Abstract
Endothelial cells assemble von Willebrand factor (VWF) multimers into ordered tubules within storage organelles called Weibel–Palade bodies, and tubular packing is necessary for the secretion of VWF filaments that can bind connective tissue and recruit platelets to sites of vascular injury. We now have recreated VWF tubule assembly in vitro, starting with only pure VWF propeptide (domains D1D2) and disulfide-linked dimers of adjacent N-terminal D′D3 domains. Assembly requires low pH and calcium ions and is reversed at neutral pH. Quick-freeze deep-etch electron microscopy and three-dimensional reconstruction of negatively stained images show that tubules contain a repeating unit of one D′D3 dimer and two propeptides arranged in a right-handed helix with 4.2 units per turn. The symmetry and location of interdomain contacts suggest that decreasing pH along the secretory pathway coordinates the disulfide-linked assembly of VWF multimers with their tubular packaging.
Keywords: electron microscopy, endothelial cell, three-dimensional reconstruction
Weibel–Palade bodies are large cigar-shaped secretory granules, unique to vascular endothelial cells of vertebrates, that are 100–200 nm in diameter and 1–5 μm in length. Lengthwise sections show longitudinal striations, and cross sections reveal closely spaced ≈20-nm diameter tubules separated by a less-dense matrix (1). Weibel–Palade bodies are composed almost entirely of von Willebrand factor (VWF) (2, 3), which is a multimeric plasma glycoprotein that can exceed 20 million Da in mass and 4 μm in length. Megakaryocytes synthesize large VWF multimers and package them into platelet α-granules that are roughly spherical rather than cigar-shaped. Nevertheless, the VWF multimers in α-granules are organized into clusters of tubules with dimensions similar to those of VWF tubules in Weibel–Palade bodies (4). VWF is the largest known protein in the blood, and the very largest VWF multimers bind connective tissue and mediate platelet adhesion at sites of vascular injury. Weibel–Palade bodies play a critical role in hemostasis by delivering VWF multimers into the circulation. Defects in VWF multimer structure cause several forms of von Willebrand disease, the most common inherited bleeding disorder worldwide (5).
VWF is synthesized as an ≈350-kDa precursor with a signal peptide and five kinds of structural domains arranged in the order D1-D2-D′-D3-A1-A2-A3-D4-B1-B2-B3-C1-C2-CK (Fig. 1) (5). In the endoplasmic reticulum (ER), proVWF dimerizes through disulfide bonds between C-terminal CK domains (6–8). The resultant “tail-to-tail” proVWF dimers are transported to the Golgi, where the propeptide (domains D1D2) is cleaved by furin and additional “head-to-head” disulfide bonds form between D3 domains, yielding multimers that condense into tubules and form Weibel–Palade bodies (6, 9).
Fig. 1.
VWF domain structure and plasmid constructs. VWF consists of five types of structural domain, as indicated. The locations of binding sites are shown for the physiological ligands factor VIII (FVIII); platelet glycoprotein Ib (GPIb); collagen types I, III, and VI; and integrin αIIbβ3. The metalloprotease ADAMTS13 cleaves the Tyr1605-Met1606 bond in domain A2. VWF multimers are held together by intersubunit disulfide bonds between CK domains, which form in the ER, and by intersubunit disulfide bonds between D3 domains, which form in the Golgi. The structures of polypeptides encoded by the plasmids used in these studies are indicated.
In fact, the expression of VWF determines the existence and also the cigar-like shape of Weibel–Palade bodies. Without VWF, endothelial cells lack Weibel–Palade bodies (10, 11), and the expression of VWF in other cell types that have a regulated secretory pathway results in the generation of organelles that are indistinguishable from Weibel–Palade bodies (12, 13). This self-organizing behavior depends on a conserved set of N-terminal domains that have the dual function of promoting multimer assembly and directing tubular storage.
