Abstract
Chromosomal genes modulate Ty retrotransposon movement in the genome of Saccharomyces cerevisiae. We have screened a collection of 4739 deletion mutants to identify those that increase Ty1 mobility (Ty1 restriction genes). Among the 91 identified mutants, 80% encode products involved in nuclear processes such as chromatin structure and function, DNA repair and recombination, and transcription. However, bioinformatic analyses encompassing additional Ty1 and Ty3 screens indicate that 264 unique genes involved in a variety of biological processes affect Ty mobility in yeast. Further characterization of 33 of the mutants identified here show that Ty1 RNA levels increase in 5 mutants and the rest affect mobility post-transcriptionally. RNA and cDNA levels remain unchanged in mutants defective in transcription elongation, including ckb2Δ and elf1Δ, suggesting that Ty1 integration may be more efficient in these strains. Insertion-site preference at the CAN1 locus requires Ty1 restriction genes involved in histone H2B ubiquitination by Paf complex subunit genes, as well as BRE1 and RAD6, histone H3 acetylation by RTT109 and ASF1, and transcription elongation by SPT5. Our results indicate that multiple pathways restrict Ty1 mobility and histone modifications may protect coding regions from insertional mutagenesis.
THE Ty1, -2, and -5 and Ty3 retrotransposons of Saccharomyces belong to the Ty1/Copia and the Ty3/Gypsy superfamilies, respectively, which are present in every eukaryotic genome examined to date (Boeke and Devine 1998; Eickbush and Malik 2002; Sandmeyer et al. 2002; Voytas and Boeke 2002; Lesage and Todeschini 2005). Ty-element structure, expression strategy, and the process of retrotransposition resemble those of retroviruses, except Ty elements lack an envelope gene and retrotransposition is not infectious. Ty elements are flanked by long terminal repeats (LTRs) and transcribed from end to end, forming a transcript that is a template for translation and reverse transcription. Translation results in the synthesis of Ty Gag, a retroviral-like capsid protein, and a Gag-Pol fusion protein containing protease (PR), integrase (IN), and reverse-transcriptase (RT) domains. Synthesis of Gag-Pol results from a specific frameshifting event within a small region of overlapping coding sequence in GAG and POL. Gag and Gag-Pol proteins form cytoplasmic virus-like particles (VLPs) within which protein maturation and reverse transcription occur, and assembly of functional Ty3 VLPs takes place in association with P-bodies (Beliakova-Bethell et al. 2006; Larsen et al. 2007). Cis-acting signals for Ty translational frameshifting, RNA packaging and dimer formation, and the initiation and strand-transfer events necessary for reverse transcription are present on Ty mRNA. Like retroviruses, a Ty preintegration complex (PIC) minimally containing Ty cDNA and IN may be released from cytoplasmic VLPs. The Ty PIC traverses the nuclear membrane in a manner similar to human immunodeficiency virus (HIV) infection of quiescent cells, via a nuclear localization signal present on IN. Most Ty1- and all Ty3-element insertions occur near genes transcribed by RNA polymerase III (Chalker and Sandmeyer 1990; Kirchner et al. 1995; Devine and Boeke 1996; Yieh et al. 2000; Bachman et al. 2005), while Ty5 inserts into subtelomeric regions and adjacent to the silent-mating cassettes (Zou et al. 1996; Zou and Voytas 1997; Xie et al. 2001). Ty1 elements are capable of insertional mutagenesis, where they preferentially insert into promoter regions; however, insertion into coding sequences also occurs (Natsoulis et al. 1989). In addition, Ty cDNA undergoes recombination with chromosomal elements, especially when transpositional integration is blocked.
The Ty life cycle's nuclear and cytoplasmic phases set the stage for interactions with a variety of cellular genes and processes. The first nuclear phase involves Ty transcription, RNA processing, and export, while the second involves nuclear import of the PIC followed by transpositional integration. Distinct cytoplasmic phases derive from the fact that Ty RNA is used for translation and is also encapsidated into VLPs for reverse transcription and formation of the PIC. Identifying cellular genes that have been co-opted by Ty1 and Ty3 elements has not only resulted in a deeper understanding of retroelement replication and control, but has also helped elucidate the normal function of these genes in the cell. Certain Ty–host cell interactions are conserved with retroviruses and, therefore, may help identify new antiviral targets for HIV (see the recent review by Maxwell and Curcio 2007). Highlighted among these are mammalian genes required for DNA repair, genome maintenance, and protein trafficking that also modulate retroviral replication and virion budding (Yoder and Bushman 2000; Li et al. 2001; Kilzer et al. 2003; Aye et al. 2004; Lau et al. 2004; Irwin et al. 2005; Beliakova-Bethell et al. 2006; Lloyd et al. 2006; Smith and Daniel 2006; Yoder et al. 2006; Larsen et al. 2007).
Minimizing Ty1 retrotransposition is important for maintaining the integrity of the yeast genome since these elements can mutate genes and are involved in genome rearrangements (Voytas and Boeke 2002; Lesage and Todeschini 2005). A variety of genetic screens have identified host genes modulating Ty1 RNA level (Winston 1992; Yamaguchi et al. 2001; Timmers and Tora 2005), translational frameshifting (Xu and Boeke 1990; Kawakami et al. 1993; Farabaugh 1995), protein processing and VLP maturation (Conte et al. 1998), RT activity (Bolton et al. 2002; Yarrington et al. 2007), cDNA level (Lee et al. 1998; Rattray et al. 2000; Scholes et al. 2001; Griffith et al. 2003) and stability (Lee et al. 2000), target-site preference (Liebman and Newnam 1993; Bachman et al. 2005; Gelbart et al. 2005; Mou et al. 2006), or the overall level of retrotransposition (Scholes et al. 2001; Griffith et al. 2003). Analyzing deletion and transposon libraries has also revealed functions that modulate Ty3 transposition involving chromatin dynamics, RNA metabolism and translation, tRNA processing, vesicular trafficking, nuclear transport, and genome integrity (Aye et al. 2004; Irwin et al. 2005; Beliakova-Bethell et al. 2006; Larsen et al. 2007).
An important cellular modulator of Ty retrotransposition, RAD6, encodes an E2 ubiquitin-conjugating enzyme involved in DNA repair and transcription elongation (Broomfield et al. 2001; Osley 2004). RAD6 inhibits Ty1 and Ty3 retrotransposition and minimizes Ty1 integration in coding sequences and transcription units (Picologlou et al. 1990; Kang et al. 1992; Liebman and Newnam 1993; Irwin et al. 2005). Rad6p acts together with the E3 ubiquitin ligases Rad18p and Bre1p to ubiquitinate Pol30p (PCNA) (Hoege et al. 2002) and histone H2B (Robzyk et al. 2000; Hwang et al. 2003; Wood et al. 2003a), respectively. The RNA polymerase II-associated Paf complex is also required for Rad6p-mediated H2B ubiquitination (Ng et al. 2003; Wood et al. 2003b; Xiao et al. 2005).
Here, we report the results of a systematic screen for Ty1 restriction genes, which complements and expands the number and functional classes of retrotransposition inhibitory genes obtained from previous studies. Bioinformatic analyses have been utilized to illustrate the various biological processes that help or restrict Ty1- and Ty3-element movement in the genome. We have also identified additional genes involved in histone transactions and transcription that minimize Ty1 insertional mutagenesis in coding sequences, as well as at their preferred targets upstream of genes transcribed by RNA polymerase III.
MATERIALS AND METHODS
Genetic techniques, media, and strains:
Yeast genetic techniques and media were used as described previously (Sherman et al. 1986; Guthrie and Fink 1991). The haploid MATα deletion collection (Giaever et al. 2002) was obtained from Invitrogen (Carlsbad, CA). A total of 4739 deletion mutants derived from BY4742 (MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0) (Brachmann et al. 1998) were transformed with pBJC573, a URA3-based integrating plasmid carrying a complete Ty1 element tagged with a modified indicator gene his3-AId1 (designated his3-AI) (Curcio and Garfinkel 1991), which cannot undergo recombination with the internal deletion of HIS3 (his3-Δ1) present in BY4742. Briefly, the his3-AId1 allele was constructed by cotransforming pBDG208, which contains the HIS3 gene oriented such that transcription of the GAL1-promoted Ty1 (pGTy1) and the HIS3 gene is in opposite directions, and a PCR-amplified disruption fragment containing the artificial intron (AI) flanked by 40 bp of HIS3 coding sequences into the Ty-less strain DG1768 (Garfinkel et al. 2003). The PCR primers used were ΔAIT2 (5′-GGATCATCTCGCAAGAGAGATCTCCTACTTTCTCCCTTTGGTATGTTAATATGGAC-3′) and ΔAIB2 (5′-TCTTTCGAACAGGCCGTACGCAGTTGTCGAACTTGGTTTGCTGTTATAAATAATACC-3′) and the AI template was pUC/intron 4 (Yoshimatsu and Nagawa 1989). The PCR primers flanked a region 400–480 bp from the start of the HIS3 coding sequence. To identify pGTy1his3-AId1 recombinants, His− Ura+ transformants identified by replica plating were spread on synthetic complete medium lacking uracil (SC −Ura) + galactose, incubated for 4 days at 20°, and then replicated to SC −His −Ura + glucose medium. Candidate pGTy1his3-AId1 plasmids were recovered from transformants that remained His− when propagated on glucose but produced many His+ papillae when induced with galactose. The presence of the AI in the expected position within HIS3 was confirmed by DNA sequencing. To construct pBJC573, the his3-AId1 gene was subcloned into a URA3-based Ty1-integrating plasmid at the BglII site in TYB1 adjacent to the 3′ LTR. Further details on pBJC573 can be found elsewhere (Bryk et al. 2001; Scholes et al. 2001). Integrants of pBJC573 upstream of the HIS4 gene were enriched for by linearizing the plasmid with PacI, which cleaves once in the HIS4 sequences adjacent to the Ty1his3-AI element. Strain DG2122 was constructed by introducing pBJC573 into the parental strain BY4742. Strain JC3787 was derived from BY4742 after transposition induction of cells harboring pBDG945 (pGTy1-H3his3-AId1) and contains a genomic Ty1his3-AId1 element (Mou et al. 2006). Strain DG3027 [MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 can1-Ty1(26) (pBJC573)] was generated by crossing BY4743 (MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0) (Brachmann et al. 1998) and DG3016 [DG2122; can1-Ty1(26) ] followed by tetrad analysis. One-step gene disruptions were performed using KanMX4-targeting fragments (Wach et al. 1994), amplified from the deletion mutants using the gene-specific flanking primers A and D (http://www-sequence.stanford.edu/group/yeast_deletion_project/Deletion_primers_PCR_sizes.txt). Mutant identity was verified by PCR using A and D primers, phenotypic analyses, and complementation tests.
Ty1 his3-AI mobility assay used for systematic screening:
To detect Ty1 mobility events, four independent transformants containing pBJC573/Ty1his3-AI from each deletion mutant along with the wild-type strain DG2122 were streaked for single colonies on SC −Ura and incubated at 20° for 5 days, which is optimal for Ty1 retrotransposition (Paquin and Williamson 1984). Ura+ cells were replica plated to SC −His plates and incubated at 30° for 3–4 days. The levels of His+ papillation from the deletion mutant and DG2122 were compared. Candidate mutants containing pBJC573 that displayed a higher level of Ty1his3-AI-mediated His+ papillation in at least three of the four transformants were retested and used for further analyses.
