Abstract
Drosophila melanogaster and D. simulans are two closely related species with a similar distribution range. Many studies suggested that D. melanogaster has a smaller effective population size than D. simulans. As most evidence was derived from non-African populations, we readdressed this question by sequencing 10 X-linked loci in five African D. simulans and six African D. melanogaster populations. Contrary to previous results, we found no evidence for higher variability, and thus larger effective population size, in D. simulans. Our observation of similar levels of variability of both species will have important implications for the interpretation of patterns of molecular evolution.
DROSOPHILA melanogaster and D. simulans are two closely related species, which have a similar, but not identical, distribution range and demographic history (David and Capy 1988; Irvin et al. 1998; Capy and Gibert 2004; Lachaise and Silvain 2004). This species pair has been used in numerous comparative studies highlighting their similarities and differences. One of the central conclusions emerging from these studies was that the effective population size (Ne) differs between the two species, with D. simulans having the larger size.
The level of neutral polymorphism which scales with effective population size was found to be higher in D. simulans than in D. melanogaster (Aquadro et al. 1988; Aquadro 1992; Moriyama and Powell 1996). In contrast, due to the higher efficacy of selection in large populations, nonneutral characters are expected to be less variable in the species with the larger population size. Consistent with D. simulans having a larger effective population size, D. melanogaster was repeatedly found to harbor higher levels of nonsynonymous polymorphism (Moriyama and Powell 1996; Morton et al. 2004). Similarly, the efficacy of selection for synonymous codon usage is expected to depend on population size, with selection being more effective in larger populations (Muto and Osawa 1987; Akashi 1995). Concordant with a smaller population size, and thus relaxed selective constraint, the D. melanogaster lineage has fixed a much higher number of unpreferred codons than the D. simulans lineage (Akashi 1995, 1996, 1997; Akashi and Schaeffer 1997; Akashi et al. 1998; McVean and Vieira 2001). Allozymes which evolve under selective constraints, have consistently been shown to be much less variable in D. simulans than in D. melanogaster (Choudhary and Singh 1987; Singh et al. 1987; Choudhary et al. 1992). Finally, morphological data that can be directly interpreted as reflecting adaptive constraints also support the picture of lower variability and less differentiation between worldwide D. simulans than D. melanogaster populations [reviewed in Capy and Gibert (2004)].
Unfortunately, the majority of data, from which evidence for different effective population sizes of D. simulans and D. melanogaster was derived, is based on comparisons of non-African populations [e.g., Moriyama and Powell (1996)]. Given that both species underwent changes in effective population size during their recent habitat expansion, these populations have not yet reached mutation-drift equilibrium. Furthermore, if the demographic past differs between both species, it is not clear how this would affect the population size estimates in the non-African populations.
African Drosophila populations were shown to harbor substantially more variation than non-African populations (Begun and Aquadro 1993; Irvin et al. 1998; Hamblin and Veuille 1999; Kauer et al. 2002), hence, they are much better suited to infer the long-term effective population size of both species. Nevertheless, it has also become apparent that even African populations are not in equilibrium, and evidence for population bottlenecks (Dieringer et al. 2005; Haddrill et al. 2005), population expansions (Glinka et al. 2003; Baudry et al. 2006; Pool and Aquadro 2006; Schöfl and Schlötterer 2006), and non-African admixture (Capy et al. 2000; Haerty et al. 2002, 2003, 2005; Kauer et al. 2003) were found. Given this complex demographic past of African populations and the difficulties to determine a distinct ancestral range for the two species (Dean and Ballard 2004; Baudry et al. 2006; Kopp et al. 2006; Pool and Aquadro 2006), we studied multiple populations covering a broad geographic range in Africa and compared the pattern of molecular variation. Contrary to the prevailing view, we found African D. melanogaster and D. simulans to harbor similar levels of variability, possibly suggesting that their ancestral population size is more similar than previously assumed.
MATERIALS AND METHODS
Fly strains:
Our study includes samples from five African D. simulans and six African D. melanogaster populations: for D. simulans, we sequenced 17 lines from Kibale (Uganda), 13 lines from Kampala (Uganda), 10 lines from Zimbabwe, 5 lines from Zomba (Malawi), and 14 lines from Madagascar.
For D. melanogaster, we used the same lines as Schöfl et al. (2005) originating from five populations: Kampala (Uganda, 7 lines), Kisoro (Uganda, 10 lines), Malindi (Kenya, 10 lines), Moribabougou (Mali, 9 lines), and Sengwa (Zimbabwe, 10 lines). In addition, we included sequence data from the Lake Kariba (Zimbabwe, 12 lines) population studied by Glinka et al. (2003). Details about the populations are provided in supplemental Tables S1 and S2 at http://www.genetics.org/supplemental/.
Loci sequenced:
We included seven loci studied by Schöfl et al. (2005) (fragments 57, 120, 139, 157, 203, 216, and 287) and added three new loci (fragments 55, 326, and 330) from Glinka et al. (2003). Primers for the new fragments were designed on the basis of release 3.2 of the D. melanogaster genome (http://www.flybase.org) (supplemental Table S3 at http://www.genetics.org/supplemental/); for the remaining seven fragments we used the primers reported in Schöfl et al. (2005). DNA was isolated from single male flies according to Miller et al. (1988), and PCR products were directly sequenced in both directions on a MegaBace 500 (Amersham Biosciences, Piscataway, NJ) using ET terminator sequencing chemistry (Amersham Biosciences).
