure 3C). However, the decrease in FLO11F expression observed for the remaining mutants did allow invasive growth to occur in flo8Δ, mss11Δ, and ash1Δ but not in msn1Δ, gal11Δ mutants, nor in rim20Δ, which showed the highest level of FLO11F expression. These observations suggest that the latter three genes affect invasive growth in a FLO11-independent manner.
Other SWI/SNF complex members are involved in FLO11F activation:
In this screen, we have identified four members of the SWI/SNF complex. Deletions of SNF2 and SNF5 in the 133d strain generate the highest decrease in FLO11F expression, while deletions of SWI1 and ARP7 were found to be lethal. The SWI/SNF complex is composed of 11 subunits (Peterson et al. 1998). SWI3 and SNF6 encode proteins that have been shown to copurify with Swi1p, Snf2p, and Snf5p (Cairns et al. 1994; Peterson et al. 1994). To determine whether other members of the SWI/SNF complex are also involved or essential for FLO11F regulation, we deleted SWI3 and SNF6 from the 133d strain. FLO11F expression level decreased in swi3Δ and snf6Δ mutants to the same level as in snf2Δ and snf5Δ mutants (Figure 4A). As with other members of this complex identified in our screen, including the isolated swi1 and arp7 mutants [swi1(T) and arp7(T)], loss of SWI3 and SNF6 resulted in the loss of all the characteristic FLO11F-dependent phenotypes (Figure 4, B–D). Thus our results clearly show that the SWI/SWF complex is essential for FLO11 activation.
Figure 4.—
FLO11F expression and related phenotypes in mutants for SWI/SNF complex members. (A) FLO11F expression. Northern blot analysis using FLO11F as a probe. ACT1 was used as a loading control. B–D represent hydrophobicity, biofilm on solid surface, and invasive growth, respectively, performed as mentioned in Figure 3. For hydrophobicity and biofilm on solid surface, data presented represent averages of three independent assays. Error bars correspond to standard deviation. swi1(T) and arp7(T) are the original transposon insertion mutants isolated during the screen, because full deletion of these genes caused lethality.
Rim20p activates FLO11F via Rim101p:
Rim20p is necessary for the proteolytic activation of Rim101p, the budding yeast homolog of the Aspergillus nidulans PacC protein (Xu and Mitchell 2001). Rim101p is a transcription factor involved in the response to alkaline pH (Lamb et al. 2001). Loss of RIM101 results in phenotypes similar to those observed in the rim20Δ mutant, including weaker invasive growth (Xu and Mitchell 2001). To establish a role for Rim101p in FLO11F activation, we deleted the RIM101 gene from the 133d strain and compared its FLO11F expression to that of the rim20Δ mutant by Northern blot analysis. We found that the loss of RIM101 resulted in a reduction in the level of FLO11F expression similar to that observed for the rim20Δ mutant (Figure 5). Moreover, we found that all of the FLO11F-related phenotypes were also affected in a similar manner (data not shown). To confirm that FLO11F activation by Rim20p occurs via Rim101p, we generated a rim20Δ rim101Δ double mutant. The double mutant showed an expression level similar to that of the single mutants (Figure 5), suggesting that the two genes act in the same pathway to activate FLO11F expression. Thus, we can conclude that the alkaline pH response pathway is a novel regulatory mechanism for FLO11 activation.
Figure 5.—
FLO11F expression in rim20Δ, rim101Δ, and rim20Δrim101Δ mutants. FLO11F expression was measured by Northern blot using ACT1 as probe for loading control.