Multimer assembly (14–16) and tubular packing (12, 17) both depend on the N-terminal D1D2D′D3 domains and require the acidic pH of the late secretory pathway (13). Furthermore, the tubular morphology of Weibel–Palade bodies is essential for the orderly secretion and hemostatic function of VWF (13). How VWF becomes organized into tubules is unknown. We have now reproduced the reversible assembly of VWF tubules in vitro from pure VWF propeptide (D1D2) and dimeric N-terminal D′D3 domains. Surprisingly, no other cellular proteins or components of Weibel–Palade bodies are required.
Results
VWF Propeptide and D′D3 Dimer Form Helical Tubules.
The smallest fragment of VWF that can direct tubular packing in storage granules consists of domains D1D2D′D3 from the N terminus of the VWF precursor (Fig. 1) (13, 17). Baby hamster kidney (BHK) cells that express this construct secrete a mixture of VWF propeptide (domains D1D2), D′D3 monomers, and D′D3 dimers (17, 18). We purified each of these proteins [supporting information (SI) Fig. 6] to assess their interactions under conditions that are thought to occur in the ER and Golgi.
When mixed at pH 7.4 with Ca2+ ions, D1D2 and D′D3 dimers form a complex with a mass of ≈280 kDa by multiangle light scattering (Fig. 2A and SI Table 1) composed of two D1D2 propeptides and one D′D3 dimer. EDTA dissociates the complex (Fig. 2B), whereas lowering the pH to 6.2, with Ca2+ ions, causes the reversible formation of enormous aggregates (>3 million Da) with the same stoichiometry (Fig. 2C and SI Fig. 7). By electron microscopy (EM) in negative stain (Fig. 2D), these aggregates consist of hollow tubules ≈25 nm in diameter and ≈40–300 nm in length. By quick-freeze deep-etch (QFDE) EM (19), the tubules are right-handed helices with a pitch of ≈11 nm (Fig. 2E). Top views of complete or partial turns (Fig. 2 D and F) suggest that tubules assemble from the ≈280-kDa complexes observed at pH 7.4 with Ca2+ (Fig. 2G).
Fig. 2.
Assembly of VWF tubules depends on pH and Ca2+. (A–C) D1D2 and D′D3 dimers were chromatographed on Superdex 200 at pH 7.4 with 10 mM CaCl2 (A) or 5 mM EDTA (B), or at pH 6.2 with 10 mM CaCl2 (C). (Insets) Column fractions were analyzed by SDS/PAGE and stained with Coomassie blue. (D and E) By EM in negative stain (D) or QFDE (E), (D1D2)2-D′D3 dimer aggregates at pH 6.2, with Ca2+, consist of hollow tubules. (F) Axial views suggest a tetragonal arrangement of the 280-kDa complexes that assemble with Ca2+ at pH 7.4. (G) QFDE EM of (D1D2)2-D′D3 dimer complex at pH 7.4. (H) D1D2 and D′D3A1 dimers (SI Fig. 8) assemble into longer right-handed helices. A left-handed actin filament is highlighted in pink. (Scale bars, 100 nm.)
VWF truncated after domain A1 forms longer Weibel–Palade body-like organelles than does VWF truncated after domain D3 (13), suggesting that adding domain A1 might stabilize tubules. In fact, D1D2 and D′D3A1 dimers (SI Fig. 8) formed longer tubules than D1D2 and D′D3 dimers (Fig. 2 H and E). QFDE images of F-actin and D1D2-D′D3A1 in the same field display the left-handed 5.9-nm helix of F-actin (19), confirming that D1D2-D′D3A1 helices are right-handed (Fig. 2H).
D1D2 Dimerization and Binding to D′D3 Domains.
The assembly of tubules depends on pH and Ca2+ sensing by the D1D2 propeptide. At neutral pH, without Ca2+, D1D2 is monomeric. Low pH or Ca2+ induces weak self-association, and the combination of pH 6.2 with Ca2+ yields ≈160-kDa D1D2 homodimers (Fig. 3A–D and SI Table 1). In contrast to D1D2, low pH and Ca2+ does not induce self-association of D′D3 dimer or monomer (Fig. 3 E–G and SI Table 1), and a mixture of D1D2 and D′D3 monomer does not form complexes larger than ≈217 kDa (Fig. 3H). Therefore, the assembly of helical tubules involves Ca2+-dependent contacts between D1D2 and D′D3 and requires covalent dimerization of D′D3.