New sequences have been submitted to GenBank under accession nos. EU278701–EU279399.
Despite the fragments being originally identified as intronic and intergenic, a later genome release indicated that five fragments contained parts of coding regions or 5′-UTR (fragments 55, 120, 139, 203, and 216). All fragments are located in regions of normal to high recombination rate. Details on the loci sequenced are provided in supplemental Table S4 at http://www.genetics.org/supplemental/.
Sequence analysis:
Sequences were assembled and edited using AutoAssembler 3.1. We used ClustalX (Thompson et al. 1994) to generate multiple alignments that were subsequently edited manually. Some regions could not be unambiguously aligned and were excluded from the interspecific comparison (≈70 bp in fragment 139, ≈30 bp in fragment 157, ≈100 bp in fragment 216, ≈30 bp in fragment 330). Summary statistics and tests of neutrality were calculated with DnaSP v.4.0 (Rozas et al. 2003) on the basis of the number of segregating sites. Since we observed three different bases per site in some populations, we also performed the analyses on the basis of the total number of mutations (η) and obtained qualitatively similar results (data not shown). For calculating Fu and Li's D (Fu and Li 1993) and Fay and Wu's H (Fay and Wu 2000) we used the published D. melanogaster (Adams et al. 2000) and the D. simulans (http://genome.wustl.edu/) sequences as an outgroup for D. simulans and D. melanogaster populations, respectively. We measured linkage disequilibrium using the ZnS statistic (Kelly 1997) on the basis of the parsimony informative sites.
Statistical significance for Tajima's D (Tajima 1989), Fu and Li's D (Fu and Li 1993), and Fay and Wu's H (Fay and Wu 2000) was assessed by coalescent simulations with 10,000 replicates as implemented in DnaSP v. 4.0 (Rozas et al. 2003). We performed the simulations with and without intragenic recombination. As the X chromosome on which the analyzed fragments are located shows similar levels of recombination in D. melanogaster and D. simulans (True et al. 1996), we used the appropriate recombination rate for each fragment in D. melanogaster as provided by Glinka et al. (2003) and used an effective population size of 106 for both species. We performed coalescent simulations with 10,000 replicates implemented in the HKA software (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm#HKA) to assess deviation from neutrality on the basis of multilocus Tajima's D and Fu and Li's D values for each population. Divergence was calculated between the pooled D. simulans and the pooled D. melanogaster samples on the basis of the average number of nucleotide substitutions per site, Dxy (Nei 1987). All statistical analyses were performed with the SPSS version 13.0 software package. We tested all data sets for normality before applying parametric tests.
Population differentiation:
For each fragment we determined pairwise FST values among D. simulans and D. melanogaster populations using Arlequin 3.0 (Excoffier et al. 2005) (http://cmpg.unibe.ch/software/arlequin3/); levels of significance for each fragment were assessed on the basis of 10,000 permutations.
For comparison, we also report differentiation based on Snn (Hudson 2000), which is a powerful method for the inference of population differentiation using the number of differences between haplotypes instead of haplotype frequencies (Hudson 2000). Pairwise Snn values and significance based on 10,000 permutations were calculated using DnaSP v. 4.0 (Rozas et al. 2003). The differentiation probabilities P from pairwise population comparisons for all fragments were combined to the χ2-distributed quantity 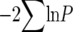 with 2k d.f. (k being the number of fragments). This method of combining probabilities allows creating an overall test for significance from a series of separate significance tests on different sets of data (Sokal and Rohlf 1995). Isolation by distance was tested using a Mantel test implemented in the Isolation By Distance (IBD) Web Service (http://ibdws.sdsu.edu/) (Jensen et al. 2005). We determined approximate geographic distance between the sampling locations using the Google Maps Distance Calculator (http://www.daftlogic.com/Projects/Google-Maps-Distance-Calculator/) and determined significance of IBD on the basis of 30,000 randomizations.
with 2k d.f. (k being the number of fragments). This method of combining probabilities allows creating an overall test for significance from a series of separate significance tests on different sets of data (Sokal and Rohlf 1995). Isolation by distance was tested using a Mantel test implemented in the Isolation By Distance (IBD) Web Service (http://ibdws.sdsu.edu/) (Jensen et al. 2005). We determined approximate geographic distance between the sampling locations using the Google Maps Distance Calculator (http://www.daftlogic.com/Projects/Google-Maps-Distance-Calculator/) and determined significance of IBD on the basis of 30,000 randomizations.
Differences in population differentiation between D. melanogaster and D. simulans were tested by averaging pairwise FST or Snn values for every locus across all analyzed populations and comparing the values with a paired t-test.
RESULTS
Population structure:
We analyzed population structure among five African D. simulans populations using 10 X-linked fragments. Pairwise FST values, averaged over all 10 fragments, ranged from 0 to 0.084. All but two of the pairwise comparisons were significant after correction for multiple testing (Table 1). Interestingly, the two nonsignificant comparisons included populations that are located in close proximity (a 300-km distance between Kampala and Kibale and a 700-km distance between Zomba and Zimbabwe), suggesting either gene flow or shared ancestry. On the basis of Snn, we obtained the same result with an additional population pair (Zomba–Madagascar) being not significantly differentiated (Table 1).
TABLE 1.