The identified genes also regulate FLO11 expression in a Σ1278b background:
The fact that the 133d strain has a higher level of FLO11 expression than the laboratory L5684 strain (Σ1278b genetic background) provided a powerful tool that permitted us to undertake this screen for FLO11 activators. However, given the differences between the flor FLO11F and laboratory FLO11L promoters and, potentially, the differences in the genetic background between 133d and L5684 strains, we extended our analysis of putative FLO11F activators to FLO11L expression in the L5684 laboratory strain. We tested the requirement for FLO11L expression by growing L5684 mutant strains on media in which the FLO11L promoter is derepressed (low glucose) (Kuchin et al. 2002) to mimic the high expression level of FLO11F observed in the 133d strain. The putative FLO11 regulators that we chose to examine were MSS11 as a member of the previously described FLO11F activators, SNF5 as a member of the SWI/SNF complex, PHO23 as a member of the Rpd3L complex, RIM101 as a component of the pH response pathway, and TUP1 because it had showed a role in FLO11 regulation in the 133d strain different from the one described in laboratory strains. As expected, the deletion of MSS11, RIM101, SNF5, and PHO23 yielded a decrease in FLO11L expression, confirming them as genes encoding general FLO11 activators, capable of regulating FLO11L in the laboratory strain (Figure 6). However, the effects of these deletions on FLO11 expression differed between L5684 and 133d yeast cells (Figure 6). This suggests that differences in the FLO11 promoter and/or the genetic background do influence the degree to which these genes are required for FLO11 activation. Deletion of TUP1 has a low but detectable role in repressing FLO11 expression in the L5684 strain, as previously described for laboratory strains (Conlan and Tzamarias 2001), while the 133d strain has an activator role (Figure 6). Further experiments to determine other factors involved in the role exchange for TUP1 are in progress.
Figure 6.—
Expression of FLO11 in L5684 (Σ1278b background) and 133d derivative strains. A gene representing any of the main group of genes identified in the screening was analyzed. Cells were grown on YEPD with a low amount of glucose to mimic the natural derepressed state of the FLO11 promoter in the 133d strain. Expression was measured by Northern blot using ACT1 as a probe for loading control.
DISCUSSION
The discovery of a specific FLO11 allele (FLO11F) in flor yeast has allowed us to perform an in-depth study of the activation mechanisms controlling FLO11 expression. The promoter that drives expression in the FLO11F allele confers the highest level of expression known for this gene (Fidalgo et al. 2006). This property has been crucial for the development of a powerful strategy for characterizing the mechanisms involved in FLO11 activation in S. cerevisiae. Using this method, we identified a number of known FLO11 activators multiple times, validating the methodology. Nevertheless, other known FLO11 activators were isolated only once, indicating that the screen may not have reached saturation (see Table 2). Among the known genes identified, it is worth highlighting the genes MSN1 and MSS11, which have been previously postulated as the major FLO11 activators (Lorenz and Heitman 1998; van Dyk et al. 2005). The fact that several activation pathways converge on Msn1p and Mss11p, combined with a requirement in the screen for a significant decrease in FLO11F expression, may explain why other proteins, previously described as FLO11 activators, have not been identified. In addition to known components, our novel approach has allowed us to identify two new mechanisms for FLO11 activation: the pH response pathway and chromatin remodeling.
Unicellular eukaryotes use complex systems to sense and adapt to extracellular environmental conditions. Flo11p is an essential protein involved in changes in cellular behavior in response to environmental alterations, such as carbon source depletion, nitrogen starvation, pheromone presence, etc. (Gagiano et al. 1999b; Pan and Heitman 1999; Lorenz et al. 2000; Gancedo 2001). It has been previously demonstrated that FLO11-dependent flocculation occurs only at acidic pH (Bayly et al. 2005). This observation, together with our results regarding the role of the Rim101p in FLO11 activation, allow us to propose pH as a new input sensed by yeast to respond to changes in external acidity by modifying FLO11 expression. Rim101p is the main transcription factor involved in the pH response of S. cerevisiae, which acts indirectly by repressing the expression of genes encoding transcriptional repressors such as NRG1 and SNP1 (Lamb and Mitchell 2003). Curiously, FLO11 expression is repressed by NRG1, so Rim101p could act on FLO11 activation via NRG1 repression. However, the expression level for FLO11F in a double rim101Δ nrg1Δ mutant is not as high as for a nrg1Δ single mutant (data not shown), indicating that Rim101p can activate FLO11 expression in an NRG1-independent way. Rim101p is proteolytically activated by Rim20p, and because the double rim101Δrim20Δ mutant has the same effect in FLO11 expression as the two single mutants (Figure 5), it suggests that the effect of Rim101p on FLO11 regulation is dependent on its activation by Rim20p.
Most of the other genes identified either can be assigned to complexes involved in chromatin remodeling, (i) the Rpd3L and (ii) SWI/SNF complexes, or (iii) are in some way related to chromatin remodeling.