Gene ontology enrichment analysis:
Gene ontology biological processes (GOBP) enrichment analysis was performed using Whole Pathway Scope (WPS) software (Yi et al. 2006). Yeast gene ontology annotations were obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/). The GOBP enrichment was performed using overrepresentation analysis. Fisher's exact test was performed on 2 × 2 contingency tables, to determine whether a gene is in a given list or not vs. whether this gene is associated with a GOBP term or not. A one-sided Fisher's exact test was used to determine which biological processes and pathways had a statistically significant enrichment within the Ty1 restriction gene list or other gene lists. The associated GOBP terms were ranked into a term enrichment list on the basis of their Fisher's exact test P-values with the most enriched terms at the top. Comparison of gene lists of Ty modulators from different sources at the GOBP level was performed using an extended version of WPS (M. Yi and R. M. Stephens, unpublished results). Briefly, enrichment levels of each GOBP term were computed in a batch mode for each of the lists and the results were merged into a Stanford format file with a matrix of enrichment scores [−Log(P-value) of Fisher's exact test P-values], which were filtered using the criteria of list hits >1 (list hit: genes from the intended list that are associated with the corresponding GOBP term) and P-values <0.05. The results were displayed in color-coded “heat maps” to reveal the patterns of significantly altered biological processes from the multiple-gene lists. The color coding of the heat maps is related to the enrichment of genes in a GOBP term [−Log(P-value) based]. The gradient of red color in the heat map indicated the enrichment levels with the maximal red color (enrichment score ≥3; P-value ≤0.001) and black denoting no enrichment (enrichment score 0; P-value >0.05). Underrepresented GOBP terms were not included in this analysis as a separate feature. Hierarchical clustering was obtained using the average linkage and Euclidean distance of the GOBP terms in the rows. All rows were organized into a binary tree as a dendrogram. The lower height of a subtree indicated a greater similarity in enrichment levels of GOBP terms across all the gene lists in the heat map. The vertical lines across the hierarchical lines determined the number of clusters at a certain depth.
Frequency of Ty1his3-AI mobility:
Each strain was streaked for single colonies on SC −Ura at 20°. A single colony was suspended in 10 ml of SC −Ura and ∼103 cells were inoculated into four individual 1-ml SC −Ura liquid cultures and grown at 20° until saturation. Aliquots of the cultures were spread on SC −Ura and SC −His −Ura plates, followed by incubation at 30° for 5 days. The frequency of Ty1his3-AI mobility was calculated by dividing the average number of His+ Ura+ cells per milliliter by the average number of Ura+ cells per milliliter.
Monitoring Ty1 insertions at SUF16 by PCR:
The deletion mutants and the wild-type strain DG2122 were grown on SC −Ura plates at 20° for 4 days. Three single colonies per strain were inoculated into individual tubes containing 10 ml of SC −Ura liquid media and grown at 20° until saturation. Total genomic DNA was isolated by glass-bead/phenol lysis (Hoffman and Winston 1987) and used as a template in a PCR reaction with oligonucleotide primers specific to Ty1 (TyB OUT: 5′-GAACATTGCTGATGTGATGACA-3′) and SNR33 (SNR33 OUT: 5′-TTTTAGAGTGACACCATCGTAC-3′), which is adjacent to the 3′ end of SUF16, as described previously (Sundararajan et al. 2003). An aliquot of the reaction was analyzed by electrophoresis on a 1% agarose gel in Tris–Borate–EDTA running buffer. The ethidium bromide-stained gel was scanned on a Typhoon Trio phosphorimager (GE Healthcare, Piscataway, NJ), and the intensity of the Ty1 insertion pattern was compared to that obtained with the wild-type strain DG2122. To ensure that the DNA samples were PCR competent, control reactions were performed with primers containing sequence from the CPR7 locus (CPR7A: 5′-GTTTGTGATTTATCTCTGGACTGCT-3′ and CPR7D: 5′-AGTTCGTCTCTCCTTCATATTCTCA-3′).
Northern blot analysis of Ty1 RNA:
A single colony from each strain was inoculated into 10 ml of SC −Ura liquid medium and grown until late log phase at 20°. Total RNA was isolated using the MasterPure yeast RNA purification kit (Epicentre, Madison, WI). RNA samples were separated on a 1.2% agarose– formaldehyde gel and transferred to Hybond-N (GE Healthcare). An RT-domain probe was obtained by purifying a 1.6-kb PvuII–ClaI fragment from pGTy1Cla (Garfinkel et al. 1988), using a QIAquick gel extraction kit (QIAGEN, Valencia, CA). The RT-domain fragment was labeled by randomly primed DNA synthesis using the Megaprime DNA labeling system and [α-32P]dCTP (GE Healthcare). The levels of Ty1 and Ty1his3-AI transcripts were normalized to the 18S and 28S rRNA bands visualized by staining with ethidium bromide. Gel electrophoresis and Northern hybridizations were performed as described previously (Lee et al. 1998). The hybridization and ethidium bromide fluorescence signals were quantified using a Typhoon Trio phosphorimager and ImageQuant TL software (GE Healthcare). Lane background was subtracted using the rolling-ball method, which calculates the background as if a disc with a radius setting of 200 (default) were rolling underneath each lane profile. We also verified the level of Ty1 RNAs for 16 (asc1Δ, asf1Δ, bre1Δ, bud27Δ, cdc40Δ, cdc73Δ, ckb2Δ, ipk1Δ, leo1Δ, paf1Δ, pbs2Δ, rpd3Δ, rtf1Δ, spt5Δ#, ssk2Δ, and ypl183cΔ/rtt10) of the 33 Ty1 restriction mutants.
Southern blot analysis of Ty1 cDNA:
The deletion strains and the wild-type strain DG2122 were grown on SC −Ura plates at 20° for 4 days. An individual colony from each strain was inoculated into 10-ml SC −Ura liquid cultures and grown until mid- to late-log phase at 20°. Total genomic DNA was isolated as described above, digested with PvuII, separated on a 0.8% agarose gel at 4°, and transferred to Hybond-N (GE Healthcare). A 32P-labeled DNA probe was derived from the Ty1 RT region as described above. Gel electrophoresis and Southern hybridizations were performed as described previously (Lee et al. 1998). The intensity of the 2-kb cDNA band was determined by phosphorimage analysis and normalized to three conserved Ty1-chromosomal junction fragments.
Monitoring Ty1 insertions at CAN1:
Spontaneous canavanine-resistant (CanR) mutants from selected deletion strains as well as DG2122 were obtained by streaking cells for single colonies on SC plates. After incubating for 4–5 days at 20°, cells were replicated to SC −Arg +Can plates and incubated at 30° until CanR papillae appeared (Rinckel and Garfinkel 1996). Independent CanR mutants were clonally purified on SC −Arg +Can plates prior to isolation of DNA. The frequency of CanR was determined by inoculating ∼103 cells from an independent colony of each mutant into four individual 1-ml SC liquid cultures followed by incubation at 20° until the cultures saturated. Dilutions of each culture were spread onto SC plates and SC −Arg +Can plates and incubated at 30° for 5 days. The frequency of CanR was calculated by dividing the average number of CanR cells per milliliter by the average number of total cells per milliliter. PCR was used to detect and map the positions of Ty1 insertions that disrupted CAN1. Total genomic yeast DNA was initially analyzed using primers CAN1(−317) (5′-GTCTCTATCAATGAAAATTTCGAGG-3′) and CAN1(+1966) (5′-GTTTCAAATGCTTCTACTCCGTCTGC-3′) that bracket CAN1 and include 317 bp of 5′-noncoding sequence, 1773 bp of coding sequence, and 193 bp of 3′-noncoding sequence. CanR mutants lacking the 2283-bp CAN1 amplification product were analyzed using Ty1-specific primers midLTR OUT (5′-ATTCATTGATCCTATTACATTATC-3′) and midLTR IN (5′-GATAATGTAATAGGATCAATGAAT-3′) and primers adjacent to the CAN1 start codon CAN1 IN (5′-ATGACAAATTCAAAAGAAGACG-3′) and CAN1 OUT (5′-CGTCTTCTTTTGAATTTGTCAT-3′). Chi-square analysis was performed to determine if the incidence of 5′-noncoding insertions vs. coding sequence insertions was significantly altered in the deletion mutants. Since the fraction of spontaneous Ty1-induced can1 mutations in wild-type cells is too low to be recapitulated here, we compiled the data from hundreds of Ty1 transposition events at CAN1 obtained in wild-type backgrounds from previous studies showing that ∼50% of Ty1 insertions occur in the CAN1 promoter, defined as a 317-bp region upstream of the initiation codon, and the rest were in the 1773-bp CAN1 coding sequence (Wilke et al. 1989; Picologlou et al. 1990; Liebman and Newnam 1993; Rinckel and Garfinkel 1996). The number of trials used for the wild-type sample was adjusted to the number of trials for a given restriction mutant in the chi-square analysis. For example, one promoter region and 19 coding sequence insertions of Ty1 were obtained at CAN1 in a paf1Δ mutant. Therefore, we assumed that 10 promoter and 10 coding sequence insertions occurred in the wild type to estimate the P-value. DNA sequencing of Ty1 or solo-LTR insertions at CAN1 was performed using Ty1 primer LTR (+89) (5′-CATTTGCGTCATCTTCTAACACCG-3′) or the midLTR primers, respectively.
RESULTS AND DISCUSSION
Identifying genes that restrict Ty1 mobility:
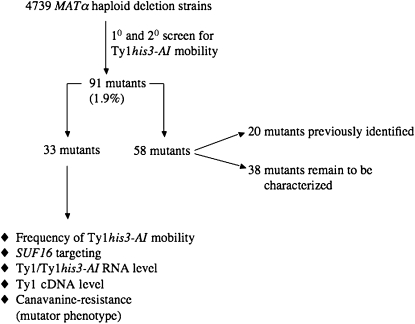
We introduced a functional Ty1 element under the control of its native promoter and carried on a URA3-integrating plasmid, pBJC573 (Bryk et al. 2001; Scholes et al. 2001), into 4739 haploid MATα deletion mutants (Figure 1). The Ty1 element contained the his3-AI (artificial intron) retroelement indicator gene, which allows Ty1 movement to be monitored by the formation of His+ colonies, following Ty1his3-AI RNA splicing and reverse transcription (Curcio and Garfinkel 1991). His+ cells usually arise by de novo retrotransposition of Ty1HIS3 to a new chromosomal location in wild-type cells, but homologous recombination between Ty1HIS3 cDNA and a resident element also occurs (Sharon et al. 1994). Therefore, the term “Ty1 mobility” is used to describe the Ty1HIS3 events that arise from Ty1 retrotransposition and cDNA recombination. Integrative recombination of pJBC573 containing Ty1his3-AI was targeted to the 5′-noncoding region of HIS4 by digestion with PacI. Four independent Ura+ transformants of each deletion mutant were tested along with a wild-type strain containing pJBC573 (DG2122) for an increase in His+ papillation to identify Ty1 restriction genes. An increase of threefold in Ty1his3-AI mobility could be reproducibly detected using this qualitative assay and at least three of the four independent transformants from a given mutant were required to show increased Ty1his3-AI mobility to be saved for further analyses. We identified 91 mutants with a higher level of Ty1his3-AI mobility when compared with DG2122 (Figure 1, supplemental Table S1 at http://www.genetics.org/supplemental/). The Ty1 restriction mutants were placed in the following categories (Figure 1): Thirty-three novel mutants were chosen for further analysis on the basis of the function of the deleted gene or their level of Ty1 mobility, 20 mutants were identified in previous screens, and 38 mutants remain to be characterized.
Figure 1.—
Characterization of Ty1 restriction mutants. Ninety-one mutants (1.9%) were identified from 4639 MATα deletion strains, on the basis of an increased level of His+ papillation mediated by a chromosomal Ty1 element containing the retrotransposon mobility marker, his3-AI. Thirty-three mutants were chosen for further analyses after considering the function of the deleted gene and the level of Ty1his3-AI mobility, 20 mutants were identified in other screens for Ty1 mobility, and 38 mutants remain to be characterized.