Pairwise FST (bottom diagonal) and Snn values (top diagonal) among D. simulans populations
| KAM_sim | KIB | Z | ZOM | M | |
|---|---|---|---|---|---|
| KAM_sim | 0.527 NS | 0.661*** | 0.699** | 0.654*** | |
| KIB | −0.006 NS | 0.689*** | 0.787*** | 0.709*** | |
| Z | 0.063*** | 0.060*** | 0.539 NS | 0.631** | |
| ZOM | 0.061** | 0.078** | −0.010 NS | 0.636 NS | |
| M | 0.062*** | 0.063*** | 0.050** | 0.084** |
NS, nonsignificant. *P < 0.05; **P < 0.01; ***P < 0.001.
We evaluated the impact of gene flow on genetic differentiation by testing for isolation by distance. On the basis of the average FST and Snn across fragments, we found an almost significant correlation between geographic and genetic distance (FST, r = 0.7005, one-sided P < 0.057, Mantel test; Snn, r = 0.7027, one-sided P < 0.059, Mantel test).
For comparison, we analyzed the same loci in six African D. melanogaster populations. Six (FST) and 7 (Snn) of 15 comparisons were nonsignificant with different population pairs being nondifferentiated depending on whether FST or the Snn statistic were used (Table 2). Interestingly, even geographically adjacent populations were highly differentiated (the two populations from Uganda), while distantly located populations were not significantly differentiated (e.g., Mali and Sengwa). Hence, no significant isolation by distance was observed (FST, r = 0.0410, P < 0.45, Mantel test; Snn, r = −0.1036, P < 0.43, Mantel test).
TABLE 2.
Pairwise FST (bottom diagonal) and Snn values (top diagonal) among D. melanogaster populations
| KAM_mel | KIS | KEN | MALI | ZS | ZBMEL | |
|---|---|---|---|---|---|---|
| KAM_mel | 0.604** | 0.595** | 0.656*** | 0.641** | 0.701*** | |
| KIS | 0.065** | 0.502 NS | 0.481 NS | 0.497 NS | 0.590* | |
| KEN | 0.037* | 0.030* | 0.524 NS | 0.496 NS | 0.512 NS | |
| MALI | 0.087** | 0.025 NS | 0.006 NS | 0.489 NS | 0.607*** | |
| ZS | 0.057*** | 0.003 NS | −0.008 NS | 0.017 NS | 0.530* | |
| ZBMEL | 0.082*** | 0.052*** | 0.040** | 0.071*** | 0.008 NS |
NS, nonsignificant. *P < 0.05; **P < 0.01; ***P < 0.001.
Mean FST and Snn values suggested a more pronounced population structure in D. simulans (mean FST = 0.0506, mean Snn = 0.6532) than in D. melanogaster (mean FST = 0.0383, mean Snn = 0.5617). We tested for statistical significance by comparing average FST and Snn values across all pairwise comparisons within each species. While we observed no significant differences in FST between D. melanogaster and D. simulans (P > 0.6, paired t-test), Snn differed significantly (P = 0.017, paired t-test).
Given that estimates of population differentiation using Snn could be influenced by the unequal sample sizes available for D. simulans and D. melanogaster populations (Hudson 2000), we repeated Snn and FST analyses using a matched sample size of n = 8 for all populations. For this analysis, we excluded the two populations with the smallest sample sizes [D. simulans, Zomba (ZOM), and D. melanogaster, Kampala (KAM_mel)] and randomly chose eight lines from each of the remaining populations. We observed the same trend of more differentiation in D. simulans (mean FST = 0.0358, mean Snn = 0.5416) than in D. melanogaster (mean FST = 0.0206, mean Snn = 0.4972), but the differences were no longer significant (Snn, P = 0.462; FST, P = 0.477) (for pairwise comparisons see supplemental Tables S5 and S6 at http://www.genetics.org/supplemental/).
Levels of variability:
Levels of variability for the 10 fragments were heterogeneous among populations in both species: in D. simulans populations, π ranged from 0.0107 (Zomba) to 0.0131 (Madagascar); in D. melanogaster populations, it ranged from 0.0114 (Kampala) to 0.0146 (Kenya) (Tables 3 and 4, and for information per population per locus see supplemental Tables S7 and S8 at http://www.genetics.org/supplemental/). Consistent with the presumed origin of D. simulans in Madagascar (Dean and Ballard 2004; Baudry et al. 2006), this population harbored more variation than the other D. simulans populations although the difference was significant only when variability was measured on the basis of the number of segregating sites (P < 0.003, ANOVA, Tukey's HSD post hoc test).
TABLE 3.
Measures of variability, neutrality tests, and linkage disequilibrium averaged across 10 loci in five D. simulans populations
| π | θW | Hd | Taj D | Fu and Li's D | Fay and Wu's H | ZnS | |
|---|---|---|---|---|---|---|---|
| KAM_sim | 0.0120 | 0.0131 | 0.89 | −0.42 | −0.46 | −1.18 | 0.26 |
| KIB | 0.0116 | 0.0121 | 0.89 | −0.13 | −0.09 | −0.08 | 0.25 |
| Z | 0.0111 | 0.0118 | 0.85 | −0.21 | −0.35 | −0.28 | 0.31 |
| ZOM | 0.0107 | 0.0111 | 0.87 | −0.36 | −0.47 | −0.14 | 0.60 |
| M | 0.0131 | 0.0181 | 0.97 | −1.19*** | −1.86*** | −0.38 | 0.12 |
π, nucleotide diversity; θW, θ = 4Neμ estimated from the number of segregating sites; Hd, haplotype diversity; Taj D, Tajima's D; ZnS, linkage disequilibrium (Kelly 1997); significance of Tajima's D and Fu and Li's D across fragments was determined as indicated in materials and methods. *P < 0.05; **P < 0.01; ***P < 0.001.