Rpd3L complex:
The regulation of FLO11 expression by histone deacetylase has been described previously (Halme et al. 2004). Hda1p was identified as an essential element of FLO11 silencing. Here we have found an activation effect directed by the Rpd3L complex. In our screen, we have isolated PHO23, SAP30, RXT2, and SDS3, all of which encode members of the Rpd3L histone deacetylase complex (Carrozza et al. 2005). A single deletion of any one of these genes almost completely abolishes FLO11F expression (Figure 2). The Rpd3L complex is also composed of Rpd3p, Sin3p, Dep1, Ash1p, Ume1p, Cti6p, Rxt3p, and Ume6p (Carrozza et al. 2005). Of these components, only Ash1p has been previously described as being involved in the control of FLO11 expression (Chandarlapaty and Errede 1998; Pan and Heitman 2000). Curiously, although ASH1 was also isolated in the screen, the FLO11F expression level in the ash1Δ deletion mutant was not as strongly diminished as in the pho23Δ, sap30Δ, rtx2Δ, and sds3Δ mutants (Figure 2). Ash1p and Ume6p are the only known sequence-specific DNA-binding protein in the Rpd3L complex (Carrozza et al. 2005), suggesting that the role for Ash1p in FLO11 activation might be the recruitment of the Rpd3L complex to the FLO11 promoter. The reason that the effect of ASH1 deletion is not as dramatic as for pho23Δ, sap30Δ, rtx2Δ, and sds3Δ could be that this recruitment activity is shared with other proteins. Rpd3p is the only protein with deacetylase activity in the Rpd3L complex, but was not identified in this screen. This might be because Rpd3p is not required for the FLO11 activation by the Rpd3L complex or because the screen did not achieve saturation. Further studies to determine the requirement of Rpd3p deacetylase activity for FLO11 activation are currently in progress.
SWI/SNF complex:
Another major class of genes identified in our screen is composed of elements of the SWI/SNF ATP-dependent remodeling complex. Deletion for several members of this complex from the 133d strain resulted in an extremely low level for FLO11F expression (Figure 2) and in the abolishment of all phenotypes in which FLO11 is involved (Figure 3).
Another promoter, almost identical to FLO11, exists in the S. cerevisiae (var. diastaticus) genome. This promoter governs the expression of STA1, STA2, and STA3, a gene family encoding for glucoamylase proteins. Most of the previously described pathways involved in FLO11 regulation are also involved in STA regulation (Gagiano et al. 1999a,b, 2003; van Dyk et al. 2003). This promoter is directly controlled by the SWI/SNF complex. (Kim et al. 2004) According to our results and considering the regulatory similarities between FLO11 and STA genes, we may assume that FLO11 activation is also directly controlled by the SWI/SNF complex. In particular, as it has been previously demonstrated for STA1 (Kim et al. 2004), we predict that certain transcription factors such as FLO8 and MSS11 recruit the SWI/SNF complex to the FLO11 promoter. The fact that deletion for FLO8 or MSS11 in the 133d strain does not produce as dramatic an effect on FLO11F expression levels as the deletion for members of the SWI/SNF complex suggests that the recruitment of this complex to the FLO11 promoter might also be mediated by other transcription factors. In this context, it is significant that a role for Ste12p and Tec1p transcription factors in recruitment of the SWI/SNF complex to the STA1 promoter has been described. However, although these transcription factors are involved in FLO11 activation in the laboratory strains (Madhani and Fink 1997; Rupp et al. 1999; Kohler et al. 2002; Zeitlinger et al. 2003), they are not required for FLO11 activation in the 133d strain (data not shown).
YTA7 is another gene encoding a protein involved in chromatin remodeling that we found in our screen for FLO11 activators. It encodes a protein with a bromo-like domain and has been found in the DPB4 complex, which is located in regions flanking transcriptionally silent areas (Tackett et al. 2005). It has been proposed that this complex is necessary to preserve the transcriptionally active state of regions adjacent to these silenced zones (Tackett et al. 2005). Significantly, the DPB4 complex has been found to bind sequences close to FLO11 as well as other FLO genes (Tackett et al. 2005). Thus, this protein might play a role in countering the silencing effect that normally occurs close to the centromere where FLO11 is located. The deletion of YTA7 did not have as drastic an effect on FLO11 expression as the loss of members of the SWI/SNF or Rpd3L complexes. This difference might make it more difficult to find other DPB4-complex-related genes using this screening methodology.
