Abstract
Targeted gene silencing by RNAi requires the RNA-induced silencing complex (RISC), whose core component is the protein Argonaute (Ago) bound to a microRNA (miRNA) or an siRNA. In humans, Ago2 is loaded with miRNAs by the action of a specialized assembly called the RISC-loading complex (RLC), comprising the proteins Ago2, Dicer, and TRBP. Here we show that the human RLC assembles spontaneously in vitro from purified components. No cofactors or chaperones are required for the complex to form. The reconstituted RLC, containing one copy of each protein, has the dicing, slicing, guide-strand selection, and Ago2-loading activities observed for the endogenous RLC. Furthermore, once Ago2 is loaded with an miRNA, it tends to dissociate from the rest of the complex. These results lay the groundwork for future structural and functional dissection of RISC loading in humans.
Keywords: Ago2, Dicer, RNAi
RNAi and related RNA-silencing pathways are widespread eukaryotic mechanisms of gene regulation. All RNA-silencing processes are carried out by large ribonucleoprotein assemblies, generically termed RNA-induced silencing complexes (RISCs) (reviewed in ref. 1). RISCs reduce expression of target genes through transcriptional, posttranscriptional, and translational regulatory mechanisms, although in at least one case components of RISC can apparently activate translation as well (2). At its functional core, every RISC contains a member of the Argonaute (Ago) family of proteins. Ago proteins bind small RNAs, 19–30 nt in length, which are used to guide the RISC to cognate RNA transcripts through base-pairing interactions.
The human genome encodes four Ago proteins and four Ago-related proteins called Piwi (3). Thus, there are potentially at least eight distinct types of RISC activity in humans. Furthermore, numerous Ago-associated proteins have been identified, raising the possibility of a high degree of combinatorial complexity in RISC composition and function. The list of Ago-associated proteins identified thus far includes helicases (4–7), nucleases (3, 8), and RNA-binding proteins (4, 8–11). However, the exact role for many Ago-associated proteins in RISC function has not yet been established, and the molecular composition of RISC has been determined in only a few instances.
One recent report isolated a stable functional human RISC composed of the three proteins Ago2, Dicer, and TRBP by immunopurification and size-exclusion chromatography (12). This complex was shown to cleave target RNA by using a precursor microRNA (pre-miRNA) hairpin as the source of guide RNA. A human complex isolated under similar conditions displayed the same activities and was renamed the RISC-loading complex (RLC) because of its ability to cleave input pre-miRNA and to selectively load a guide miRNA into Ago2 (13). This activity is functionally similar to RLC activity observed in Drosophila embryo extracts (6).
To date, all studies of large RISC assemblies have been carried out by using either crude cell extracts or immunopurified endogenous proteins. Here we show that it is possible to reconstitute the human RLC in vitro entirely from recombinant components. We find that the reconstituted complex has similar, but not identical, biochemical properties to the endogenous complex.
Results
The Human RLC Assembles Spontaneously in Vitro.
The human RLC has been isolated from HEK293-derived cell lines expressing FLAG-tagged Dicer by immunopurification (12, 13). In each case, FLAG-Dicer coeluted with the proteins TRBP and Ago2, suggesting that this macromolecular complex is biologically relevant. We wondered whether the RLC assembly is an intrinsic property of these three proteins or whether other cellular factors, such as the Ago2-associated protein Hsp90 (14), are required to form the complex. To establish a biochemical system for functional and structural studies of the human RLC, we attempted to reconstitute the complex in vitro from recombinant components. Initial attempts to assemble human RISC by coexpression of Dicer, Ago2, and TRBP in triply baculovirus-infected insect (Sf9) cells resulted in low yields of the trimeric complex. To increase production, Dicer and Ago2 were each expressed separately in Sf9 cells by using the baculovirus system. TRBP was expressed in Sf9 cells or as a maltose-binding protein fusion in Escherichia coli. The individual proteins were purified separately and then mixed together and applied to a Superose 6 size-exclusion column.