TABLE 4.
Measures of variability, neutrality tests, and linkage disequilibrium averaged across 10 loci in six D. melanogaster populations
| π | θW | Hd | Taj D | Fu and Li's D | Fay and Wu's H | ZnS | |
|---|---|---|---|---|---|---|---|
| KAM_mel | 0.0114 | 0.0112 | 0.81 | 0.01 | 0.18 | −0.05 | 0.57 |
| KIS | 0.0129 | 0.0138 | 0.97 | −0.38 | −0.48 | −0.97 | 0.19 |
| KEN | 0.0146 | 0.0157 | 0.98 | −0.33 | −0.30 | −0.31 | 0.18 |
| MALI | 0.0139 | 0.0140 | 0.95 | −0.09 | −0.15 | −0.49 | 0.26 |
| ZS | 0.0141 | 0.0151 | 0.97 | −0.35 | −0.57 | 0.32 | 0.19 |
| ZBMEL | 0.0127 | 0.0140 | 0.97 | −0.48 | −0.59 | −0.68 | 0.16 |
π, nucleotide diversity; θW, θ = 4Neμ estimated from the number of segregating sites; Hd, haplotype diversity; Taj D, Tajima's D; ZnS, linkage disequilibrium (Kelly 1997); significance of Tajima's D and Fu and Li's D across fragments was determined as indicated in materials and methods. All values were nonsignificant.
Most D. melanogaster populations were more variable than D. simulans populations (Table 5). To quantify this difference, we averaged the variability for each locus across populations and tested for significance. We observed a marginally significant difference only for haplotype diversity (P = 0.046, paired t-test), but not for π and θW. According to release 5.1 of the D. melanogaster genome, our data set contained loci from both intergenic and intronic regions some of which partially overlap with transcribed or coding regions. Hence, we repeated the analyses on the basis of only noncoding regions, but obtained similar results (supplemental Table S9 at http://www.genetics.org/supplemental/).
TABLE 5.
Average values of different variability estimators (π, θW, and haplotype diversity) and linkage disequilibrium (ZnS) compared between five D. simulans and six D. melanogaster populations
| Mean π
|
Mean θW
|
Mean Hd
|
Mean ZnS
|
|||||
|---|---|---|---|---|---|---|---|---|
| Locus | D. sim | D. mel | D. sim | D. mel | D. sim | D. mel | D. sim | D. mel |
| 55 | 0.0144 | 0.0176 | 0.0152 | 0.0186 | 0.93 | 0.98 | 0.39 | 0.23 |
| 57 | 0.0059 | 0.0095 | 0.0076 | 0.0104 | 0.83 | 0.96 | 0.37 | 0.22 |
| 120 | 0.0123 | 0.0122 | 0.0133 | 0.0126 | 0.96 | 0.95 | 0.19 | 0.26 |
| 139 | 0.0098 | 0.0136 | 0.0129 | 0.0149 | 0.91 | 0.89 | 0.32 | 0.39 |
| 157 | 0.0060 | 0.0073 | 0.0059 | 0.0093 | 0.74 | 0.89 | 0.30 | 0.18 |
| 203 | 0.0143 | 0.0164 | 0.0180 | 0.0161 | 0.88 | 0.95 | 0.37 | 0.23 |
| 216 | 0.0165 | 0.0192 | 0.0180 | 0.0203 | 0.95 | 0.94 | 0.28 | 0.30 |
| 287 | 0.0036 | 0.0083 | 0.0049 | 0.0095 | 0.83 | 0.95 | 0.23 | 0.14 |
| 326 | 0.0073 | 0.0052 | 0.0078 | 0.0057 | 0.94 | 0.93 | 0.16 | 0.26 |
| 330 | 0.0268 | 0.0231 | 0.0291 | 0.0221 | 0.97 | 0.98 | 0.24 | 0.25 |
| mean | 0.0117 | 0.0133 | 0.0132 | 0.0140 | 0.89 | 0.94 | 0.31 | 0.26 |
π, nucleotide diversity; θW, θ = 4Neμ estimated from the number of segregating sites; Hd, haplotype diversity; ZnS, linkage disequilibrium (Kelly 1997).
The lower variability of D. simulans populations collected in continental Africa may have obscured the pattern of more variability in D. simulans collected at its presumed origin in Madagascar. Hence, we compared the variability of D. simulans from Madagascar to each of the African D. melanogaster populations and found no significant difference for π (P > 0.177, ANOVA, Tukey's HSD post hoc test). Using θW or haplotype diversity, significantly more variability was detected only in comparison to a single D. melanogaster population collected in Kampala (Uganda) (P < 0.002, ANOVA, Tukey's HSD post hoc test). No significant difference was found between D. simulans and all remaining five D. melanogaster populations (P > 0.138, ANOVA, Tukey's HSD post hoc test). These results show that even the most variable D. simulans population collected at the putative geographic origin of the species does not harbor significantly more variation than D. melanogaster.