Genes related to chromatin remodeling:
Another gene identified in our screen as a positive regulator of FLO11F in the 133d strain is TUP1, which is related to chromatin remodeling. Tup1p is a component of the Tup1p-Cyc8p corepressor, which binds to underacetylated histone tails (Edmondson et al. 1996; Huang et al. 1997) and requires deacetylase activity for its repression function (Watson et al. 2000; Davie et al. 2002, 2003). Tup1p-Cyc8p has been described as an important repressor of a wide variety of genes (Keleher et al. 1992), although it has also been described as an activator in some cases (Conlan et al. 1999; Proft and Struhl 2002). In relation to FLO11 regulation, Tup1p-Cyc8p has been described as a transcriptional repressor (Conlan and Tzamarias 2001). In contrast, here we have found that Tup1p can act as an activator of FLO11 in the 133d strain (Figure 2). This suggests that differences in the FLO11 promoter region or the genetic background of the 133d strain affect Tup1 function and result in conversion from transcription repressor to transcription activator for FLO11. However, a laboratory strain transformed with GFP under the control of the FLO11 promoter from the 133d strain did not show an increase in GFP expression when TUP1 was deleted (data not shown). These results suggest that changes in the promoter region may be responsible for modifying the activity of Tup1p-Cyc8p. The differential distribution of Tup1p within the SUC2 promoter has been linked to changes in its activation or repression state (Boukaba et al. 2004). Moreover, the phosphorylation of additional elements that bind to Tup1p-Cyc8 may help maintain an activating state by promoting the recruitment of SAGA and SWI/SNF complexes as has been previously described (Proft and Struhl 2002). It remains to be determined if the changes in the FLO11F promoter result directly in an altered distribution of Tup1p-Cyc8p or indirectly, for example, via changes in the binding of proteins involved in the modification of Tup1p-Cyc8p. Another interesting finding regarding Tup1p activity is the observation that when FLO11F expression levels are decreased as a consequence of TUP1 deletion in the 133d strain, all phenotypes related to FLO11 activity are lost, except for cellular hydrophobicity, which is maintained at wild-type levels (Figure 3). This suggests that, in 133d, Tup1p might regulate the expression of other proteins involved in cellular hydrophobicity, which, when altered in a tup1Δ strain, can maintain cellular hydrophobicity in a FLO11-independent manner. One of these proteins could be Flo1p, a protein involved in flocculation. The gene encoding this protein is located in a region repressed by Tup1p (Fleming and Pennings 2001). Thus, Tup1p appears to act via at least two mechanisms to promote the floating capability of 133d yeast, maintaining the increased expression of FLO11 while repressing other genes, such as those involved in flocculation.
Adhesin expression is essential for fungal pathogenesis. Our screen has revealed novel activators required for FLO11 adhesin expression, which may provide new targets and strategies to overcome fungal infections. Of these novel regulators, our data suggest that chromatin remodeling plays a fundamental role in controlling FLO11 expression. Further studies of FLO11 regulation by these chromatin activators may help reveal underlying principles of gene regulation by chromatin modification.
Acknowledgments
We thank Anabel Lopez and especially Victor Carranco for excellent technical assistance; Manuel Fidalgo for useful discussions and technical advice; and John R. Pearson for critical reading of the manuscript. This work was supported by Ministerio de Educacion y Ciencia grants BMC2003-05495 and VIN1-043 and by OSBORNE and Cia SA. R.R.B. was awarded a Postgraduate Fellowship from the Junta de Andalucia. The support of Centro Andaluz de Biología del Desarrollo by the Junta de Andalucía is acknowledged.