Bioinformatic analyses reveal functional relationships between Ty modulators:
Computational approaches were used to determine if the 91 Ty1 restriction genes were functionally related (Table 1, supplemental Table S1) and the degree of overlap with previous genetic screens for Ty1 restriction genes (Scholes et al. 2001), Ty1 helper genes (Griffith et al. 2003), and Ty3 restriction and helper genes (Irwin et al. 2005) (Figures 2 and 3, supplemental Figure S1 at http://www.genetics.org/supplemental/). Eighty-five of the 91 Ty1 restriction genes identified here were associated with at least one GOBP (Table 1, list total), while six ORFs remained uncharacterized (YBR239C, YGR110W, YJR142W, YLR282C, YML009W-B, and YPL183C). Most of the genes described here contain multiple GOBP terms, ranging from a specific term such as telomerase-independent telomere maintenance to a general term such as nucleic acid metabolism. The number of Ty1 restriction genes (list hits) associated with at least one GOBP term ranged from 5 (telomerase-independent telomere maintenance) to 51 (nucleic acid metabolism), while the number of unique yeast genes (population hits) annotated to a GOBP ranged from 8 (negative regulation of transposition) to 1490 (nucleic acid metabolism). When the list hits of Ty1 restriction genes and population hits of unique yeast genes were compared, the top-scoring GOBP terms were enriched for DNA repair and recombination, regulation of transposition, transcription, the cell cycle, cell proliferation, and chromatin transactions, with P-values ranging from 3.58 × 10−14 to 4.9 × 10−7. Furthermore, 80% of the 85 annotated genes identified here are involved in nuclear processes. Together, these results suggest that a limited number of cellular functions inhibit Ty1 movement in the yeast genome.
TABLE 1.
Overrepresentation analysis of Ty1 restriction mutants
| Gene ontology biological process (GOBP) | List hits | List total | Population hits | Population total | P-value (×10−10) |
|---|---|---|---|---|---|
| Double-strand break repair | 12 | 85 | 39 | 6455 | 0.000358 |
| Nucleic acid metabolism | 51 | 85 | 1490 | 6455 | 0.00223 |
| Nonrecombinational repair | 10 | 85 | 27 | 6455 | 0.00641 |
| Response to DNA damage stimulus | 19 | 85 | 194 | 6455 | 0.0374 |
| Double-strand repair via homologous recombination | 8 | 85 | 17 | 6455 | 0.143 |
| Recombinational repair | 8 | 85 | 17 | 6455 | 0.143 |
| Chromosome organization and biogenesis (sensu Eukaryota) | 20 | 85 | 237 | 6455 | 0.15 |
| Negative regulation of DNA transposition | 6 | 85 | 8 | 6455 | 1.2 |
| Nuclear organization and biogenesis | 20 | 85 | 297 | 6455 | 8.91 |
| DNA recombination | 16 | 85 | 187 | 6455 | 17.7 |
| Transcription/DNA dependent | 23 | 85 | 441 | 6455 | 58.2 |
| Meiotic recombination | 8 | 85 | 39 | 6455 | 286 |
| Cell cycle | 23 | 85 | 530 | 6455 | 1830 |
| Establishment and maintenance of chromatin architecture | 14 | 85 | 202 | 6455 | 2950 |
| Telomerase-independent telomere maintenance | 5 | 85 | 13 | 6455 | 4170 |
| Cell proliferation | 24 | 85 | 601 | 6455 | 4360 |
| Chromatin modification | 13 | 85 | 178 | 6455 | 4490 |
List hits, number of Ty1 restriction genes associated with the designated GOBP term; list total, number of Ty1 restriction genes associated with at least one GOBP term; population hits, the number of unique genes annotated for the designated GOBP term; population total, the number of unique genes annotated for a GOBP term; P-value, one-sided Fisher's exact test.
Figure 2.—
Heat map of enrichment scores of GO biological process terms (GOBPs) for gene lists derived from independent screens for Ty1 (Scholes et al. 2001; Griffith et al. 2003) and Ty3 modulators (Irwin et al. 2005). The gradient of red color indicates the enrichment levels with black representing no enrichment [maximum enrichment ≥3 (P-value ≤0.001), no enrichment = 0 (P-value >0.05)]. Rows of the heat map are GOBPs and columns are the genetic screens for Ty modulators. A-R: more detailed GOBPs associated with Ty1 restriction genes identified in this work (supplemental Table S1). A heat map containing all GOBPs associated with Ty1 restriction genes identified here is shown in supplemental Figure S1. B-R: GOBPs associated with Ty1 restriction genes identified by transposon mutagenesis (Scholes et al. 2001). C-H: GOBPs associated with Ty1 helper genes identified by systematic screening of a diploid deletion library (Griffith et al. ;2003). D-R: GOBPs associated with Ty3 restriction genes identified by systematic screening of a haploid deletion library (Irwin et al. 2005). D-H: GOBPs associated with Ty3 helper genes identified by systematic screening of a haploid deletion library (Irwin et al. 2005). Hierarchical clustering of the GOBPs for Ty modulators is shown on the left.
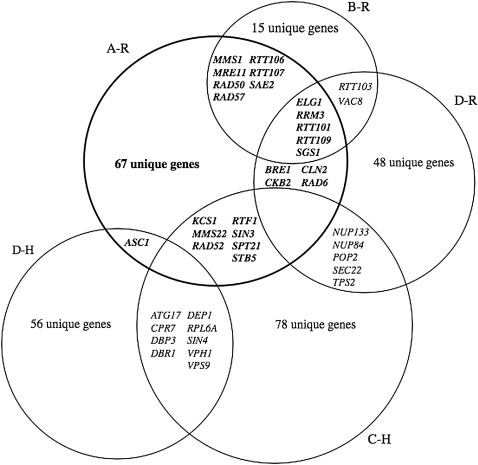
Figure 3.—
Identification of common and unique Ty modulators. Abbreviations are defined in supplemental Figure S1 and in Figure 2. The shaded circle represents Ty1 restriction genes identified in this work (A-R). No genes in common were found when B-R and C-H, and B-R and D-H, were compared. Also refer to Maxwell and Curcio (2007) for further comparative analyses of Ty1 (B-R and C-H) and Ty3 (D-R and D-H) modulators.
To compare the enrichment levels and patterns of GOBP terms associated between the gene list obtained in our screen for Ty1 restriction genes and the terms obtained in previous screens for Ty1 and Ty3 cellular modulators, GOBP heat maps (supplemental Figure S1 and Figure 2) were generated using the data from our systematic screen (A-R), a subgenomic screen for Ty1 restriction genes using transposon mutagenesis (B-R) (Scholes et al. 2001), a systematic screen for Ty1 helper genes (C-H) (Griffith et al. 2003), and a systematic screen for Ty3 restriction (D-R) and helper genes (D-H) (Irwin et al. 2005). Briefly, enrichment levels of each GOBP term were computed for gene lists from each Ty modulator screen, and the results were combined into a data matrix used for the comparison. The matrix was displayed in color-coded heat maps to reveal the patterns of related biological processes identified by comparing different Ty modulator gene lists. The color coding of the heat maps is related to the enrichment of genes with specific GOBP terms. Increasing shades of red indicate higher enrichment and black indicates no enrichment. Two heat maps are included. The first (supplemental Figure S1) compares all GOBP terms associated with the 85 Ty1 restriction genes identified here (supplemental Table S1) and the second (Figure 2) compares more detailed GOBP terms from these restriction genes. Also note that novel genes and GOBP terms obtained in the other screens are not represented in the heat maps because comparisons were made with the GOBP terms identified in our screen for Ty1 restriction genes.
The systematic screen for Ty1 restriction genes presented here verified and expanded the number of GOBP terms identified by Scholes et al. (2001) (compare A-R with B-R) (supplemental Figure S1 and Figure 2). There was significant overlap between a subset of GOBP terms, because we recovered 12/21 mutants (Figure 3) previously identified as nonmarginal inhibitors of Ty1 mobility. These included general nuclear processes such as DNA replication, repair, and transcription, as well as the regulation of transposition (supplemental Figure S1 and Figure 2). We identified genes with novel GOBP terms for stress responses and signaling, transcriptional regulation and elongation, and several aspects of chromatin structure and function.
Several common GOBP clusters were also shared between the screen reported here and the systematic screen for Ty1 helper genes performed by Griffith et al. (2003) (supplemental Figure S1 and Figure 2; compare A-R and C-H). The GOBP terms tended to be more general (refer to the extreme top and bottom clusters of the heat map, supplemental Figure S1), although more specific processes such as negative regulation of transcription, RNA elongation, and DNA double-strand break repair were identified in both screens (supplemental Figure S1 and Figure 2). However, an apparent conflict was created by common GOBP terms associated with several genes identified in both screens, because the Griffith et al. (2003) systematic screen identified Ty1 helper genes, whereas our screen identified Ty1 restriction genes (see below, Figure 3). Genes involved in RNA processing and turnover, translation, and protein folding and trafficking are required for Ty1 mobility (Griffith et al. 2003), but were not highly represented in our screen for restriction genes. Together, these results suggest that common as well as distinct processes help or restrict Ty1 mobility, a property that may reflect the interactive capacity of regulatory networks in yeast (Harrison et al. 2007).
Although Ty1 and Ty3 are both LTR retrotransposons inhabiting the Saccharomyces genome, they are distantly related (Eickbush and Malik 2002) and, therefore, may have unique interactions with their host cells. Consistent with this idea, the heat map analyses (supplemental Figure S1 and Figure 2) show several processes that restrict Ty1 but not Ty3 mobility (compare A-R and D-R), such as stress responses and DNA double-strand break repair. However, several closely related processes restricted Ty1 and Ty3 mobility, including chromatin transactions and chromatin-based gene silencing at telomeres and the response to DNA damage. There were few cellular processes that restricted Ty1 but were required for Ty3 mobility (compare A-R and D-H). This probably reflects the large number of genes involved in RNA metabolism and protein trafficking that are required for Ty3 and in certain instances Ty1 mobility (Griffith et al. 2003; Irwin et al. 2005; Maxwell and Curcio 2007). Together, these results indicate that both element-specific and shared cellular processes affect Ty1 and Ty3 movement. Our results also suggest the possibility that similar element–host interactions exist with Ty1/Copia and Ty3/Gypsy family members present in other organisms.
Common and unique cellular modulators obtained in different genetic screens:
We cross-referenced the four screens for Ty1 or Ty3 modulators to determine the number of identical genes identified in each screen (Figure 3). As mentioned above, over half of the restriction genes identified by Scholes et al. (2001) were reisolated here, including RTT101, RTT106, RTT107, and RTT109. BRE1, CKB2, CLN2, ELG1, RAD6, RRM3, RTT101, RTT103, RTT109, SGS1, and VAC8 restricted both Ty1 and Ty3 mobility, forming a core set of retrotransposon restriction genes and functions. In particular, BRE1 and RAD6 are responsible for ubiquitinating histone H2B, additional histone modifications (Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002), and transcription elongation by RNA polymerase II (Xiao et al. 2005). RAD6 is also implicated in determining Ty1 insertion-site preference at several loci (Kang et al. 1992; Liebman and Newnam 1993; Huang et al. 1999). RRM3 may inhibit Ty1 cDNA recombination (Scholes et al. 2001), and SGS1 minimizes multimeric Ty1-integration events (Bryk et al. 2001). Therefore, it will be important to determine whether a given restriction gene affects the same process during Ty1 and Ty3 retrotransposition.