Fig. 3.
Noncovalent D1D2 dimerization and D1D2-D′D3 monomer heterodimerization require Ca2+ and depend on pH. Proteins were dialyzed against 20 mM Hepes, pH 7.4, or 20 mM Mes, pH 6.2, containing 150 mM NaCl, and 10 mM CaCl2 or 5 mM EDTA, as indicated, before chromatography on Superdex 200. (A) pH 7.4 with EDTA, ≈87-kDa D1D2 monomer. (B) pH 7.4 with Ca2+, D1D2 associates weakly. (C) pH 6.2 with EDTA, D1D2 associates weakly. (D) pH 6.2 with Ca2+, ≈160-kDa D1D2 homodimer. (E) pH 7.4 with EDTA, ≈114 kDa D′D3 dimer. (F) pH 6.2 with Ca2+, D′D3 dimer does not self-associate. (G) pH 6.2 with Ca2+, ≈57-kDa D′D3 does not self-associate. (H) pH 6.2 with Ca2+, mixing D1D2 with D′D3 monomer causes some D′D3 monomer to elute with D1D2 at 60.3 ml. (Inset) SDS/PAGE suggests the species eluting at 60.3 ml contains two D1D2 and one D′D3 monomers (217 kDa total).
Three-Dimensional Reconstruction of VWF Tubules.
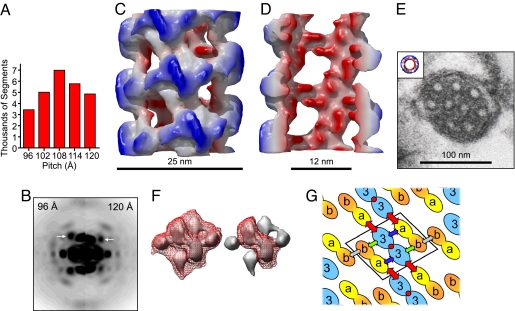
We used the iterative helical real space reconstruction (IHRSR) method (20, 21) to reconstruct VWF tubules from negative-stained EM images. The IHRSR approach is especially powerful when applied to helical filaments that are flexible and heterogeneous in axial rise, which are features of VWF tubules. We extracted 26,131 overlapping tubule segments (each 290 Å in length) and combined the power spectrum from each image. The resulting average power spectrum showed four layer lines consistent with ≈4.2 units per turn and a pitch of ≈105 Å. This finding agreed with the appearance of flat rings composed of D1D2-D′D3 by negative stain (Fig. 2D) and QFDE EM (Fig. 2F), which show approximately fourfold rotational symmetry. However, the broad n = 1 layer line on the averaged power spectrum indicated heterogeneity in pitch. We therefore used reference helices with different pitches to sort the segments into five bins (Fig. 4A). The separate power spectra for segments classified as ≈96 Å or ≈120 Å in pitch gave the expected shifts in layer line spacing (Fig. 4B), indicating that sorting into bins accurately reflected the variability in the data. The layer line corresponding to the 1-start helix remained rather broad, indicating that the bins are still somewhat heterogeneous in pitch.
Fig. 4.
Reconstruction of VWF tubules. (A) Distribution of segments from negative-stain images classified into bins of differing pitch. (B) Power spectra for the 96-Å bin and 120-Å bin differ in the position of the 1-start helix layer line (arrows). (C) Surface of the reconstruction colored red to blue based on distance from the central axis. The outside diameter is ≈25 nm. (D) Cutaway view. The cavity diameter is ≈12 nm. (E) Transmission EM of a cross-section of a Weibel–Palade body compared with the reconstruction at the same scale. (F) One repeating unit (Left) contoured at the same level as in C (red mesh) occupies ≈340 nm3. The central D′D3 dimer (Right) occupies ≈140 nm3. (G) Packing of repeating units composed of disulfide-linked D′D3 polypeptides (labeled with “3”s), and two D1D2 propeptides with D domains (labeled with “a”s and “b”s). In one arrangement, D1 = D“a” and D2 = D“b.” Alternatively, D2 = D“a” and D1 = D“b.” Arrows indicate interdomain contacts: blue, Da-D′D3 within a helical turn; green, Db-D′D3 within a helical turn; gray, Db-Db within a helical turn; red, Da-D′D3 between successive turns. Red circles indicate the local symmetry axis in the D′D3 dimer.