References
- Bayly, J. C., L. M. Douglas, I. S. Pretorius, F. F. Bauer and A. M. Dranginis, 2005. Characteristics of Flo11-dependent flocculation in Saccharomyces cerevisiae. FEMS Yeast Res. 5: 1151–1156. [DOI] [PubMed] [Google Scholar]
- Bjorklund, S., and C. M. Gustafsson, 2005. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30: 240–244. [DOI] [PubMed] [Google Scholar]
- Boukaba, A., E. I. Georgieva, F. A. Myers, A. W. Thorne, G. Lopez-Rodas et al., 2004. A short-range gradient of histone H3 acetylation and Tup1p redistribution at the promoter of the Saccharomyces cerevisiae SUC2 gene. J. Biol. Chem. 279: 7678–7684. [DOI] [PubMed] [Google Scholar]
- Boysen, J. H., and A. P. Mitchell, 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell 17: 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braus, G. H., O. Grundmann, S. Bruckner and H. U. Mosch, 2003. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 4272–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg et al., 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105. [DOI] [PubMed] [Google Scholar]
- Cairns, B. R., Y. J. Kim, M. H. Sayre, B. C. Laurent and R. D. Kornberg, 1994. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 91: 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza, M. J., L. Florens, S. K. Swanson, W. J. Shia, S. Anderson et al., 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731: 77–87; discussion 75–76. [DOI] [PubMed] [Google Scholar]
- Chandarlapaty, S., and B. Errede, 1998. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 2884–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan, R. S., and D. Tzamarias, 2001. Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J. Mol. Biol. 309: 1007–1015. [DOI] [PubMed] [Google Scholar]
- Conlan, R. S., N. Gounalaki, P. Hatzis and D. Tzamarias, 1999. The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J. Biol. Chem. 274: 205–210. [DOI] [PubMed] [Google Scholar]
- Davie, J. K., R. J. Trumbly and S. Y. Dent, 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22: 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie, J. K., D. G. Edmondson, C. B. Coco and S. Y. Dent, 2003. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 278: 50158–50162. [DOI] [PubMed] [Google Scholar]
- de la Cruz, X., S. Lois, S. Sanchez-Molina and M. A. Martinez-Balbas, 2005. Do protein motifs read the histone code? BioEssays 27: 164–175. [DOI] [PubMed] [Google Scholar]
- De Las Penas, A., S. J. Pan, I. Castano, J. Alder, R. Cregg et al., 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17: 2245–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nadal, E., M. Zapater, P. M. Alepuz, L. Sumoy, G. Mas et al., 2004. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374. [DOI] [PubMed] [Google Scholar]
- Edmondson, D. G., M. M. Smith and S. Y. Roth, 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10: 1247–1259. [DOI] [PubMed] [Google Scholar]
- Fidalgo, M., R. R. Barrales, J. I. Ibeas and J. Jimenez, 2006. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. USA 103: 11228–11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A. B., and S. Pennings, 2001. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 20: 5219–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano, M., D. van Dyk, F. F. Bauer, M. G. Lambrechts and I. S. Pretorius, 1999. a Divergent regulation of the evolutionarily closely related promoters of the Saccharomyces cerevisiae STA2 and MUC1 genes. J. Bacteriol. 181: 6497–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano, M., D. van Dyk, F. F. Bauer, M. G. Lambrechts and I. S. Pretorius, 1999. b Msn1p/Mss10p, Mss11p and Muc1p/Flo11p are part of a signal transduction pathway downstream of Mep2p regulating invasive growth and pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Microbiol. 31: 103–116. [DOI] [PubMed] [Google Scholar]
- Gagiano, M., M. Bester, D. van Dyk, J. Franken, F. F. Bauer et al., 2003. Mss11p is a transcription factor regulating pseudohyphal differentiation, invasive growth and starch metabolism in Saccharomyces cerevisiae in response to nutrient availability. Mol. Microbiol. 47: 119–134. [DOI] [PubMed] [Google Scholar]
- Gancedo, J. M., 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 107–123. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., R. H. Schiestl, A. R. Willems and R. A. Woods, 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360. [DOI] [PubMed] [Google Scholar]
- Gimeno, C. J., P. O. Ljungdahl, C. A. Styles and G. R. Fink, 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090. [DOI] [PubMed] [Google Scholar]
- Guldener, U., S. Heck, T. Fielder, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B., C. A. Styles, Q. Feng and G. R. Fink, 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97: 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme, A., S. Bumgarner, C. Styles and G. R. Fink, 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- Hoyer, L. L., 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9: 176–180. [DOI] [PubMed] [Google Scholar]
- Huang, L., W. Zhang and S. Y. Roth, 1997. Amino termini of histones H3 and H4 are required for a1-alpha2 repression in yeast. Mol. Cell. Biol. 17: 6555–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N., A. W. Martinez, E. C. Robert, N. B. Agochukwu, M. E. Ibos et al., 2005. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171: 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh, D., and K. Struhl, 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371. [DOI] [PubMed] [Google Scholar]