Linkage disequilibrium:
Under equilibrium conditions, a species with larger Ne is expected to show a lower degree of linkage disequilibrium. Hence, we compared levels of linkage disequilibrium for the 10 fragments sequenced in D. melanogaster and D. simulans. Average ZnS values were slightly lower for African D. melanogaster (mean ZnS = 0.26) than for D. simulans (mean ZnS = 0.31) populations (Tables 3 and 4). This difference was, however, not significant when average ZnS values were compared for each locus (P > 0.2, paired t-test). The ZnS value for the D. simulans population from Madagascar was lower than for the other D. simulans populations, but only significant in comparison to Zimbabwe and Zomba (P < 0.046, ANOVA, Tukey's HSD post hoc test). In comparison to D. melanogaster, the linkage disequilibrium was lower in the D. simulans population from Madagascar, but this difference was statistically significant only in two populations (Kampala, P < 0.001 and Mali, P = 0.006).
Deviation from mutation-drift equilibrium:
For African D. melanogaster and D. simulans populations, a deviation from mutation-drift equilibrium has been described (Dieringer et al. 2005; Ometto et al. 2005; Schöfl et al. 2005; Baudry et al. 2006; Schöfl and Schlötterer 2006). Consistent with these results, we found a negative Tajima's D, Fu and Li's D, and Fay and Wu's H in almost all populations of the two species (Tables 3 and 4). Interestingly, the D. simulans population from Madagascar was the only population showing both a significantly negative Tajima's D and Fu and Li's D value when considering all loci jointly, suggesting a more pronounced deviation from mutation-drift equilibrium despite being the D. simulans population with the highest level of sequence variability.
DISCUSSION
Our survey of sequence variation at 10 X-chromosomal regions in multiple African D. melanogaster and D. simulans populations resulted in similar levels of variability in the two species. This observation contrasts previous results, which found D. simulans to be more variable than D. melanogaster. Given the important implications that potential differences in effective population sizes of the two species could have for the interpretation of patterns of molecular evolution, we discuss in the following how our data can be reconciled with previous results.
Microsatellite variation:
Like the African sequence data, microsatellites support similar levels of variability in both species rather than more variation in D. simulans. Harr and Schlötterer (2004) compared a set of microsatellites in the two species and found D. melanogaster to be more variable than D. simulans. Given that the majority of loci used in this study was isolated in D. melanogaster, the higher variation of D. melanogaster could be explained by an ascertainment bias. Nevertheless, after a correction for the difference in repeat count, both species harbored similar levels of variability (Harr and Schlötterer 2004). Similarly, a comparison of microsatellite variability in African D. melanogaster and D. simulans that was based on microsatellites derived from both species found a significant effect for the focal species, but all measures of variability (gene diversity, variance in repeat number, and number of alleles) were higher in D. melanogaster than in D. simulans (Hutter et al. 1998).
DNA sequence polymorphism:
The majority of sequence polymorphism data supporting a larger effective population size of D. simulans were collected in non-African populations (Aquadro et al. 1988, 1992; Moriyama and Powell 1996). Given that the colonization histories of the two species may differ (David and Capy 1988; Irvin et al. 1998; Capy and Gibert 2004; Lachaise and Silvain 2004), it is possible that the higher variability of D. simulans is limited to non-African populations, and it may, therefore, be preferable to consider non-African and African data sets separately.
For comparison of our data with previous polymorphism studies we used the data compilation of Andolfatto (2001) and extracted variability data under the strict criterion of using only those genes for which data from African samples of both species are available.
Contrary to our results, levels of nucleotide variability (θ) as well as synonymous site diversity (π, θ) are higher in African D. simulans than in African D. melanogaster on both the X chromosome and autosomes (supplemental Tables S10 and S11 at http://www.genetics.org/supplemental/), although the differences are not statistically significant (P > 0.068, paired t-test).
When the ratio of synonymous (S) and replacement substitutions (R) is analyzed X-chromosomal data show an only slightly lower S/R ratio in D. melanogaster than in D. simulans as shown by Andolfatto (2001) (supplemental Table S12 at http://www.genetics.org/supplemental/). Contrary to previous analyses (Andolfatto 2001; Mousset and Derome 2004), which did not apply our stringent criterion of using the same genes in both species, the autosomes show a lower S/R ratio in D. simulans, thereby suggesting a larger population size of D. melanogaster. A larger set of loci sequenced in both species is required to determine if the discrepancy between these analyses is biologically meaningful.
Kern and Begun (2005) compared levels of variability among D. melanogaster and D. simulans for intronic, intergenic, and synonymous sites. After correcting for multiple testing, the only significant difference was observed for intronic regions, with D. simulans being more variable than D. melanogaster. The same pattern was observed when African D. melanogaster was compared against a D. simulans sample containing flies from cosmopolitan populations. The interpretation of these results strongly depends on the functional constraint of intronic sequences. While it has been assumed for a long time that introns are evolving under low functional constraint, recent results (Halligan et al. 2004; Andolfatto 2005) suggest that introns are highly constrained in length and sequence. Hence, depending on the assumed model of intron evolution, the higher variability in D. simulans introns suggests a larger effective population size either in D. simulans (low functional constraint) or in D. melanogaster (high functional constraint).