Seven genes were identified that restricted Ty1 mobility in our systematic screen yet were required for Ty1 mobility in the screen performed by Griffith et al. (2003) (Figure 3; KCS1, MMS22, RAD52, RTF1, SIN3, SPT21, and STB5). Although there were several differences in the way the screens were performed, including cell type and incubation temperature, a major reason for this discrepancy in the data was that different Ty1 mobility assays and secondary tests were employed. Griffith et al. (2003) used a Ty1HIS3 element expressed at a high level from the GAL1 promoter carried on an episomal plasmid (termed a pGTy1 element), whereas we used a chromosomal Ty1his3-AI element expressed from its natural promoter that will yield a phenotypic signal only if reverse transcription of Ty1HIS3 mRNA occurs. Expression of a pGTy1 element overrides post-translational and copy number control mechanisms (Curcio and Garfinkel 1992; Garfinkel et al. 2003) and, therefore, may have biased the genes identified in the Griffith et al. (2003) screen. Conversely, the Ty1his3-AI mobility assay is very sensitive with a dynamic range over several orders of magnitude (Curcio and Garfinkel 1991). Since GAL1-promoted Ty1 and native Ty1 RNA levels were not determined in the helper mutants identified by Griffith et al. (2003), Ty1 expression may have been altered in certain mutants, even though GAL1 expression apparently remained unchanged, as judged by a qualitative assay using a GAL1-lacZ reporter. A control for the level of DNA recombination between pGTy1HIS3 and chromosomal Ty1 sequences or the internally deleted his3-Δ1 locus was also not included, which could be the reason for identifying the DNA repair and recombination gene, RAD52, as a Ty1 helper (Griffith et al. 2003). Finally, four of the Ty1 restriction genes in conflict have been described independently (RAD52) (Rattray et al. 2000) or confirmed by gene disruption (KCS1, RTF1, and SIN3) in a strain (JC3787) containing a different chromosomal Ty1his3-AI insertion (supplemental Table S1).
Although 31 genes were isolated in more than one of the Ty modulator screens, a large number of unique genes were identified in the various screens (Figure 3). Isolation of 160 and 104 unique modulators of Ty1 and Ty3 retrotransposition, respectively, suggests that the life cycles of these elements may indeed have different genetic requirements, which is consistent with several differences in their mode of retrotransposition. For example, Ty1 elements can mutate cellular genes and their transcripts accumulate to very high levels (Voytas and Boeke 2002), whereas Ty3 elements do not mutate cellular genes and Ty3 transcription and retrotransposition are induced by mating pheromones (Sandmeyer et al. 2002). Ty1 and Ty3 also utilize different targeting strategies at their preferred sites of insertion upstream of tRNA genes. Another possibility is that different factors involved in the same biological process (supplemental Figure S1 and Figure 2) modulate Ty1 and Ty3 retrotransposition. For example, BRE1 and RAD6 restrict Ty1 and Ty3 mobility; however, several genes composing the Paf complex, which interact with BRE1 and RAD6, restrict Ty1 but apparently do not modulate Ty3 mobility (Aye et al. 2004; Irwin et al. 2005). GAL-Ty3 expression may have also biased the mutants identified in the screen performed by Irwin et al. (2005).
Alternatively, the large number of different Ty modulators identified in the various screens may act through a few common pathways having many inputs. Support for this idea is evident from recent results indicating that loss of any one of 19 genome integrity genes stimulates Ty1 transposition by activating S-phase checkpoints caused by an increase in intrinsic DNA-damage or -replication blocks (Curcio et al. 2007). Therefore, epistasis analysis between additional Ty1 and Ty3 restriction genes and S-phase DNA checkpoint genes will further define the genetic pathways restricting transposition. How S-phase checkpoint activation stimulates Ty retrotransposition remains to be determined, but multiple steps in the process of retrotransposition and its control may be involved.
Cellular genes restrict Ty1his3-AI mobility to varying degrees: the chromatin/transcription group:
The frequency of Ty1his3-AI-mediated His+ events was determined for 33 Ty1 restriction mutants chosen for further analysis (Figure 1) that have defects in chromatin/transcription, stress response, and miscellaneous functions (Table 2, supplemental Table S1). The chromatin/transcription gene deletions conferred an increase in Ty1his3-AI mobility ranging from 5- to 275-fold. Interestingly, several members of the Paf transcription complex were identified as Ty1 restriction genes. Deletion of CDC73, LEO1, PAF1, and RTF1 enhanced Ty1his3-AI mobility 16- to 101-fold. The Paf complex is required for transcription elongation, 3′-end formation (Squazzo et al. 2002; Rondon et al. 2004; Penheiter et al. 2005; Sheldon et al. 2005), and histone H2B ubiquitination (Ng et al. 2003; Wood et al. 2003b; Xiao et al. 2005), where it acts in concert with Bre1p and Rad6p (Hwang et al. 2003; Wood et al. 2003a). RAD6 was previously identified as a Ty1 modulator (Picologlou et al. 1990) and was also detected in our screen (supplemental Table S1). In addition, Ty1his3-AI mobility increased 34-fold in a bre1Δ mutant (Table 2), which was also detected in our screen. Genes required for histone acetylation by the NuA4 complex (EAF3) (Reid et al. 2004), histone chaperone activity (ASF1) (Schwabish and Struhl 2006), histone deacetylation by the Rpd3-Sin3 complex (Kadosh and Struhl 1998), histone gene transcription by SPT21 (Dollard et al. 1994), and one of the histone H3 subunit genes, HHT1, restricted Ty1 mobility from 5- to 131-fold. It is surprising that deleting RPD3 or SIN3 increased Ty1 mobility 131- or 5-fold, respectively, since mutations in these genes usually confer similar phenotypes (Stillman et al. 1994; Kasten et al. 1996). Disrupting RPD3 and SIN3 in the wild-type strain JC3787 (supplemental Table S1) recapitulated the different levels of Ty1 mobility observed in the original rpd3Δ and sin3Δ strains, suggesting that suppressor mutations in additional Ty1 modulator genes were not present in the original mutants.
TABLE 2.
Effects of deleting Ty1 restriction genes on Ty1 mobility and CAN1 mutagenesis
| Gene deleteda | Ty1his3-AI mobility × 10−5 (SD) | Ty1 mobility, fold increase | SUF16 insertionsa | Ty1 RNA, fold change | Ty1his3-AI RNA, fold change | Ty1 cDNA, fold change | CanR levelb |
|---|---|---|---|---|---|---|---|
| Wild type | 0.16 (0.03)c | 1 | d | 1 | 1 | 1 | 0 |
| Chromatin/transcription | |||||||
| ARG82 | 23 (11) | 144 | e | 4.4 | 16 | 13.9 | 0 |
| ASF1 | 5 (0.8) | 31 | e | 5.2 | 1.6 | 5 | + |
| BRE1 | 5.4 (0.6) | 34 | f | 1.8 | 1.4 | 4.3 | + |
| CDC73 | 2.6 (0.3) | 16 | f | 1.2 | 1.4 | 2 | 0 |
| CSE2 | 5 (0.2) | 31 | e | 1.7 | 1.3 | 4.2 | 0 |
| EAF3 | 1 (0.1) | 6 | d | 1 | 1.5 | 1 | 0 |
| ELF1 | 4 (0.4) | 25 | e | 1 | 1 | 1 | + |
| HHT1 | 1.2 (0.1) | 8 | f | 3 | 2.6 | 1 | 0 |
| LEO1 | 4.2 (0.4) | 26 | f | 1 | 1 | 3 | + |
| PAF1 | 16.2 (7.0) | 101 | e | 2.8 | 2.8 | 4.3 | + |
| RPD3 | 21 (1.4) | 131 | <WTg | 0.6 | 1.3 | 1 | 0 |
| RTF1 | 2.8 (0.6) | 17.5 | e | 1.5 | 1.3 | 2.5 | + |
| SIN3 | 0.8 (0.1) | 5 | <WTg | 0.5 | 1 | 1.4 | 0 |
| SOH1 | 5.6 (0.9) | 35 | d | 2 | 2.6 | 2.7 | 0 |
| SPT21 | 2.7 (0.5) | 17 | f | 3.4 | 3 | 3 | 0 |
| SPT5#h | 44 (10) | 275 | e | 2.8 | 8.4 | 2.5 | + |
| SRB5 | 7 (0.4) | 44 | e | 2.8 | 4.2 | 5 | + |
| Stress response | |||||||
| ASC1 | 4.4 (3.1) | 27.5 | d | 1.5 | 1.5 | 4.8 | 0 |
| IPK1 | 3.4 (0.3) | 21.3 | e | 5 | 2.8 | 4 | 0 |
| KCS1 | 2.4 (0.43) | 15 | f | 3.7 | 2.4 | 3.8 | 0 |
| MMS22 | 1.5 (0.3) | 9.4 | f | 2 | 1 | 4.2 | 0 |
| PBS2 | 0.5 (0.11) | 3 | d | 1 | 1.3 | 2 | + |
| SSK2 | 0.7 (0.02) | 4.4 | d | 1 | 1.3 | 2 | 0 |
| SSK22 | 0.6 (0.08) | 3.8 | d | 1 | 2 | 1 | 0 |
| Miscellaneous | |||||||
| AGP3 | 1.3 (1.0) | 8 | f | 1 | 1.7 | 1 | 0 |
| ALR2 | 0.7 (0.13) | 4.4 | d | 0.5 | 1 | 0.6 | 0 |
| BEM4 | 2.2 (0.6) | 13.8 | f | 2 | 2.6 | 1 | 0 |
| BUD27 | 38 (1.4) | 237.5 | f | 0.8 | 6.3 | 0.5 | 0 |
| CKB2 | 9.7 | 60.6 | f | 1.6 | 1.8 | 0.7 | 0 |
| CLN2 | 1 (0.2) | 6.3 | d | 1.4 | 2.6 | 1.7 | 0 |
| SIC1 | 1.6 (0.01) | 10 | f | 2 | 2.4 | 2.4 | 0 |
| Uncharacterized | |||||||
| YPL183C (RTT10) | 5 (0.2) | 31.3 | f | 1.7 | 2.4 | 1.9 | 0 |
Mutants analyzed in parallel for insertions at SUF16 are underlined (supplemental Figure S2).
Mutations that confer a mutator phenotype at the CAN1 locus. 0, wild-type level of CanR; +, increased level of CanR. Also refer to Figures 4 and 5 and to supplemental Tables S4–S6.
Average of 13 trials.
Wild-type level of insertions as judged by intensity of the banding pattern upstream of SUF16.
Relative increase in the intensity of the banding pattern.
Relative decrease in the intensity of the banding pattern. WT, wild type (also refer to supplemental Figure S2).
YML009W-B overlaps SPT5, designated as SPT5#.
Functional relationships between members of the chromatin/transcription genes identified here and in other studies were also observed. For example, Rtt109p, which was initially identified by Scholes et al. (2001), has recently been shown to promote genome stability by acetylating histone H3 K56 in association with Asf1p (Driscoll et al. 2007), a Ty1 modulator identified in our screen. Asf1p also interacts with members of the chromatin assembly factor (CAF-1) and the HIR complex (Green et al. 2005), both of which have been implicated in restricting Ty1 transposition (supplemental Table S1) (Qian et al. 1998). In addition, a previously identified Ty1 restriction gene, RTT106 (Scholes et al. 2001), encodes another histone chaperone involved in heterochromatin silencing along with CAF-1 (Huang et al. 2007).
Genes encoding subunits of the RNA polymerase II Mediator complex (Dotson et al. 2000; Lewis and Reinberg 2003; Guglielmi et al. 2004) modulate Ty1 mobility. We identified three Mediator genes, CSE2, SOH1, and SRB5 that restrict Ty1 mobility from 31- to 44-fold (Table 2), while Scholes et al. (2001) identified MED1 and a viable allele of NUT2 that also restrict Ty1 mobility. Griffith et al. (2003) identified three other Mediator subunit genes, SIN4, SRB8, and SSN2 that are required for Ty1 mobility. The Mediator subunits that help or restrict Ty1 mobility are correlated with Mediator modules that activate or repress transcription (Dotson et al. 2000; Lewis and Reinberg 2003; Guglielmi et al. 2004). Ty1 helper proteins Srb8p and Ssn2p, and Sin4p contribute to the Cdk8 and Tail modules, respectively; the Ty1 restriction protein Srb5p is present in the Head module; and the restriction proteins Cse2p, Med1p, Nut2p, and Soh1p are associated with the Middle module. Together our results show that genes involved in histone dynamics and transcription restrict Ty1 mobility.