- Kadosh, D., and K. Struhl, 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson and A. D. Johnson, 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68: 709–719. [DOI] [PubMed] [Google Scholar]
- Kim, T. S., H. Y. Kim, J. H. Yoon and H. S. Kang, 2004. Recruitment of the Swi/Snf complex by Ste12-Tec1 promotes Flo8-Mss11-mediated activation of STA1 expression. Mol. Cell. Biol. 24: 9542–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, T., S. Wesche, N. Taheri, G. H. Braus and H. U. Mosch, 2002. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot. Cell 1: 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin, S., V. K. Vyas and M. Carlson, 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22: 3994–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, T. M., and A. P. Mitchell, 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, T. M., W. Xu, A. Diamond and A. P. Mitchell, 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276: 1850–1856. [DOI] [PubMed] [Google Scholar]
- Lambrechts, M. G., F. F. Bauer, J. Marmur and I. S. Pretorius, 1996. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 93: 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, W. S., and A. M. Dranginis, 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., and J. Heitman, 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150: 1443–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., R. S. Muir, E. Lim, J. McElver, S. C. Weber et al., 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158: 113–117. [DOI] [PubMed] [Google Scholar]
- Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue et al., 2000. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani, H. D., and G. R. Fink, 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275: 1314–1317. [DOI] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 2000. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell. Biol. 20: 8364–8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22: 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C. L., and J. L. Workman, 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10: 187–192. [DOI] [PubMed] [Google Scholar]
- Peterson, C. L., A. Dingwall and M. P. Scott, 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91: 2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C. L., Y. Zhao and B. T. Chait, 1998. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J. Biol. Chem. 273: 23641–23644. [DOI] [PubMed] [Google Scholar]
- Proft, M., and K. Struhl, 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9: 1307–1317. [DOI] [PubMed] [Google Scholar]
- Puig, S., M. Lau and D. J. Thiele, 2004. Cti6 is an Rpd3-Sin3 histone deacetylase-associated protein required for growth under iron-limiting conditions in Saccharomyces cerevisiae. J. Biol. Chem. 279: 30298–30306. [DOI] [PubMed] [Google Scholar]
- Reynolds, T. B., and G. R. Fink, 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291: 878–881. [DOI] [PubMed] [Google Scholar]
- Roberts, R. L., and G. R. Fink, 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8: 2974–2985. [DOI] [PubMed] [Google Scholar]
- Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner et al., 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93: 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, S., E. Summers, H. J. Lo, H. Madhani and G. Fink, 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18: 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D.W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sidorova, J., and L. Breeden, 1999. The MSN1 and NHP6A genes suppress SWI6 defects in Saccharomyces cerevisiae. Genetics 151: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett, A. J., D. J. Dilworth, M. J. Davey, M. O'Donnell, J. D. Aitchison et al., 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyk, D., G. Hansson, I. S. Pretorius and F. F. Bauer, 2003. Cellular differentiation in response to nutrient availability: the repressor of meiosis, Rme1p, positively regulates invasive growth in Saccharomyces cerevisiae. Genetics 165: 1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyk, D., I. S. Pretorius and F. F. Bauer, 2005. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics 169: 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen, K. J., and F. M. Klis, 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60: 5–15. [DOI] [PubMed] [Google Scholar]
- Verstrepen, K. J., T. B. Reynolds and G. R. Fink, 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2: 533–540. [DOI] [PubMed] [Google Scholar]
- Vidal, M., and R. F. Gaber, 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. Yu et al., 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14: 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman et al., 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400: 784–787. [DOI] [PubMed] [Google Scholar]
- Xu, W., and A. P. Mitchell, 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183: 6917–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. J., 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. BioEssays 26: 1076–1087. [DOI] [PubMed] [Google Scholar]
- Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert et al., 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404. [DOI] [PubMed] [Google Scholar]
- Zeng, L., and M. M. Zhou, 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513: 124–128. [DOI] [PubMed] [Google Scholar]