Using an experimental design in which a consistent set of populations was sequenced for the same loci in both species, Baudry et al. (2004, 2006) studied four X-linked genes in African D. melanogaster (Kenya, Zimbabwe, Ivory Coast, and Niger) and D. simulans populations (Madagascar, Mayotte, Tanzania, and Kenya). Although it is not discussed by the authors, these studies revealed similar levels of variability in both species, even if only the most variable populations from the “Eastern group” (Madagascar, Mayotte, Tanzania, and Kenya) are considered for D. simulans: D. simulans populations θ = 0.0041, π = 0.0031; D. melanogaster populations θ = 0.0037, π = 0.0038.
Demography:
The inference of effective population sizes based on standing levels of variation is problematic as demographic events such as population bottlenecks, admixture, and population subdivision strongly affect levels of variability. While the pronounced impact of demography on non-African Drosophila populations has been widely discussed (Hamblin and Veuille 1999; Kauer et al. 2002; Haddrill et al. 2005; Ometto et al. 2005; Schlötterer et al. 2006; Schöfl and Schlötterer 2006), less is known about African populations. Recent studies showed that also in Africa both species are not in mutation-drift equilibrium (Dieringer et al. 2005; Haddrill et al. 2005; Ometto et al. 2005; Baudry et al. 2006; Pool and Aquadro 2006; Schöfl and Schlötterer 2006). Our result of a more pronounced population structure in African D. simulans than D. melanogaster implies that the demographic history may differ between both species. In D. melanogaster, several studies found that African populations were recently experiencing admixture from non-African populations (Capy et al. 2000; Haerty et al. 2002, 2003; Kauer et al. 2003). In D. simulans, both subdivision of the ancestral populations and more recent admixture of divergent lineages during the out-of-Africa expansion of this species have been discussed to explain the observed strong haplotype structure of cosmopolitan D. simulans populations (Hamblin and Veuille 1999). Given these demographic complications, a larger number of loci is required to estimate the effective population size of D. melanogaster and D. simulans jointly with the demography.
We note that our data are limited to X-linked loci, but striking differences in the relative levels of autosomal and X-linked variability have been observed in D. melanogaster and D. simulans. While in D. melanogaster, X-linked microsatellite variability was higher than the autosomal (Kauer et al. 2002), in D. simulans, the X chromosome and autosomes harbored similar levels of variability (Irvin et al. 1998; Schöfl and Schlötterer 2004, 2006) [see supplemental Table S13 at http://www.genetics.org/supplemental/ for the reanalysis of previously unmapped microsatellites from Irvin et al. (1998)].
Our results may therefore have implications only for the interpretation of evolutionary forces operating on the X chromosome of the two species. For example, the difference in the patterns of codon usage between D. melanogaster and D. simulans has frequently been attributed to a reduced efficacy of selection in D. melanogaster due to a smaller Ne. Consistent with this explanation, a recent study focusing on X-chromosomal data confirmed a strong decline of major codon usage in the D. melanogaster lineage, whereas no such trend in D. simulans was observed, at least in nontelomeric regions on the X chromosome (Ko et al. 2006). Given the highly similar levels of X-linked variability in African D. melanogaster and D. simulans populations, it remains an open question which differences in Ne are compatible with our polymorphism data and could still cause the dissimilar patterns of selection intensity (Nes) operating on codon usage on the X chromosome as observed by Ko et al. (2006).
Our finding of similar levels of variability in ancestral populations of both species suggests that a difference in Ne may be limited to derived populations, which implies that it arose only recently. Given that selection on codon usage is weak and thus requires long-term evolutionary differences, it appears unlikely that recently developed differences in Ne account for differences in codon usage.
Our analysis is limited in that the polymorphism data do not necessarily reflect the long-term Ne of the two species. On the basis of patterns of synonymous codon usage it has been suggested that the long-term Ne of D. simulans was more stable than that of D. melanogaster (Akashi 1995; Akashi and Schaeffer 1997; McVean and Vieira 2001), with its current Ne being similar to the long-term Ne (Bierne and Eyre-Walker 2004). Hence, our results could be reconciled with this suggestion by assuming a recent decrease of Ne in the D. simulans lineage or a recent increase of Ne in the D. melanogaster lineage.
Whether the observed strong decline of selection on synonymous codon usage in the D. melanogaster lineage can be sufficiently explained using the simple correlation with a decline in its long-term effective population size remains unclear. Akashi et al. (2006) showed that the effective population size of different Drosophila species (as inferred from their current habitat range) is correlated with the patterns of protein evolution but not with codon usage bias. Similarly, Begun and Whitley (2002) found that higher levels of codon usage bias in the Xdh gene are not necessarily observed in the species with a large Ne as inferred from levels of silent heterozygosity; instead, codon usage bias in the most variable species D. willistoni and D. equinoxialis was low. In light of our results of similar variability and thus effective population size in the African Drosophila populations, alternative, probably more complex explanations for the observed differences in codon usage may have to be invoked, e.g., unequal selection coefficients between species (Bierne and Eyre-Walker 2006) or frequent changes and heterogeneous patterns of mutation or recombination rates (Ko et al. 2003).