Two genes chosen for further study are involved in transcription elongation and pre-mRNA processing. Deletion of the transcription elongation gene ELF1 (Prather et al. 2005) increased Ty1 mobility 25-fold (Table 2, supplemental Table S1). Deletion of the dubious ORF, YML009W-B, probably created a viable deletion allele of SPT5 (designated SPT5#), an essential gene involved in transcription elongation of RNA polymerase I and II and processing of pre-mRNA and rRNA (Hartzog et al. 1998; Lindstrom et al. 2003; Schneider et al. 2006). This deletion elevated Ty1 mobility 275-fold. ELF1 is synthetically lethal with SPT4, -5, and -6, as well as with members of the Paf complex (Prather et al. 2005). Elf1p also associates with casein kinase 2 and a regulatory subunit of casein kinase 2, Ckb2p, was identified in our screen. Therefore, several of the Ty1 restriction genes identified in our screen have genetic or physical interactions with ELF1, including SPT5, genes composing the Paf complex, and CKB2.
We chose one pathway-specific gene regulator, ARG82, to analyze further because Ty1 mobility increased 144-fold in an arg82Δ mutant. Arg82p is a multifunctional protein with inositol polyphosphate multikinase activity and is a component of the ArgR repressor that cooperates with diverse sequence-specific transcription factors to control transcription of arginine-, phosphate-, and nitrogen-responsive genes (Odom et al. 2000; Yoon et al. 2004; York 2006). However, the Arg82p kinase activity is not required for regulation of arginine gene expression in yeast (Dubois et al. 2000).
Ty1 restriction genes involved in stress responses:
Genes involved in various types of stress responses restricted Ty1 mobility 3- to 144-fold, including those affecting osmotic challenge through the high-osmolarity glycerol (HOG) pathway (PBS2, SSK2, and SSK22) (Hohmann 2002), ribosome-associated signal transduction (ASC1) (Nilsson et al. 2004; Valerius et al. 2007), ionizing radiation (MMS22) (Baldwin et al. 2005), and inositol signaling (ARG82, IPK1, and KCS1) (Odom et al. 2000; Shears 2000; Dubois et al. 2002; Auesukaree et al. 2005; York 2006) (Table 2, supplemental Table S1). Although genes composing the HOG pathway modestly restricted Ty1 mobility, our results extend the work of Conte et al. (2000) who showed that inactivation of the HOG pathway stimulates Ty1 transposition by precociously activating the haploid invasive-growth pathway. Genes involved in inositol phosphate metabolism also restricted Ty1 mobility. As mentioned above, ARG82 is a potent Ty1 restriction gene and has roles in both gene regulation and inositol metabolism. Deletion of IPK1, which encodes a nuclear inositol 1,3,4,5,6-pentakisphosphate 2-kinase involved in a variety of cellular processes including mRNA export and telomere maintenance (York et al. 2005; Alcazar-Roman et al. 2006; York 2006), increased Ty1 mobility ∼21-fold. Deletion of KCS1, which encodes an inositol hexakisphosphate required for resistance to salt stress, cell wall integrity, vacuolar morphogenesis, and phosphate regulation (Dubois et al. 2002; Auesukaree et al. 2005), enhanced Ty1 mobility 15-fold. A variety of genes involved in DNA double-strand break repair and genome maintenance restrict Ty1 retrotransposition (Maxwell and Curcio 2007). Here we analyzed MMS22, a gene that interacts with previously identified Ty1 modulators MMS1 (RTT108), RTT101, and RTT107 to repair DNA damage associated with DNA replication (supplemental Table S1) (Scholes et al. 2001; Baldwin et al. 2005). Ty1 mobility increased >9-fold when MMS22 was deleted, which is comparable to the Ty1 mobility observed in an rtt107 mutant (11-fold), but is lower than that obtained in mms1 (75-fold) or rtt101 (60-fold) mutants (Scholes et al. 2001).
Ty1 restriction genes involved in miscellaneous functions:
We chose genes involved in several additional cellular processes such as transport of amino acids (AGP3) and magnesium ions (ALR2), cell polarity (BEM4 and BUD27), cell-cycle progression (CDC40, CLN2, and SIC1), and protein phosphorylation by casein kinase 2 (CKB2) to learn more about the diversity of pathways that restrict Ty1 mobility (Table 2, supplemental Table S1). In particular, deletion of BUD27, CDC40, or CKB2 dramatically increased Ty1 mobility ∼237-, 87-, and 60-fold, respectively. BUD27 encodes a prefoldin protein chaperone involved in bud-site selection, nutrient signaling, and gene expression controlled by the TOR kinase (Gstaiger et al. 2003). CDC40 encodes a pre-mRNA splicing factor required for cell-cycle progression at the G1/S and G2/M transitions (Kaplan and Kupiec 2007). As mentioned above, CKB2 encodes a β-regulatory subunit of casein kinase 2, a Ser/Thr protein kinase with wide-ranging roles in cell growth, the cytoskeleton, DNA checkpoint activation, and transcription (Ghavidel et al. 1999; Ahmed et al. 2002; Prather et al. 2005; Guillemain et al. 2007).
Although we recovered several uncharacterized Ty1 restriction genes (supplemental Table S1; YLR282C, YPL183C, YGR110W, YJR142W, YBR239C, and YEL008W), only YPL183C (RTT10) was analyzed further because deleting RTT10 increased Ty1 mobility 31-fold (Table 2), which is more than that from the other uncharacterized genes recovered in our screen. Rtt10p may be present in a complex with the tRNA methyltransferase Trm7p (Pintard et al. 2002; Krogan et al. 2004, 2006), which was also identified here (supplemental Table S1).
Integration at preferred sites upstream of SUF16 increases in many Ty1 restriction mutants:
To determine whether deletion of a Ty1 restriction gene influenced de novo retrotransposition events, spontaneous Ty1 insertions upstream of a preferred tRNA target, the SUF16 locus on chromosome III (Ji et al. 1993), were monitored using a qualitative PCR assay (Table 2, supplemental Figure S2 at http://www.genetics.org/supplemental/). Three independent colonies per strain from 33 Ty1 restriction mutants were chosen for PCR analysis using one primer containing Ty1 RT sequence and one primer from SNR33, which is adjacent to SUF16 (supplemental Figure S2). Ty1 transposition events several hundred base pairs upstream of and in the same transcriptional orientation as SUF16 were detected after separation of the PCR products by agarose-gel electrophoresis, staining with ethidium bromide, and phosphorimage analysis. The intensity of the PCR products indicated that de novo Ty1-integration events were elevated when compared with the pattern and intensity observed with the wild-type strain DG2122. To assess the variation inherent in this Ty1-integration assay, 12 representative restriction mutants were analyzed in parallel, and comparable integration patterns and intensities were obtained when compared with those of the original mutants (supplemental Figure S2). Amplification of the CPR7 gene served as a PCR control (data not shown).
Most Ty1 restriction mutants showed a concomitant increase in Ty1his3-AI mobility and integration events upstream of SUF16, with arg82Δ, asf1Δ, cse2Δ, elf1Δ, paf1Δ, rtf1Δ, spt5Δ#, and srb5Δ mutants in the chromatin/transcription group and an ipk1Δ mutant in the stress-response group showing the largest increases (Table 2 and supplemental Figure S2). We did not observe a change in the pattern of Ty1-integration events upstream of SUF16 in any of the restriction mutants, suggesting that Ty1his3-AI mobility faithfully monitored Ty1 retrotransposition and that normal targeting preferences at SUF16 were maintained. Since CDC40 encodes a splicing factor, the increase in integration events at SUF16 in this mutant also suggests that alterations in splicing of the Ty1his3-AI intron cannot account for the increase in Ty1 mobility. However, Ty1his3-AI mobility increased much more than the level of Ty1 integration at the SUF16 locus in the prefoldin mutant bud27Δ, the histone deacetylase subunit mutant rpd3Δ, the ribosome-associated regulatory mutant asc1Δ, and the transcriptional Mediator subunit mutant soh1Δ (Table 2 and supplemental Figure S2). As mentioned above, homologous recombination between Ty1 cDNA and chromosomal elements may increase in these restriction mutants and, if so, is consistent with the hyperrecombination phenotype observed in a soh1 mutant (Fan and Klein 1994). Other possibilities are that the differences in Ty1his3-AI mobility and SUF16 insertion levels reflect a biased insertion orientation or novel insertion sites in the genome.
Most restriction genes act post-transcriptionally to inhibit Ty1 mobility:
Total RNA isolated from the 33 mutants as well as from the wild-type strain DG2122 was subjected to Northern hybridization using a 32P-labeled probe from the Ty1 RT region (Table 2, supplemental Figure S3 and supplemental Table S2 at http://www.genetics.org/supplemental/). The Ty1 probe also detects the Ty1his3-AI transcript from pBJC573 due to the size increase resulting from the presence of his3-AI (data not shown). The level of Ty1 transcripts was normalized to that of the 18S and 28S rRNAs, as determined by phosphorimage analysis. Ty1 RNA increased less than threefold in 28 of the 33 restriction mutants while the level of Ty1 RNA increased threefold or more in 5 mutants. Surprisingly, the level of Ty1 RNA increased less than threefold in cells lacking the Mediator subunit genes identified in our screen (CSE2, SOH1, and SRB5) or elsewhere (MED1 and NUT2) (Scholes et al. 2001). These results suggest that the Mediator complex may act indirectly by influencing expression of other Ty1 modulator genes or restricts Ty1 mobility post-transcriptionally. The levels of Ty1 and Ty1his3-AI transcripts usually showed similar changes in abundance in most restriction mutants; however, the levels of Ty1 and Ty1his3-AI RNAs were markedly different in arg82Δ, asf1Δ, srb5Δ, spt5Δ#, bud27Δ, and ipk1Δ mutants. Two altered patterns were observed. The level of the Ty1his3-AI transcript increased more than the level of Ty1 RNA in strains lacking ARG82, SPT5#, SRB5, or BUD27, while the converse occurred in the absence of ASF1 or IPK1. The differences in Ty1 and Ty1his3-AI transcript levels in these mutants were not pursued further, but may result from exogenous signals from the HIS4 promoter region that influence transcription of the Ty1his3-AI element integrated at HIS4 (Silverman and Fink 1984). In addition, both element-based sequence polymorphisms and chromosomal context differences can influence transcription of resident Ty1 elements (Morillon et al. 2002).
The level of Ty1 RNA increased 3.7- to 5-fold in arg82Δ, ipk1Δ, and ksc1Δ mutants, suggesting that transcription or stability of Ty1 RNA is regulated by inositol phosphate metabolism or signaling to a downstream process. Alternatively, Ty1 transcription may be under the control of the ArgR repressor. Deletion of ASF1, the histone H3 subunit gene, HHT1, or a regulator of histone gene transcription, SPT21, increased the levels of Ty1 RNA 3- to 5.2-fold. These results suggest that genes involved in chromatin transactions negatively regulate the level of Ty1 RNA. Relief of chromatin-based repression of Ty1 transcription also occurs in cells lacking histone 2A subtype genes HTA1 and HTB1 (Hirschhorn et al. 1992; Morillon et al. 2002; Todeschini et al. 2005). Taken together, our results suggest that although most Ty1 restriction mutations affect Ty1 mobility at a post-transcriptional level, deletion of certain genes increases the level of genomic Ty1 RNA or affects the levels of Ty1 and Ty1his3-AI differentially.
Diverse cellular genes affect the level of Ty1 cDNA:
We determined the level of unincorporated Ty1 cDNA in the 33 restriction mutants and the wild-type strain DG2122 by Southern hybridization (Table 2, supplemental Figure S4 and supplemental Table S3 at http://www.genetics.org/supplemental/). Digestion of total DNA with PvuII generates a characteristic 2-kb fragment containing sequences from a conserved internal PvuII restriction site in Ty1 (nucleotide 3944) to the end of the linear unincorporated cDNA (nucleotide 5918) (Conte et al. 1998; Lee et al. 1998) (supplemental Figure S4). A 32P-labeled probe spanning part of this region of Ty1 was hybridized with the resulting membranes, and the 2-kb Ty1 cDNA fragment was quantified by phosphorimage analysis. The PvuII fragments containing preexisting Ty1-genomic DNA junctions provide internal loading controls.