VWF tubules were reconstructed for the three bins of segments with a mean pitch of ≈102 Å (containing 19% of the segments), ≈114 Å (containing 22% of the segments), and ≈108 Å (the central bin containing 27% of the segments). The hand of the reconstruction was determined by using the QFDE results (Fig. 2H), which showed that the 1-start helix was right-handed. In the absence of such information, reconstructions from either negatively stained samples or cryo-EM suffer from an enantiomorphic ambiguity. By using the 0.5 Fourier shell correlation criterion, the resolution of each reconstruction is ≈22 Å. The main limitation on resolution appears to be the continuous variability of pitch.
The reconstruction from the central bin (Fig. 4C) shows that tubules consist of a right-handed 1-start helix with an axial rise of 26.2 Å and twist of 85.6° per subunit, corresponding to ≈4.2 subunits per turn of a 110-Å helix. A view of the hollow interior (Fig. 4D) emphasizes the open mesh-like arrangement of subunits. Reconstructions from other bins had a similar surface appearance, diameter, and twist (0.2% difference) but a different axial rise (SI Fig. 9). Other dimensions of the reconstruction include an outside diameter of ≈25 nm, inside diameter of ≈12 nm, and wall thickness of ≈6.5 nm.
The dimensions of the reconstruction are similar to the dimensions of VWF tubules in Weibel–Palade bodies of human endothelial cells (1) and in the α-granules of platelets (4). For example, across several transmission EM images of Weibel–Palade bodies (Fig. 4E), the tubules have an outside diameter of 24 ± 2.5 nm, inside diameter of 10.1 ± 0.9 nm, and wall thickness of 6.8 ± 1.3 nm (n = 12). A recent study by electron tomography shows that VWF tubules in Weibel–Palade bodies have an inside diameter of 12 ± 2 nm. Interestingly, tubules also display a subtle helical twist over a length scale of ≈100–200 nm (22). In addition, longitudinal sections of tubules within immature Weibel–Palade bodies sometimes display regular cross striations with a spacing of ≈14 nm (22, 23). It is possible that these cross striations correspond to the ≈11-nm helical turns of our three-dimensional reconstruction (Fig. 4C).
One repeating unit of the reconstruction can be contained within the volume of ≈340 nm3 (1.23 nm3/kDa) (24) calculated for an ≈280-kDa complex of one D′D3 dimer and two D1D2 polypeptides. The ≈114-kDa D′D3 dimer (≈140 nm3) corresponds to the central density of the repeating unit (Fig. 4F). The smaller flanking densities therefore correspond to domains D1 and D2 of the ≈87-kDa D1D2 polypeptides (≈100 nm3).
The reconstruction is consistent with two possibilities for the location of the D1 and D2 domains (Fig. 4G), but both involve similar protein–protein interactions that could depend on Ca2+ concentration and pH. Each D1D2 polypeptide makes (at least) three contacts with D′D3 and one end-to-end homotypic interaction, which is consistent with the fact that D1D2 alone forms homodimers and not larger complexes.
Discussion
Tubular Packing Implies a Model for Multimer Assembly.
The helical packing of VWF tubules suggests a mechanism for how changes in pH regulate the assembly and subsequent deployment of disulfide-linked VWF multimers (Fig. 5). Although the ER provides a good environment for disulfide bond formation, intersubunit disulfide bonds between the VWF D3 domains do not form in the ER. Based on the structure of the D1D2-D′D3 tubule (Fig. 4), a propeptide dimer could adopt one of two arrangements in the context of a proVWF dimer, depending on whether the homotypic interaction between propeptides involves the D1 domains (Fig. 5A) or D2 domains (Fig. 5B). Either pattern physically separates the D3 domains and could prevent inappropriate disulfide bond formation in the ER.