Sampling variance:
As in previous studies, we relied on variability estimates to draw conclusions about the relative population sizes of D. melanogaster and D. simulans. This procedure does not, however, account for sampling variance and the associated statistical power. The variance of sequence polymorphism among loci in combination with the moderate number of loci studied may result in a low statistical power to detect differences in population size. Hence, it is possible that previous observations of more variation in derived D. simulans populations are compatible with no difference in population size between D. simulans and D. melanogaster. Alternatively, the similar level of variability in African D. melanogaster and D. simulans populations could also be consistent with a larger long-term effective population size of D. simulans. With the new sequencing technologies it is possible to obtain sufficient statistical power through the analysis of multiple loci or even entire genomes in multiple populations from both species to resolve the controversy about the difference in population sizes.
Acknowledgments
We are thankful to all collaborators who provided us with D. melanogaster and D. simulans samples, as well as to CSlab members, C. Vogl, and two anonymous reviewers for their helpful comments on earlier versions of the manuscript. The work was supported by Fonds zur Förderung der wissenschaftlichen Forschung grants to C.S.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Akashi, H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 1996. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics 144: 1297–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., 1997. Codon bias evolution in Drosophila. Population genetics of mutation-selection drift. Gene 205: 269–278. [DOI] [PubMed] [Google Scholar]
- Akashi, H., and S. W. Schaeffer, 1997. Natural selection and the frequency distributions of “silent” DNA polymorphism in Drosophila. Genetics 146: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., R. M. Kliman and A. Eyre-Walker, 1998. Mutation pressure, natural selection, and the evolution of base composition in Drosophila. Genetica 102/103: 49–60. [PubMed] [Google Scholar]
- Akashi, H., W. Y. Ko, S. Piao, A. John, P. Goel et al., 2006. Molecular evolution in the Drosophila melanogaster species subgroup: frequent parameter fluctuations on the timescale of molecular divergence. Genetics 172: 1711–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Aquadro, C. F., 1992. Why is the genome variable? Insights from Drosophila. Trends Genet. 8: 355–362. [DOI] [PubMed] [Google Scholar]
- Aquadro, C. F., K. M. Lado and W. A. Noon, 1988. The rosy region of Drosophila melanogaster and Drosophila simulans. I. Contrasting levels of naturally occurring DNA restriction map variation and divergence. Genetics 119: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro, C. F., R. M. Jennings, Jr., M. M. Bland, C. C. Laurie and C. H. Langley, 1992. Patterns of naturally occurring restriction map variation, dopa decarboxylase activity variation, and linkage disequilibrium in the Ddc gene region of Drosophila melanogaster. Genetics 132: 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry, E., B. Viginier and M. Veuille, 2004. Non-African populations of Drosophila melanogaster have a unique origin. Mol. Biol. Evol. 21: 1482–1491. [DOI] [PubMed] [Google Scholar]
- Baudry, E., N. Derome, M. Huet and M. Veuille, 2006. Contrasted polymorphism patterns in a large sample of populations from the evolutionary genetics model Drosophila simulans. Genetics 173: 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and P. Whitley, 2002. Molecular population genetics of Xdh and the evolution of base composition in Drosophila. Genetics 162: 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, N., and A. Eyre-Walker, 2004. The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol. 21: 1350–1360. [DOI] [PubMed] [Google Scholar]
- Bierne, N., and A. Eyre-Walker, 2006. Variation in synonymous codon use and DNA polymorphism within the Drosophila genome. J. Evol. Biol. 19: 1–11. [DOI] [PubMed] [Google Scholar]
- Capy, P., and P. Gibert, 2004. Drosophila melanogaster, Drosophila simulans: so similar yet so different. Genetica 120: 5–16. [DOI] [PubMed] [Google Scholar]
- Capy, P., M. Veuille, M. Paillette, J. M. Jallon, J. Vouidibio et al., 2000. Sexual isolation of genetically differentiated sympatric populations of Drosophila melanogaster in Brazzaville, Congo: The first step towards speciation? Heredity 84(Pt 4): 468–475. [DOI] [PubMed] [Google Scholar]
- Choudhary, M., and R. S. Singh, 1987. Historical effective size and the level of genetic diversity in Drosophila melanogaster and Drosophila pseudoobscura. Biochem. Genet. 25: 41–51. [DOI] [PubMed] [Google Scholar]
- Choudhary, M., M. B. Coulthart and R. S. Singh, 1992. A comprehensive study of genic variation in natural populations of Drosophila melanogaster. VI. Patterns and processes of genic divergence between D. melanogaster and its sibling species, Drosophila simulans. Genetics 130: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, J. R., and P. Capy, 1988. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4: 106–111. [DOI] [PubMed] [Google Scholar]
- Dean, M. D., and J. W. Ballard, 2004. Linking phylogenetics with population genetics to reconstruct the geographic origin of a species. Mol. Phylogenet. Evol. 32: 998–1009. [DOI] [PubMed] [Google Scholar]
- Dieringer, D., V. Nolte and C. Schlötterer, 2005. Population structure in African Drosophila melanogaster revealed by microsatellite analysis. Mol. Ecol. 14: 563–573. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., G. Laval and S. Schneider, 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka, S., L. Ometto, S. Mousset, W. Stephan and D. De Lorenzo, 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multilocus approach. Genetics 165: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15: 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty, W., J. M. Jallon, J. Rouault, C. Bazin and P. Capy, 2002. Reproductive isolation in natural populations of Drosophila melanogaster from Brazzaville (Congo). Genetica 116: 215–224. [PubMed] [Google Scholar]