When the levels of unincorporated Ty1 cDNA in the restriction mutants and wild type were compared, there was <3-fold increase in Ty1 cDNA in 20 of the 33 restriction mutants, while a ≥3-fold increase was observed in 13 mutants (Table 2, supplemental Figure S4 and supplemental Table S3). The relatively modest elevation in Ty1 cDNA exhibited in a variety of restriction mutants suggests that the increase in cDNA may not entirely account for the change in Ty1 mobility. For example, when the Paf subunit genes CDC73, RTF1, and LEO1 or the functionally related transcription elongation gene ELF1 were deleted, Ty1 RNA and cDNA levels increased ≤3-fold, yet Ty1his3-AI mobility increased 16- to 26-fold, and insertions at SUF16 were also more frequent. Perhaps Ty1 cDNA is more efficiently utilized for retrotransposition in these mutants.
The 13 Ty1 restriction mutants showing threefold or more additional cDNA covered a variety of cellular functions. Certain mutants, such as arg82Δ, asf1Δ, ipk1Δ, kcs1Δ, and spt21Δ, had an elevated level of Ty1 RNA and cDNA, suggesting that an increase in Ty1 gene expression results in a higher level of Ty1 mobility. Several Ty1 restriction mutants, such as bre1Δ and paf1Δ, showed moderate increases in Ty1 RNA or cDNA and large increases in Ty1 mobility and insertions at SUF16 (Table 2, supplemental Figure S2), again raising the possibility that cDNA utilization might be enhanced during the process of retrotransposition. Conversely, deletion of ASC1 resulted in an increase in Ty1 cDNA and Ty1his3-AI mobility, but SUF16 insertions and Ty1 RNA remained at wild-type levels. These results can be explained by an elevation in cDNA recombination in the asc1Δ mutant, although a change in his3-AI splicing and Ty1 insertion preference remain formal possibilities.
Ty1 restriction genes that alter insertional mutagenesis and target-site preference at CAN1:
Certain Ty1 restriction genes minimize transposition into coding sequences of genes, as first illustrated by the E2 ubiquitin conjugating gene, RAD6 (Liebman and Newnam 1993). Therefore, we determined whether any of the 33 Ty1 restriction mutants as well as several additional candidates from the larger collection (supplemental Table S1) possessed a mutator phenotype at the arginine permease gene CAN1 (Table 2), where resistance to the amino acid analog canavanine (CanR) occurs when CAN1 is defective. Deletion of the Paf complex subunit genes PAF1, LEO1, and RTF1; the histone chaperone ASF1; the transcription elongation genes ELF1 and SPT5#; the HOG pathway protein kinase gene PBS2; and the ubiquitin-metabolism genes BRE1, RAD6, and RAD18 increased the frequency of CanR from 2- to 11-fold when cells were grown at a permissive temperature for Ty1 retrotransposition (20°) (Paquin and Williamson 1984) (Figure 4, supplemental Table S4 at http://www.genetics.org/supplemental/). Unlike in the Paf complex mutants leo1Δ, paf1Δ, and rtf1Δ, the frequency of CanR remained unchanged in a cdc73Δ mutant. Deleting RTT109, which encodes the histone H3 K56 acetylase and interacts with Asf1p (Driscoll et al. 2007), also did not alter the frequency of CanR.
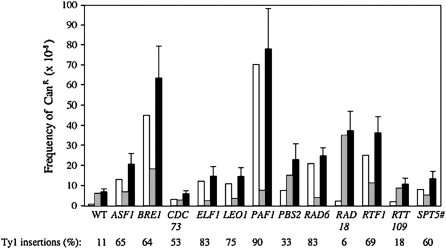
Figure 4.—
Frequency estimate of Ty1- and non-Ty1-induced CanR mutations. Open bars, frequency of CanR mutations caused by Ty1 insertion; cross-hatched bars, frequency of CanR mutations caused by other mutational events; solid bars, overall frequency of CanR mutations. Standard deviations are above the solid bars. On the bottom is the fraction of CanR mutations caused by Ty1. Also refer to supplemental Tables S4 and S5.
The fraction of Ty1-induced can1 mutations was determined by PCR analysis using oligonucleotide primers specific to CAN1 and Ty1 (Figure 4, supplemental Table S4). Reactions with primers that flank CAN1 either amplified a 2283-bp product indicative of the wild-type CAN1 locus or did not amplify a wild-type product. The mutant DNA samples that failed to amplify a wild-type CAN1 product were analyzed with a primer specific to Ty1 and overlapping primers in opposite orientations located at the start of the CAN1 coding region. In the wild-type strain DG2122, ∼11% (4/36) of the can1 mutations were caused by Ty1 insertion while the rest were caused by mutations that did not dramatically alter the size of the 2283-bp PCR product (supplemental Table S4), which was comparable to the fraction of can1 mutations resulting from endogenous Ty1 insertions obtained previously in wild-type strains (Wilke et al. 1989; Picologlou et al. 1990; Liebman and Newnam 1993; Qian et al. 1998). The efficiency of Ty1 insertional mutagenesis at CAN1 ranged between 6.25 and 90% in different restriction mutants. There was a dramatic increase in the fraction of Ty1-induced can1 mutations in the strains lacking PAF1 (90%), ELF1 (83%), or RAD6 (83%). Solo-LTR insertions, most likely caused by a Ty1 insertion followed by intraelement LTR–LTR recombination (Sutton and Liebman 1992), and putative deletion events occurred in the Ty1 restriction mutants at about the same level as had been observed in wild-type strains (Wilke et al. 1989; Picologlou et al. 1990; Liebman and Newnam 1993; Rinckel and Garfinkel 1996; Qian et al. 1998) (supplemental Table S5 at http://www.genetics.org/supplemental/).
Knowing the fraction of can1 mutations resulting from Ty1 retrotransposition events allowed us to estimate the increase in Ty1 mutagenesis vs. non-Ty1 mutagenesis in the restriction mutants (Figure 4, supplemental Table S4). Although several mutational spectra are observed, the results suggest that most of the mutator activity observed at CAN1 in the Ty1 restriction mutants is caused by Ty1 insertional mutagenesis and is not due to other mutational events. For example, Ty1-induced mutations at CAN1 increased 90-fold in a paf1Δ mutant, whereas other mutational events remained at the wild-type level. Deletion of BRE1 showed a modest elevation in non-Ty1 events of ∼3-fold when compared to the >57-fold increase in Ty1 mutagenesis, while deletion of PBS2 showed 2.5-fold more non-Ty1 events and a moderate ∼10-fold increase in Ty1 insertions. Deletion of ELF1 increased Ty1 mutagenesis by almost 16-fold, but surprisingly, non-Ty1-induced mutations decreased 5-fold when compared with the wild-type strain DG2122. It will be interesting to determine whether elf1Δ's non-Ty1 “antimutator” phenotype occurs elsewhere in the genome. Conversely, deleting RAD18 increased the frequency of non-Ty1 can1 mutations 5.6-fold, which is expected from previous work (Quah et al. 1980), while Ty1 insertions increased only 3-fold. Ty1his3-AI mobility was also higher in a rad18Δ mutant (supplemental Table S1), suggesting that insertions into preferred targets upstream of genes transcribed by RNA polymerase III or cDNA recombination may occur more often when RAD18 is deleted.
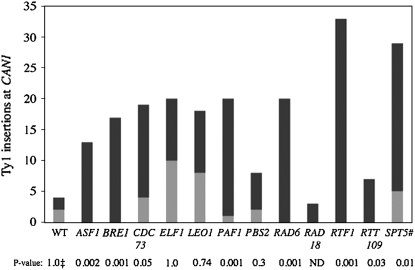
Ty1 insertions at CAN1 show a strong preference for the CAN1 promoter region, with ∼50% of Ty1 insertions occurring in a 317-bp window upstream and 50% in the 1773-bp open reading frame (Wilke et al. 1989; Picologlou et al. 1990; Liebman and Newnam 1993; Rinckel and Garfinkel 1996; Qian et al. 1998), which is similar to the distribution of Ty1 insertions at other genes such as LYS2 and URA3 (Eibel and Philippsen 1984; Simchen et al. 1984; Natsoulis et al. 1989). PCR analysis using CAN1 primers specific to the beginning of the coding sequence allowed us to determine if the Ty1 insertion-site preference for the CAN1 promoter region observed in wild-type cells was altered in the Ty1 restriction mutants (Figure 5, supplemental Table S5). There was a striking change in insertion-site preference in strains lacking ASF1, BRE1, CDC73, PAF1, RTF1, RTT109, and SPT5#, as well as RAD6. Between 78 and 100% of the Ty1 insertions occurred in the coding sequence of CAN1 in these mutants, suggesting that Ty1 targeting was now random within the CAN1 interval monitored in our analysis. Similar insertion patterns have also been observed in strains lacking RAD6 (Liebman and Newnam 1993) and also when CAC3, which encodes a CAF-1 subunit (Ach et al. 1997), and HIR3 were both deleted (Huang et al. 1999). The sizes of Ty1-CAN1 PCR products suggested that Ty1 insertions occurred throughout CAN1 (data not shown). In addition, sequence analysis of 15 Ty1 transposition events obtained in a paf1Δ mutant confirmed the change in Ty1 target-site preference (supplemental Figure S5 at http://www.genetics.org/supplemental/). Although the fraction of Ty1-induced can1 mutations increased from 3- to 7.5-fold in strains lacking ELF1, LEO1, or PBS2, the insertion-site preference remained the same (Figure 5). Taken together, these results suggest that certain defects in Paf complex function or transcriptional elongation fail to protect CAN1 coding sequence from Ty1 insertional mutagenesis.
Figure 5.—
Ty1 insertions in the CAN1 promoter region vs. the coding sequence. Shaded bars, Ty1 insertions in the CAN1 promoter region; solid bars, Ty1 insertions in the CAN1 coding sequence. On the bottom are Ty1 restriction genes that were analyzed. P-values were obtained by comparing the distribution of promoter vs. coding sequence insertions in WT and Ty1 restriction mutants (‡, refer to materials and methods). Also refer to supplemental data for more information on the spectrum of CanR mutations (supplemental Tables S4 and S5), the orientation of the Ty1 insertions (supplemental Table S6), and DNA sequence analysis of Ty1 insertions when PAF1 was deleted (supplemental Figure S5).
Almost all of the Ty1 insertions (28/29, supplemental Table S6 at http://www.genetics.org/supplemental/) within the promoter region were oriented such that Ty1 and CAN1 transcription were in the same direction, as expected from previous work showing that adjacent gene activation occurs when Ty1 and target gene transcription occur in opposite directions (see review by Voytas and Boeke 2002). To address the possibility that a selection bias occurred with the coding-sequence insertions, tetrad analysis was performed after mating a given Ty1 restriction mutant with a wild-type strain (DG3027) containing a spontaneous Ty1 insertion in the promoter region of CAN1 [can1-26(Ty1), 126 bp from the start of the CAN1 coding sequence], which is a common site for Ty1 integration in wild-type cells (Rinckel and Garfinkel 1996). Tetrad analysis (7–11 tetrads/Ty1 restriction mutant) of diploids derived from DG3027 and asf1Δ∷KanMX, cdc73Δ∷KanMX, leo1Δ∷KanMX, paf1Δ∷KanMX, rtf1Δ∷KanMX, rtt109Δ∷KanMX, and spt5#Δ∷KanMX mutants showed 2:2 segregation for CanR and G418R typical of unlinked markers, indicating that the restriction mutations do not suppress the Ty1-induced promoter mutation can1-26(Ty1). The orientation of the Ty1 insertions in the CAN1 coding sequence was not affected in the restriction mutants (supplemental Table S6).