Fig. 5.
Model for the assembly, tubular packing, and secretion of VWF multimers. (A and B) In the ER, proVWF subunits dimerize through disulfide bonds between C-terminal CK domains (small gray spheres). The propeptide consists of domains D1 (“1,” yellow) and D2 (“2,” orange) and is linked to D′D3 of the mature subunit (“3,” blue) by a furin cleavage site (green). Two potential orientations of the propeptide are distinguished by whether homodimer contacts involve domain D1 (A) or D2 (B). In either case, pH-dependent interactions bring two D′D3 domains together and facilitate intersubunit disulfide bond formation. (C) The propeptide (yellow) and dimeric D′D3 domains (blue) pack into a helix surrounded by the C-terminal remainder of the VWF subunits (gray). (D) Secretion exposes the tubules to blood at pH 7.4, which destabilizes the contacts between propeptide and D′D3 domains and allows extension of the multimer.
In contrast to the ER, the Golgi is unfavorable for disulfide chemistry because it lacks disulfide oxidoreductases and the acidic pH inhibits the deprotonation of thiols. However, the low pH and high Ca2+ concentration (9, 13) also increase the affinity of binding between D1D2 and D′D3, which would facilitate intersubunit disulfide bond formation by juxtaposing two D3 domains (Fig. 5 A and B). The D1D2 propeptide and D′D3 domains pack into a helical tubule (Fig. 5C), and the remaining ≈200-kDa C-terminal region of the mature VWF subunits presumably decorate the outside and comprise the matrix that surrounds the denser tubules within Weibel–Palade bodies (Fig. 4E).
The storage of VWF in Weibel–Palade bodies resembles the reversible concentration of cargo proteins within the secretory granules of other cells, particularly in neuroendocrine tissues (25, 26). Protein self-aggregation in the trans-Golgi network is also the initial step in the biogenesis of these granules.
Within the Weibel–Palade body, a network of Ca2+-dependent and pH-dependent contacts between the D1D2 propeptide and dimeric D′D3 domains maintains the tubular packing of VWF multimers, which may be thought of as constrained springs (13). Upon secretion, the increase in pH weakens these constraints and permits the helical tubules to unfurl into the flowing blood without tangling (Fig. 5D).
Tubule Length and VWF Multimer Length.
Weibel–Palade bodies are 1–5 μm long (1), and tubules usually extend their entire length (22, 23). The helical packing of the dimeric D′D3 domains, with 4.2 repeating units per 11-nm turn, suggests that every 1 μm of tubule length might correspond to a disulfide-linked multimer of ≈780 VWF subunits (2 subunits per repeating unit), having a mass of ≈195 million Da and an extended length of ≈47 μm (≈60 nm per subunit) (27). The largest plasma VWF multimers observed by gel electrophoresis or EM are perhaps one-tenth this size (27), but filaments of VWF secreted acutely by endothelial cells in culture or in vivo are often several hundred micrometers long (13, 28, 29). The extreme length of these structures suggests that one VWF tubule may consist of one VWF multimer.
VWF Tubules and the Assembly of Homologous Multimeric Proteins.
Weibel–Palade bodies are found in endothelial cells of all vertebrates examined, including hagfish (30), which suggests that the helical packing of VWF is conserved and critical for hemostasis. Other proteins also exploit pH-dependent interactions between VWF D domains for similar ends. For example, vertebrates have a family gel-forming mucins that make disulfide-linked dimers between C-terminal CK domains in the ER, and disulfide-linked multimers between N-terminal D1D2D′D3 domains in the Golgi (31). The helical packing of VWF D1D2 and D′D3 proteins provides a framework to investigate the biogenesis of Weibel–Palade bodies and the assembly of homologous multimeric proteins.
Materials and Methods
Constructs and Cell Culture.