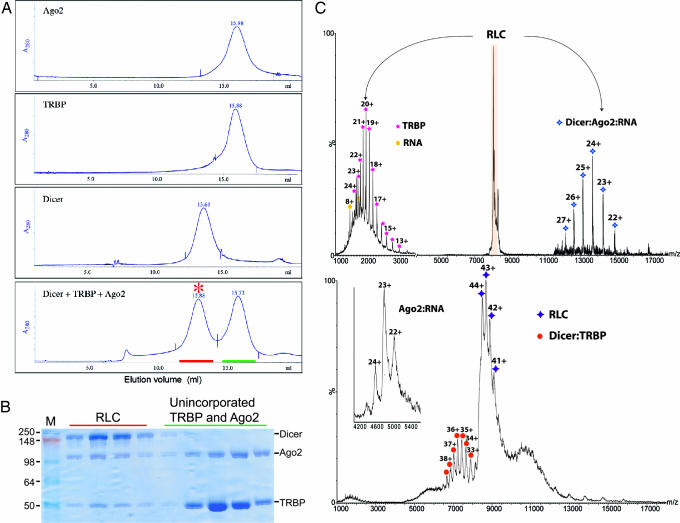
Size-exclusion chromatography indicated that human Dicer, TRBP, and Ago2 assemble into a stable complex rapidly and spontaneously in vitro. When Ago2 is preincubated with Dicer and TRBP, it elutes significantly earlier from the Superose 6 column than does Ago2 alone, demonstrating that Ago2 is assembled into a larger complex in the presence of the other two proteins (Fig. 1A). Likewise, TRBP, which exists as a dimer (15), elutes earlier from the Superose 6 column when preincubated with Dicer and Ago2. Dicer, which is monomeric (16), runs anomalously fast through size-exclusion columns, eluting at approximately the same volume as the 2-fold larger protein ferritin (440 kDa) (10). Therefore, it was not possible to separate the reconstituted RLC from free Dicer. To circumvent this problem, reconstitution was performed with a 2- to 3-fold molar excess of TRBP and Ago2 over Dicer. SDS/PAGE analysis of Superose 6 fractions from the reconstitution indicates that all three proteins are present in a peak that elutes with an apparent molecular mass of ≈500 kDa (Fig. 1B).
Fig. 1.
In vitro assembly of the human RLC. (A) Superose 6 size-exclusion chromatography elution profiles are shown for individual samples of human Dicer, Ago2, and TRBP or a preincubated mixture of the three proteins containing an ≈2-fold molar excess of Ago2 and TRBP. The chromatogram is labeled to indicate RLC (red asterisk), fractions of the RLC peak (red line), and fractions containing unincorporated Ago2 and TRBP (green line). (B) SDS/PAGE gel analysis of fractions from A and labeled as in A. In some preparations, we observed a contaminating band at ≈70 kDa that was identified as insect Hsp70-4 by MS. (C) Electrospray MS of the intact RLC complex. (Lower) Mass spectrum of the intact heterotrimeric RLC complex (purple star) confirms the presence of Dicer, Ago2, and TRBP subunits with unit stoichiometry. The charge-state series allows the molecular mass to be measured as 371.129 ± 95 Da, indicating the presence of an additional component with a molecular mass 11 kDa higher than expected. (Inset) This larger mass is likely due to an RNA bound to Ago 2, because the Ago 2 alone has a molecular mass of 109.950 ± 53 Da. At m/z ≈7,500, the Dicer:TRBP (orange circle) heterodimer also was observed (molecular mass of 259.212 ± 62 Da), indicating its formation in solution. (Upper) MS/MS analysis confirms that the RLC complex is composed of TRBP (pink star), an 11-kDa RNA fragment (yellow circle), Dicer, and Ago2 (blue star) subunits. The different heterodimers formed in these experiments are rationalized because the Dicer:TRBP complex forms in solution, whereas in the gas-phase loss of the smallest subunit, TRBP is favored, giving rise to a stripped complex Dicer:Ago2 (25).
To determine the Dicer:Ago2:TRBP stoichiometry in the reconstituted RLC, we subjected samples to nondenaturing electrospray MS analysis. This analysis revealed that the native molecular mass of the complex is 371.129 kDa, which is slightly larger than the sum of the molecular masses of the individual proteins (355.056 kDa) [Fig. 1C Lower and supporting information (SI) Table 1]. However, in the low-m/z region, we also observed the presence of an 11-kDa species that was tightly associated with Ago2 in isolation (Fig. 1C Inset). We suspected the 11-kDa species to be a contaminating small RNA that is bound to the Ago2 subunit of the complex. Consistent with this idea, Ago2 preparations had an A260/A280 absorbance ratio of ≈1.3, which indicated the presence of contaminating nucleic acids. Indeed, ethidium bromide-stained denaturing PAGE revealed that the Ago2 preparations are contaminated with RNase-sensitive nucleic acids, including a band at ≈11 kDa (data not shown). The identity of the 11-kDa RNA and its effects on RLC assembly function have not yet been determined. Accounting for the extra mass from the contaminating RNA results in a calculated mass of 366.898 kDa for the RLC; the extra 1.15% value of the observed mass was likely because of retained water molecules and buffer ions under the soft ionization conditions used, as evidenced by the broadening of the peaks. This value agrees well with previous calculated-to-observed mass ratios obtained by using a similar approach (17) human Dicer, TRBP, and Ago2 self-assemble into a single stable molecular complex in vitro. The three proteins are present in the complex at a 1:1:1 ratio, although the isolated TRBP component exists as a dimer in solution (15), suggesting that RLC assembly requires dissociation of the TRBP dimer. Also, the native molecular mas of the complex is significantly less than that estimated by size-exclusion chromatography, indicating that the complex takes on an irregular, nonspherical shape.