Protecting the yeast genome from Ty1 insertional mutagenesis:
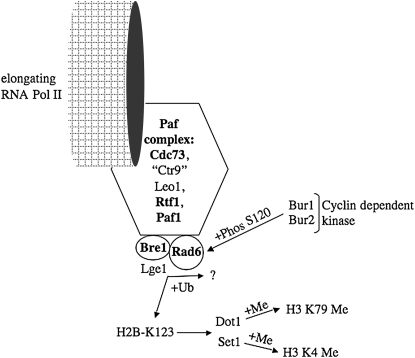
The analyses of Ty1 insertions at SUF16 and CAN1 in different restriction mutants suggest that multiple pathways protect the yeast genome from insertional mutagenesis (Figures 4 and 5, Table 2, supplemental Figure S2). However, a prominent pathway identified in this work involves genes encoding the Paf complex subunits Cdc73p, Paf1p, Leo1p, and Rtf1p and Rad6p and Bre1p (Figure 6). Analyses of these Ty1 restriction genes suggests that H2B ubiquitination or possibly additional but as yet unidentified Rad6p substrates are required to minimize Ty1 insertions not only at sites upstream of genes transcribed by RNA polymerase II or III but also within protein-coding sequences. Therefore, it will be interesting to determine whether the process of Ty1 retrotransposition is restricted by htb-K123R, a mutation in histone H2B that prevents ubiquitination by Rad6p/Bre1p (Robzyk et al. 2000; Hwang et al. 2003; Wood et al. 2003a). Since H2B-K123 ubiquitination is required for H3-K4 methylation by Set1p and H3-K79 methylation by Dot1p (Ng et al. 2002; Sun and Allis 2002), Ty1 target-site preference may be modulated by histone H3 methylation events. SET1 is also required to repress Ty1 transcription of elements inserted in silent regions of the genome (Bryk et al. 2002). Rad6p undergoes phosphorylation by the Bur1p-Bur2p cyclin-dependent protein kinase on S120, which is required for full Rad6p-Bre1 ubiquitination of H2B (Wood et al. 2005), and raises the possibility that Rad6p 120S phosphorylation restricts Ty1 retrotransposition. In addition, our results suggest that Rad6p-Rad18p-mediated ubiquitination of PCNA (Bailly et al. 1997; Hoege et al. 2002) does not protect yeast coding sequences from Ty1 transposition, because deleting RAD18 does not markedly alter the level of Ty1 insertional mutagenesis or preference at CAN1. Proteins functionally related to the Paf1 complex were also detected in our screen. In particular, the transcription elongation proteins Spt4p/Spt5p stimulate association of the Paf complex with elongating RNA polymerase II (Qiu et al. 2006). Therefore, partially deleting SPT5 may increase Ty1 retrotransposition and insertions into CAN1 coding sequences by inhibiting Paf complex function.
Figure 6.—
Relationship between ubiquitination of proteins involved in transcription elongation by RNA polymerase II and by Ty1 transposition. Protein names (in boldface type) required for restricting Ty1 transposition and maintaining target site preference at CAN1 include the Paf complex subunits Cdc73, Paf1, and Rtf1 and the Rad6-Bre1 ubiquitination (+Ub) complex that modifies histone H2B on K123. Rad6-Bre1 may also ubiquitinate additional proteins that have not been identified. The Paf1 complex subunit protein Leo1 is required for restricting Ty1 transposition but is not required for target-site preference at CAN1, and Ctr9 was not analyzed. The Bur1-Bur2 cyclin-dependent protein kinases (+Phos) and the histone H3 methylases (+Me) Dot1 and Set1 were not identified in our screen for Ty1 restriction genes, but may affect Ty1 target preference.
How does disrupting the Paf complex/Bre1p-Rad6p pathway stimulate Ty1 retrotransposition and mutagenesis throughout the genome? Our results suggest that the Paf/Bre1p-Rad6p pathway restricts Ty1 transposition at multiple steps. Since the level of Ty1 RNA increases less than threefold in strains lacking BRE1, CDC73, LEO1, PAF1, and RTF1 as well as remaining unchanged in a RAD6 mutant (Picologlou et al. 1990), the Paf/Bre1p-Rad6p pathway may restrict Ty1 transposition post-transcriptionally. However, the cDNA level increases more than fourfold in a PAF1 or a BRE1 mutant, suggesting that an increase in Ty1 reverse transcription or cDNA stability contributes to the increase in Ty1 transposition. It will be interesting to determine whether defects in the Paf/Bre1p-Rad6p pathway increase Ty1 transposition via activation of an S-phase checkpoint (Curcio et al. 2007). Target-site selection is another step in the process of Ty1 retrotransposition that is restricted by the Paf/Bre1p-Rad6p pathway. We propose that even though regions upstream of genes transcribed by RNA polymerase III are considered hotspots for Ty1 integration, favorable insertion sites remain limiting in wild-type cells. In the absence of Paf/Bre1p-Rad6p, aberrant histone transactions create more favorable target sites on a genomic scale. However, the nature of the target sites revealed by compromising the Paf complex/Bre1p-Rad6p function remains unknown. Determining the full spectrum of Ty1-integration sites throughout the genome when histone H2B ubiquitination is blocked may provide further insights on target-site preference and genome protection.
Acknowledgments
We thank Ching-yun Wang, Angela Biggus, Tory Ellis, Jeffrey Hordoz, Jennifer Dorfman, and Amelia Hinnebusch for technical assistance; Joan Curcio for plasmids and strains; and Sharon Moore and Philip Farabaugh for helpful comments. This research was sponsored in part by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), Center for Cancer Research and with federal funds from the NCI, under contract no. N01-CO-12400. We also thank the Howard Hughes Medical Institute and the NIH Foundation for Advanced Education in the Sciences High School Teacher Summer Research Program for their support. The contents of this publication do not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
References
- Ach, R. A., P. Taranto and W. Gruissem, 1997. A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell 9: 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, K., D. A. Gerber and C. Cochet, 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol. 12: 226–230. [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman, A. R., E. J. Tran, S. Guo and S. R. Wente, 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8: 711–716. [DOI] [PubMed] [Google Scholar]
- Auesukaree, C., H. Tochio, M. Shirakawa, Y. Kaneko and S. Harashima, 2005. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 280: 25127–25133. [DOI] [PubMed] [Google Scholar]
- Aye, M., B. Irwin, N. Beliakova-Bethell, E. Chen, J. Garrus et al., 2004. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics 168: 1159–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman, N., M. E. Gelbart, T. Tsukiyama and J. D. Boeke, 2005. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 19: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, V., S. Lauder, S. Prakash and L. Prakash, 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272: 23360–23365. [DOI] [PubMed] [Google Scholar]
- Baldwin, E. L., A. C. Berger, A. H. Corbett and N. Osheroff, 2005. Mms22p protects Saccharomyces cerevisiae from DNA damage induced by topoisomerase II. Nucleic Acids Res. 33: 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell, N., C. Beckham, T. H. Giddings, Jr., M. Winey, R. Parker et al., 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., and S. E. Devine, 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93: 1087–1089. [DOI] [PubMed] [Google Scholar]
- Bolton, E. C., A. S. Mildvan and J. D. Boeke, 2002. Inhibition of reverse transcription in vivo by elevated manganese ion concentration. Mol. Cell 9: 879–889. [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Broomfield, S., T. Hryciw and W. Xiao, 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486: 167–184. [DOI] [PubMed] [Google Scholar]
- Bryk, M., M. Banerjee, D. Conte, Jr. and M. J. Curcio, 2001. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol. Cell. Biol. 21: 5374–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis et al., 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12: 165–170. [DOI] [PubMed] [Google Scholar]
- Chalker, D. L., and S. B. Sandmeyer, 1990. Transfer RNA genes are genomic targets for de novo transposition of the yeast retrotransposon Ty3. Genetics 126: 837–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte, Jr., D., and M. J. Curcio, 2000. Fus3 controls Ty1 transpositional dormancy through the invasive growth MAPK pathway. Mol. Microbiol. 35: 415–427. [DOI] [PubMed] [Google Scholar]
- Conte, Jr., D., E. Barber, M. Banerjee, D. J. Garfinkel and M. J. Curcio, 1998. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol. Cell. Biol. 18: 2502–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio, M. J., and D. J. Garfinkel, 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88: 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio, M. J., and D. J. Garfinkel, 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 12: 2813–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio, M. J., A. E. Kenny, S. P. Moore, D. J. Garfinkel, M. Weintraub et al., 2007. S-phase checkpoints stimulate the activity of the retrovirus-like transposon, Ty1, when genome integrity is compromised. Mol. Cell. Biol. 27: 8874–8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine, S. E., and J. D. Boeke, 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10: 620–633. [DOI] [PubMed] [Google Scholar]
- Dollard, C., S. L. Ricupero-Hovasse, G. Natsoulis, J. D. Boeke and F. Winston, 1994. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 5223–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson, M. R., C. X. Yuan, R. G. Roeder, L. C. Myers, C. M. Gustafsson et al., 2000. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97: 14307–14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean et al., 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368–28371. [DOI] [PubMed] [Google Scholar]
- Driscoll, R., A. Hudson and S. P. Jackson, 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, E., V. Dewaste, C. Erneux and F. Messenguy, 2000. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 486: 300–304. [DOI] [PubMed] [Google Scholar]
- Dubois, E., B. Scherens, F. Vierendeels, M. M. Ho, F. Messenguy et al., 2002. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 277: 23755–23763. [DOI] [PubMed] [Google Scholar]
- Eibel, H., and P. Philippsen, 1984. Preferential integration of yeast transposable element Ty into a promoter region. Nature 307: 386–388. [DOI] [PubMed] [Google Scholar]
- Eickbush, T. H., and H. S. Malik, 2002. Origins and evolution of retrotransposons, pp. 1111–1144 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Fan, H. Y., and H. L. Klein, 1994. Characterization of mutations that suppress the temperature-sensitive growth of the hpr1 delta mutant of Saccharomyces cerevisiae. Genetics 137: 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, P. J., 1995. Post-transcriptional regulation of transposition by Ty retrotransposons of Saccharomyces cerevisiae. J. Biol. Chem. 270: 10361–10364. [DOI] [PubMed] [Google Scholar]
- Garfinkel, D. J., M. F. Mastrangelo, N. J. Sanders, B. K. Shafer and J. N. Strathern, 1988. Transposon tagging using Ty elements in yeast. Genetics 120: 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel, D. J., K. Nyswaner, J. Wang and J. Y. Cho, 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165: 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart, M. E., N. Bachman, J. Delrow, J. D. Boeke and T. Tsukiyama, 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19: 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavidel, A., D. J. Hockman and M. C. Schultz, 1999. A review of progress towards elucidating the role of protein kinase CK2 in polymerase III transcription: regulation of the TATA binding protein. Mol. Cell. Biochem. 191: 143–148. [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu et al., 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, J. L., L. E. Coleman, A. S. Raymond, S. G. Goodson, W. S. Pittard et al., 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger, M., B. Luke, D. Hess, E. J. Oakeley, C. Wirbelauer et al., 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302: 1208–1212. [DOI] [PubMed] [Google Scholar]
- Guglielmi, B., N. L. van Berkum, B. Klapholz, T. Bijma, M. Boube et al., 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32: 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemain, G., E. Ma, S. Mauger, S. Miron, R. Thai et al., 2007. Mechanisms of checkpoint kinase Rad53 inactivation after a double-strand break in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 3378–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Harrison, R., B. Papp, C. Pal, S. G. Oliver and D. Delneri, 2007. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl. Acad. Sci. USA 104: 2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog, G. A., T. Wada, H. Handa and F. Winston, 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12: 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn, J. N., S. A. Brown, C. D. Clark and F. Winston, 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6: 2288–2298. [DOI] [PubMed] [Google Scholar]
- Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis and S. Jentsch, 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. [DOI] [PubMed] [Google Scholar]
- Hohmann, S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66: 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., J. Y. Hong, C. L. Burck and S. W. Liebman, 1999. Host genes that affect the target-site distribution of the yeast retrotransposon Ty1. Genetics 151: 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., H. Zhou, J. Tarara and Z. Zhang, 2007. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 26: 2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone et al., 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11: 261–266. [DOI] [PubMed] [Google Scholar]
- Irwin, B., M. Aye, P. Baldi, N. Beliakova-Bethell, H. Cheng et al., 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15: 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H., D. P. Moore, M. A. Blomberg, L. T. Braiterman, D. F. Voytas et al., 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73: 1007–1018. [DOI] [PubMed] [Google Scholar]