Plasmids encoding human VWF fragments (Fig. 1) were derived from pSVHVWF1.1 (32). Plasmid pSVH-D1D2D′D3-His encodes the VWF signal peptide (codons 1–22) and domains D1D2D′D3 (codons 23–1241). Plasmid pSVH-D1D2-His encodes the signal peptide and domains D1D2 (codons 1–745). A His × 6 tag was inserted at the C-termini of both constructs. Plasmid pSVH-D′D3A1-His encodes the signal peptide (codons 1–22), domains D′D3A1 (codons 764-1464), and a C-terminal His × 7 tag. Plasmid pSVH-D1D2-wt encodes the signal peptide and domains D1D2 (codons 1–763).
BHK cells were grown in DMEM (Invitrogen) with heat-inactivated FBS and 2 mM glutamine. BHK-fur4 cells that express human furin (33) were used to produce D′D3 monomer and dimer. Plasmids pRSV-neo and pSVH-D1D2D′D3-His or pSVH-D1D2-His were cotransfected to produce D′D3 monomer and dimer or D1D2, respectively. Three plasmids, pRSV-neo, pSVH-D′D3A1-His, and pSVH-D1D2-wt, were cotransfected to produce D′D3A1 monomer and dimer. Cells were transfected by using LipofectAMINE PLUS (Invitrogen) and selected with 700 μg/ml Geneticin (G418).
Protein Expression and Purification.
Cells were cultured in FreeStyle 293 Expression Medium (Invitrogen) in roller bottles. Media were filtered and applied to a HiTrap Q column (5 ml; GE Healthcare) in 20 mM Hepes (pH 7.4). Proteins were eluted with a 0–1 M NaCl gradient over 60 min. Imidazole (20 mM) was added to pooled fractions, which were applied to a HisTrap column (5 ml; GE Healthcare) in 20 mM Hepes (pH 7.4), 20 mM imidazole, and 500 mM NaCl. Proteins were eluted with 20 mM Hepes (pH 7.4), 500 mM imidazole, 500 mM NaCl, and chromatographed on Superdex 200 (16/60; GE Healthcare) in buffers indicated in the text. Column fractions were analyzed by 4–15% gradient SDS/PAGE (Bio-Rad) and staining with Coomassie blue or by Western blotting with HRP-conjugated rabbit anti-human VWF (Dako) and development with ECL-Plus (GE Healthcare).
Laser Light Scattering Determination of Protein Masses and Hydrodynamic Radii.
Laser light scattering analyses (34) were performed at the Biophysics Resource of the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. Samples were chromatographed on a Superdex 200 column (10/30; GE Healthcare) with detectors for 18-angle static laser light scattering (DAWN-EOS; Wyatt Technology), dynamic light scattering (QELS; Wyatt Technology), refractive index (OPTILAB rEX; Wyatt Technology), and UV absorbance. Protein concentration was determined by refractive index and confirmed by UV absorbance (280 nm). Molecular masses were calculated by using ASTRA software (Wyatt Technology) to solve the equation relating scattered light at several angles, concentration, and weight-average molar mass by using the Zimm method (34). The method is accurate to ≈3%, based on analysis of standard proteins from 6.5 to 476 kDa. Diffusion coefficients (DT) were determined by dynamic light scattering. Hydrodynamic radii (Rh) were calculated from values for DT by using the Stokes–Einstein equation, Rh = kbT/6πηDT, where kb is Boltzmann's constant, T is temperature, and η is solvent viscosity. For some species, Rh was determined by gel filtration chromatography on columns of Superdex 200 (16/60; GE Healthcare) or Superose 6 (10/300; GE Healthcare) calibrated with standard proteins of known Rh (SI Fig. 10) (35).
Electron Microscopy.
Deep-etch replicas were prepared by adsorbing proteins on mica flakes in buffer at pH 7.4 (20 mM Hepes, pH 7.4/150 mM NaCl/10 mM CaCl2) or pH 6.2 (20 mM Mes, pH 6.2/150 mM NaCl/10 mM CaCl2) and then quick-freezing, freeze-fracturing, deep-etching, and platinum-replicating the mica flakes (19, 36). Alternatively, proteins were adsorbed on acid-cleaned glass chips and fixed with 2% glutaraldehyde before quick-freezing and freeze-drying directly on glass.