- Haerty, W., P. Gibert, P. Capy, B. Moreteau and J. R. David, 2003. Microspatial structure of Drosophila melanogaster populations in Brazzaville: evidence of natural selection acting on morphometrical traits. Heredity 91: 440–447. [DOI] [PubMed] [Google Scholar]
- Haerty, W., M. Lesbats and P. Capy, 2005. Pre-reproductive isolation as a consequence of allopatric differentiation between populations of Drosophila melanogaster. Mol. Ecol. 14: 3801–3807. [DOI] [PubMed] [Google Scholar]
- Halligan, D. L., A. Eyre-Walker, P. Andolfatto and P. D. Keightley, 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin, M. T., and M. Veuille, 1999. Population structure among African and derived populations of Drosophila simulans: evidence for ancient subdivision and recent admixture. Genetics 153: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr, B., and C. Schlötterer, 2004. Patterns of microsatellite variability in the Drosophila melanogaster complex. Genetica 120: 71–77. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 2000. A new statistic for detecting genetic differentiation. Genetics 155: 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter, C. M., M. D. Schug and C. F. Aquadro, 1998. Microsatellite variation in Drosophila melanogaster and Drosophila simulans: a reciprocal test of the ascertainment bias hypothesis. Mol. Biol. Evol. 15: 1620–1636. [DOI] [PubMed] [Google Scholar]
- Irvin, S. D., K. A. Wetterstrand, C. M. Hutter and C. F. Aquadro, 1998. Genetic variation and differentiation at microsatellite loci in Drosophila simulans. Evidence for founder effects in new world populations. Genetics 150: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J. L., A. J. Bohonak and S. T. Kelley, 2005. Isolation by distance, web service. BMC Genet. 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., B. Zangerl, D. Dieringer and C. Schlötterer, 2002. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics 160: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., D. Dieringer and C. Schlötterer, 2003. Nonneutral admixture of immigrant genotypes in African Drosophila melanogaster populations from Zimbabwe. Mol. Biol. Evol. 20: 1329–1337. [DOI] [PubMed] [Google Scholar]
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, A. D., and D. J. Begun, 2005. Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes. Mol. Biol. Evol. 22: 51–62. [DOI] [PubMed] [Google Scholar]
- Ko, W. Y., R. M. David and H. Akashi, 2003. Molecular phylogeny of the Drosophila melanogaster species subgroup. J. Mol. Evol. 57: 562–573. [DOI] [PubMed] [Google Scholar]
- Ko, W. Y., S. Piao and H. Akashi, 2006. Strong regional heterogeneity in base composition evolution on the Drosophila X chromosome. Genetics 174: 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, A., A. Frank and J. Fu, 2006. Historical biogeography of Drosophila simulans based on Y-chromosomal sequences. Mol. Phylogenet. Evol. 38: 355–362. [DOI] [PubMed] [Google Scholar]
- Lachaise, D., and J. F. Silvain, 2004. How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120: 17–39. [DOI] [PubMed] [Google Scholar]
- McVean, G. A., and J. Vieira, 2001. Inferring parameters of mutation, selection, and demography from patterns of synonymous site evolution in Drosophila. Genetics 157: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. A., D. D. Dykes and H. F. Polesky, 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. R. Powell, 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277. [DOI] [PubMed] [Google Scholar]
- Morton, R. A., M. Choudhary, M. L. Cariou and R. S. Singh, 2004. A reanalysis of protein polymorphism in Drosophila melanogaster, D. simulans, D. sechellia and D. mauritiana: effects of population size and selection. Genetica 120: 101–114. [DOI] [PubMed] [Google Scholar]
- Mousset, S., and N. Derome, 2004. Molecular polymorphism in Drosophila melanogaster and D. simulans: What have we learned from recent studies? Genetica 120: 79–86. [DOI] [PubMed] [Google Scholar]
- Muto, A., and S. Osawa, 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. USA 84: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Ometto, L., S. Glinka, D. De Lorenzo and W. Stephan, 2005. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 22: 2119–2130. [DOI] [PubMed] [Google Scholar]
- Pool, J. E., and C. F. Aquadro, 2006. History and structure of sub-Saharan populations of Drosophila melanogaster. Genetics 174: 915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schlötterer, C., H. Neumeier, C. Sousa and V. Nolte, 2006. Highly structured Asian Drosophila melanogaster populations: A new tool for hitchhiking mapping? Genetics 172: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöfl, G., and C. Schlötterer, 2004. Patterns of microsatellite variability among X chromosomes and autosomes indicate a high frequency of beneficial mutations in non-African D. simulans. Mol. Biol. Evol. 21: 1384–1390. [DOI] [PubMed] [Google Scholar]
- Schöfl, G., and C. Schlötterer, 2006. Microsatellite variation and differentiation in African and non-African populations of Drosophila simulans. Mol. Ecol. 15: 3895–3905. [DOI] [PubMed] [Google Scholar]
- Schöfl, G., F. Catania, V. Nolte and C. Schlötterer, 2005. African sequence variation accounts for most of the sequence polymorphism in non-African Drosophila melanogaster. Genetics 170: 1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. S., M. Choudhary and J. R. David, 1987. Contrasting patterns of geographic variation in the cosmopolitan sibling species Drosophila melanogaster and Drosophila simulans. Biochem. Genet. 25: 27–40. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry, Ed. 3. W. H. Freeman, New York.
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, J. R., J. M. Mercer and C. C. Laurie, 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]