- Kadosh, D., and K. Struhl, 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18: 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, X. L., F. Yadao, R. D. Gietz and B. A. Kunz, 1992. Elimination of the yeast RAD6 ubiquitin conjugase enhances base-pair transitions and G.C–T.A transversions as well as transposition of the Ty element: implications for the control of spontaneous mutation. Genetics 130: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, Y., and M. Kupiec, 2007. A role for the yeast cell cycle/splicing factor Cdc40 in the G1/S transition. Curr. Genet. 51: 123–140. [DOI] [PubMed] [Google Scholar]
- Kasten, M. M., D. E. Ayer and D. J. Stillman, 1996. SIN3-dependent transcriptional repression by interaction with the Mad1 DNA-binding protein. Mol. Cell. Biol. 16: 4215–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K., S. Pande, B. Faiola, D. P. Moore, J. D. Boeke et al., 1993. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilzer, J. M., T. Stracker, B. Beitzel, K. Meek, M. Weitzman et al., 2003. Roles of host cell factors in circularization of retroviral DNA. Virology 314: 460–467. [DOI] [PubMed] [Google Scholar]
- Kirchner, J., C. M. Connolly and S. B. Sandmeyer, 1995. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science 267: 1488–1491. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw et al., 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13: 225–239. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo et al., 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. [DOI] [PubMed] [Google Scholar]
- Larsen, L. S., M. Zhang, N. Beliakova-Bethell, V. Bilanchone, A. Lamsa et al., 2007. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 81: 6957–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, A., R. Kanaar, S. P. Jackson and M. J. O'Connor, 2004. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 23: 3421–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. S., C. P. Lichtenstein, B. Faiola, L. A. Rinckel, W. Wysock et al., 1998. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 148: 1743–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.-S., L. Bi, D. J. Garfinkel and A. M. Bailis, 2000. Nucleotide excision repair/TFIIH helicases Rad3 and Ssl2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol. Cell. Biol. 20: 2436–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage, P., and A. L. Todeschini, 2005. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 110: 70–90. [DOI] [PubMed] [Google Scholar]
- Lewis, B. A., and D. Reinberg, 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116: 3667–3675. [DOI] [PubMed] [Google Scholar]
- Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler et al., 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20: 3272–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman, S. W., and G. Newnam, 1993. A ubiquitin-conjugating enzyme, RAD6, affects the distribution of Ty1 retrotransposon integration positions. Genetics 133: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter et al., 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23: 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A. G., S. Tateishi, P. D. Bieniasz, M. A. Muesing, M. Yamaizumi et al., 2006. Effect of DNA repair protein Rad18 on viral infection. PLoS Pathog. 2: e40. [DOI] [PMC free article] [PubMed]
- Maxwell, P. H., and M. J. Curcio, 2007. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot. Cell 6: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon, A., L. Benard, M. Springer and P. Lesage, 2002. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol. Cell. Biol. 22: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., A. E. Kenny and M. J. Curcio, 2006. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172: 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis, G., W. Thomas, M. C. Roghmann, F. Winston and J. D. Boeke, 1989. Ty1 transposition in Saccharomyces cerevisiae is nonrandom. Genetics 123: 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., R. M. Xu, Y. Zhang and K. Struhl, 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277: 34655–34657. [DOI] [PubMed] [Google Scholar]
- Ng, H. H., S. Dole and K. Struhl, 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278: 33625–33628. [DOI] [PubMed] [Google Scholar]
- Nilsson, J., J. Sengupta, J. Frank and P. Nissen, 2004. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 5: 1137–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom, A. R., A. Stahlberg, S. R. Wente and J. D. York, 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287: 2026–2029. [DOI] [PubMed] [Google Scholar]
- Osley, M. A., 2004. H2B ubiquitylation: the end is in sight. Biochim. Biophys. Acta 1677: 74–78. [DOI] [PubMed] [Google Scholar]
- Paquin, C. E., and V. M. Williamson, 1984. Temperature effects on the rate of Ty transposition. Science 226: 53–55. [DOI] [PubMed] [Google Scholar]
- Penheiter, K. L., T. M. Washburn, S. E. Porter, M. G. Hoffman and J. A. Jaehning, 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20: 213–223. [DOI] [PubMed] [Google Scholar]
- Picologlou, S., N. Brown and S. W. Liebman, 1990. Mutations in RAD6, a yeast gene encoding a ubiquitin-conjugating enzyme, stimulate retrotransposition. Mol. Cell. Biol. 10: 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard, L., F. Lecointe, J. M. Bujnicki, C. Bonnerot, H. Grosjean et al., 2002. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 21: 1811–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather, D., N. J. Krogan, A. Emili, J. F. Greenblatt and F. Winston, 2005. Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 25: 10122–10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Z., H. Huang, J. Y. Hong, C. L. Burck, S. D. Johnston et al., 1998. Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins. Mol. Cell. Biol. 18: 4783–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, H., C. Hu, C. M. Wong and A. G. Hinnebusch, 2006. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol. Cell. Biol. 26: 3135–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah, S. K., R. C. von Borstel and P. J. Hastings, 1980. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 96: 819–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A. J., B. K. Shafer and D. J. Garfinkel, 2000. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics 154: 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, J. L., Z. Moqtaderi and K. Struhl, 2004. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinckel, L. A., and D. J. Garfinkel, 1996. Influences of histone stoichiometry on the target site preference of retrotransposons Ty1 and Ty2 in Saccharomyces cerevisiae. Genetics 142: 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk, K., J. Recht and M. A. Osley, 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504. [DOI] [PubMed] [Google Scholar]
- Rondon, A. G., M. Gallardo, M. Garcia-Rubio and A. Aguilera, 2004. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 5: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer, S. B., M. Aye and T. Menees, 2002. Ty3, a position specific, gypsy-like element in Saccharomyces cerevisiae, pp. 663–683 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Schneider, D. A., S. L. French, Y. N. Osheim, A. O. Bailey, L. Vu et al., 2006. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. USA 103: 12707–12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes, D. T., M. Banerjee, B. Bowen and M. J. Curcio, 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish, M. A., and K. Struhl, 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22: 415–422. [DOI] [PubMed] [Google Scholar]
- Sharon, G., T. J. Burkett and D. J. Garfinkel, 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 14: 6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears, S. B., 2000. Transcriptional regulation: A new dominion for inositol phosphate signaling? BioEssays 22: 786–789. [DOI] [PubMed] [Google Scholar]
- Sheldon, K. E., D. M. Mauger and K. M. Arndt, 2005. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Silverman, S. J., and G. R. Fink, 1984. Effects of Ty insertions on HIS4 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 4: 1246–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen, G., F. Winston, C. A. Styles and G. R. Fink, 1984. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc. Natl. Acad. Sci. USA 81: 2431–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. A., and R. Daniel, 2006. Following the path of the virus: the exploitation of host DNA repair mechanisms by retroviruses. ACS Chem. Biol. 1: 217–226. [DOI] [PubMed] [Google Scholar]
- Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic et al., 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21: 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman, D. J., S. Dorland and Y. Yu, 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SW15 transcriptional activator. Genetics 136: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z. W., and C. D. Allis, 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108. [DOI] [PubMed] [Google Scholar]
- Sundararajan, A., B. S. Lee and D. J. Garfinkel, 2003. The Rad27 (Fen-1) nuclease inhibits Ty1 mobility in Saccharomyces cerevisiae. Genetics 163: 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, P. R., and S. W. Liebman, 1992. Rearrangements occurring adjacent to a single Ty1 yeast retrotransposon in the presence and absence of full-length Ty1 transcription. Genetics 131: 833–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers, H. T., and L. Tora, 2005. SAGA unveiled. Trends Biochem. Sci. 30: 7–10. [DOI] [PubMed] [Google Scholar]
- Todeschini, A. L., A. Morillon, M. Springer and P. Lesage, 2005. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 25: 7459–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerius, O., M. Kleinschmidt, N. Rachfall, F. Schulze, S. Lopez Marin et al., 2007. The Saccharomyces homolog of mammalian RACK1, Cpc2/Asc1p, is required for FLO11 dependent adhesive growth and dimorphism. Mol. Cell. Proteomics 6: 1968–1979. [DOI] [PubMed] [Google Scholar]
- Voytas, D. F., and J. D. Boeke, 2002. Ty1 and Ty5 of Sacharomyces cerevisiae, pp. 614–630 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. ASM Press, Washington, DC.
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wilke, C. M., S. H. Heidler, N. Brown and S. W. Liebman, 1989. Analysis of yeast retrotransposon Ty insertions at the CAN1 locus. Genetics 123: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., 1992. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast, pp. 1271–1293 in Transcriptional Regulation, edited by S. L. McKnight and K. R. Yamamoto. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt et al., 2003. a Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11: 267–274. [DOI] [PubMed] [Google Scholar]
- Wood, A., J. Schneider, J. Dover, M. Johnston and A. Shilatifard, 2003. b The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278: 34739–34742. [DOI] [PubMed] [Google Scholar]
- Wood, A., J. Schneider, J. Dover, M. Johnston and A. Shilatifard, 2005. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell 20: 589–599. [DOI] [PubMed] [Google Scholar]
- Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt et al., 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25: 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, W., X. Gai, Y. Zhu, D. C. Zappulla, R. Sternglanz et al., 2001. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21: 6606–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., and J. D. Boeke, 1990. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc. Natl. Acad. Sci. USA 87: 8360–8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y., T. Narita, N. Inukai, T. Wada and H. Handa, 2001. SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J. Biochem. 129: 185–191. [DOI] [PubMed] [Google Scholar]
- Yarrington, R. M., J. Chen, E. C. Bolton and J. D. Boeke, 2007. Mn2+ suppressor mutations and biochemical communication between Ty1 reverse transcriptase and RNase H domains. J. Virol. 81: 9004–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, M., J. D. Horton, J. C. Cohen, H. H. Hobbs and R. M. Stephens, 2006. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics 7: 30. [DOI] [PMC free article] [PubMed]
- Yieh, L., G. Kassavetis, E. P. Geiduschek and S. B. Sandmeyer, 2000. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem. 275: 29800–29807. [DOI] [PubMed] [Google Scholar]
- Yoder, K. E., and F. D. Bushman, 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74: 11191–11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, K., A. Sarasin, K. Kraemer, M. McIlhatton, F. Bushman et al., 2006. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc. Natl. Acad. Sci. USA 103: 4622–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, S., C. K. Govind, H. Qiu, S. J. Kim, J. Dong et al., 2004. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Proc. Natl. Acad. Sci. USA 101: 11713–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, J. D., 2006. Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761: 552–559. [DOI] [PubMed] [Google Scholar]
- York, S. J., B. N. Armbruster, P. Greenwell, T. D. Petes and J. D. York, 2005. Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 280: 4264–4269. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu, T., and F. Nagawa, 1989. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science 244: 1346–1348. [DOI] [PubMed] [Google Scholar]
- Zou, S., and D. F. Voytas, 1997. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc. Natl. Acad. Sci. USA 94: 7412–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, S., N. Ke, J. M. Kim and D. F. Voytas, 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 10: 634–645. [DOI] [PubMed] [Google Scholar]