For thin-section EM, human umbilical vein endothelial cells were grown on Thermanox plastic coverslips, fixed in 30 mM Hepes (pH 7.4), 100 mM NaCl, 2 mM CaCl2, 2% glutaraldehyde, postfixed with 0.5% OsO4, and block stained with 1% uranyl acetate. Samples were dehydrated in alcohols, infiltrated with Araldite epoxy resin, inverted onto BEEM capsules filled with Araldite, and cured at 70°C overnight. Coverslips were removed, and blocks were sectioned at 100- to 200-nm thickness with a Porter-Blum MT-1 ultramicrotome. Sections were picked up on formvar/carbon-coated 400 mesh copper grids and stained with 4% uranyl acetate in water followed by 0.3% lead citrate in 0.1 M NaOH before examination at 80 kV in a JEOL 200CX electron microscope.
Samples for negative-stain EM were applied to lightly glow-discharged carbon-coated grids and stained with 2% (wt/vol) uranyl acetate. Grids were examined in a Tecnai-12 electron microscope (FEI) under minimal-dose conditions at an accelerating voltage of 80 kV and a nominal magnification of ×30,000. Negatives were scanned with a Nikon Coolscan 8000 as 16-bit images with a raster of 4.2 Å per pixel.
EM Three-Dimensional Reconstruction.
The SPIDER software package (37) was used for image processing. Initially, all of the segments were used for a multiple-reference alignment against reference volumes with five different pitches (from 96–120 Å), which sorted 26,131 segments into five bins. The central bin is the largest group, with 6,296 overlapping boxes (each 290 Å long) from 49 images, and was selected for the three-dimensional reconstruction. Additional reconstructions (SI Fig. 9) were generated from the two surrounding bins (n = 5,024 and n = 5,796, respectively). Molecular graphics images were produced by using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081) (38).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Daved Fremont (Washington University) for advice on dynamic light scattering measurements, Ewa Folta-Stogniew (Keck Foundation, Yale University, New Haven, CT) for size-exclusion chromatography/laser light-scattering analyses, and Elodee A. Tuley and Milan Kapadia for preparing recombinant proteins. This study was supported in part by National Institutes of Health Grants HL72917 (to J.E.S.), EB001567 (to E.H.E.), and GM029647 (to J.E.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710079105/DC1.
References
- 1.Weibel ER, Palade GE. J Cell Biol. 1964;23:101–112. doi: 10.1083/jcb.23.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner DD, Olmsted JB, Marder VJ. J Cell Biol. 1982;95:355–360. doi: 10.1083/jcb.95.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewenstein BM, Warhol MJ, Handin RI, Pober JS. J Cell Biol. 1987;104:1423–1433. doi: 10.1083/jcb.104.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cramer EM, Meyer D, le Menn R, Breton-Gorius J. Blood. 1985;66:710–713. [PubMed] [Google Scholar]
- 5.Sadler JE. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 6.Wagner DD, Marder VJ. J Cell Biol. 1984;99:2123–2130. doi: 10.1083/jcb.99.6.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voorberg J, Fontijn R, Calafat J, Janssen H, van Mourik JA, Pannekoek H. J Cell Biol. 1991;113:195–205. doi: 10.1083/jcb.113.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsumi A, Tuley EA, Bodó I, Sadler JE. J Biol Chem. 2000;275:25585–25594. doi: 10.1074/jbc.M002654200. [DOI] [PubMed] [Google Scholar]
- 9.Vischer UM, Wagner DD. Blood. 1994;83:3536–3544. [PubMed] [Google Scholar]
- 10.Denis CV, Andre P, Saffaripour S, Wagner DD. Proc Natl Acad Sci USA. 2001;98:4072–4077. doi: 10.1073/pnas.061307098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haberichter SL, Merricks EP, Fahs SA, Christopherson PA, Nichols TC, Montgomery RR. Blood. 2005;105:145–152. doi: 10.1182/blood-2004-02-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner DD, Saffaripour S, Bonfanti R, Sadler JE, Cramer EM, Chapman B, Mayadas TN. Cell. 1991;64:403–413. doi: 10.1016/0092-8674(91)90648-i. [DOI] [PubMed] [Google Scholar]
- 13.Michaux G, Abbitt KB, Collinson LM, Haberichter SL, Norman KE, Cutler DF. Dev Cell. 2006;10:223–232. doi: 10.1016/j.devcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Wagner DD, Mayadas T, Urban-Pickering M, Lewis BH, Marder VJ. J Cell Biol. 1985;101:112–120. doi: 10.1083/jcb.101.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner DD, Mayadas T, Marder VJ. J Cell Biol. 1986;102:1320–1324. doi: 10.1083/jcb.102.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purvis AR, Sadler JE. J Biol Chem. 2004;279:49982–49988. doi: 10.1074/jbc.M408727200. [DOI] [PubMed] [Google Scholar]
- 17.Voorberg J, Fontijn R, Calafat J, Janssen H, van Mourik JA, Pannekoek H. EMBO J. 1993;12:749–758. doi: 10.1002/j.1460-2075.1993.tb05709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purvis AR, Gross J, Dang LT, Huang R-H, Kapadia M, Townsend RR, Sadler JE. Proc Natl Acad Sci USA. 2007;104:15647–15652. doi: 10.1073/pnas.0705175104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuser JE. J Mol Biol. 1983;169:155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- 20.Egelman EH. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 21.Egelman EH. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Valentijn KM, Valentijn JA, Jansen KA, Koster AJ. J Struct Biol. 2007 doi: 10.1016/j.jsb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Zenner HL, Collinson LM, Michaux G, Cutler DF. J Cell Sci. 2007;120:2117–2125. doi: 10.1242/jcs.007781. [DOI] [PubMed] [Google Scholar]
- 24.Fischer H, Polikarpov I, Craievich AF. Protein Sci. 2004;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arvan P, Zhang BY, Feng L, Liu M, Kuliawat R. Curr Opin Cell Biol. 2002;14:448–453. doi: 10.1016/s0955-0674(02)00344-7. [DOI] [PubMed] [Google Scholar]
- 26.Sobota JA, Ferraro F, Back N, Eipper BA, Mains RE. Mol Biol Cell. 2006;17:5038–5052. doi: 10.1091/mbc.E06-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fowler WE, Fretto LJ, Hamilton KK, Erickson HP, McKee PA. J Clin Invest. 1985;76:1491–1500. doi: 10.1172/JCI112129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong JF, Moake JL, Nolasco L, Bernardo A, Arceneaux W, Shrimpton CN, Schade AJ, McIntire LV, Fujikawa K, López JA. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 29.Motto DG, Chauhan AK, Zhu G, Homeister J, Lamb CB, Desch KC, Zhang W, Tsai HM, Wagner DD, Ginsburg D. J Clin Invest. 2005;115:2752–2761. doi: 10.1172/JCI26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yano K, Gale D, Massberg S, Cheruvu PK, Monahan-Earley R, Morgan ES, Haig D, von Andrian UH, Dvorak AM, Aird WC. Blood. 2007;109:613–615. doi: 10.1182/blood-2006-05-026401. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Vilar J. Am J Respir Cell Mol Biol. 2007;36:183–190. doi: 10.1165/rcmb.2006-0291TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsushita T, Sadler JE. J Biol Chem. 1995;270:13406–13414. doi: 10.1074/jbc.270.22.13406. [DOI] [PubMed] [Google Scholar]
- 33.Bodó I, Katsumi A, Tuley EA, Eikenboom JC, Dong Z, Sadler JE. Blood. 2001;98:2973–2979. doi: 10.1182/blood.v98.10.2973. [DOI] [PubMed] [Google Scholar]
- 34.Folta-Stogniew E. Methods Mol Biol. 2006;328:97–112. doi: 10.1385/1-59745-026-X:97. [DOI] [PubMed] [Google Scholar]
- 35.Ackers GK. J Biol Chem. 1967;242:3237–3238. [PubMed] [Google Scholar]
- 36.Heuser J. J Electron Microsc Tech. 1989;13:244–263. doi: 10.1002/jemt.1060130310. [DOI] [PubMed] [Google Scholar]
- 37.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.