The Reconstituted RLC Is Catalytically Active.
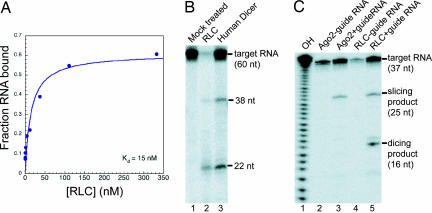
We wondered whether the reconstituted human RLC has the same biochemical activities as the endogenous RLC isolated by immunoprecipitation. The first known function of the human RLC is to recognize and bind to pre-miRNA hairpins. Filter-binding experiments demonstrated that the reconstituted RLC has a high affinity for Drosophila pre-let7 (Dmpre-let7) RNA hairpin, binding with a dissociation constant (Kd)of 15 nM (Fig. 2A). We do not know how the contaminating 11-kDa RNA species present in RLC preparations influences the measured Kd for the pre-let7A, but if there is an effect, it is likely to be competitive. Therefore, 15 nM may be an underestimate of the true Kd of the RLC for pre-let7A RNA hairpins.
Fig. 2.
RNA-binding and catalytic activity of the reconstituted RLC. (A) Nitrocellulose filter-binding assay: ≈1 nM radiolabeled Dmpre-let7 hairpin RNA was incubated with indicated concentrations of RLC, yielding a measured Kd of 15 nM. (B) Dicing assay: 200 nM RLC or 200 nM human Dicer (as a positive control) was incubated with ≈10 nM 5′ end 32P-labeled Dmpre-let7 RNA. (C) Slicing assay: 200 nM RLC or 200 nM human Ago2 (as a positive control) was incubated with 250 nM guide RNA and then was added to ≈10 nM 5′ end 32P-labeled 37-nt A RNA annealed with 37-nt B. Slicing product, 25 nt; dicing product, 16 nt.
The second function of the RLC is to process pre-miRNA into a miRNA duplex by the catalytic activity of the Dicer subunit. Subjecting the reconstituted RLC to a dicing assay by using 5′ 32P-radiolabled Dmpre-let7 RNA hairpin demonstrated that Dicer activity is retained in the complex (Fig. 2B). Moreover, the size of the miRNA produced by the RLC (lane 2) is the same as that generated by Dicer alone (lane 3), indicating that the presence of the other two proteins does not influence cleavage site selection by Dicer. We noted that RNA samples incubated with the RLC showed more degradation than those incubated with Dicer alone. This observation may indicate that the RLC has a nonspecific RNase activity or, more likely, that there is a low level of contaminating RNase in our RLC preparation.
The third established activity of the RLC is the ability to cleave or slice an RNA substrate that is base-paired to the miRNA in the complex. It was previously shown that Ago2 alone can be programmed for targeted RNA cleavage by preincubation with a single-stranded guide RNA (18). To test for slicer activity in the reconstituted RLC, samples were subjected to a two-step slicing assay. The 200 nM RLC was first incubated with a 200 nM single-stranded 21-nt guide RNA, which allows the RNA to be bound by the Ago2 subunit of the complex. After loading the complex with the guide RNA, 5′ end 32P-radiolabeled target RNA (37-nt A, 10 nM) was introduced into the reaction. The single-stranded target RNA contains a 21-nt sequence complementary to the guide RNA, which should be recognized and cleaved by the endonuclease activity in Ago2. The expected cleavage removed 12 nt from the 3′ end of the target RNA (Fig. 2C). As a positive control, 200 nM recombinant Ago2 was loaded with the same guide RNA and incubated with the radiolabeled target RNA. These results show that the recombinant RLC possesses guide RNA-dependent slicing activity on a targeted RNA. RLC samples generate a distinct slicing product that is identical in size to the product made by Ago2 alone. In the absence of a guide RNA, there is no position-specific endonucleolytic cleavage of the target by the RLC, although there is nonspecific target RNA degradation. Lower levels of nonspecific degradation in reactions containing the guide RNA (Fig. 2C, lanes 3 and 5) are likely due to a protective effect of unincorporated, unlabeled guide RNA.
Intriguingly, we saw an additional smaller product in the RLC reaction that is not present in the reaction with Ago2 alone (Fig. 2C, compare lanes 3 and 5). The size of this product is consistent with a cleavage 21 nt from the 5′ end of the target RNA, suggesting that it is generated by Dicer. We suspect that this cleavage is an in vitro artifact, which is the result of Dicer recognizing a dsRNA region formed by excess guide strand annealing with some of the target RNA.
Reconstituted RLC Displays RISC-Loading Activity.
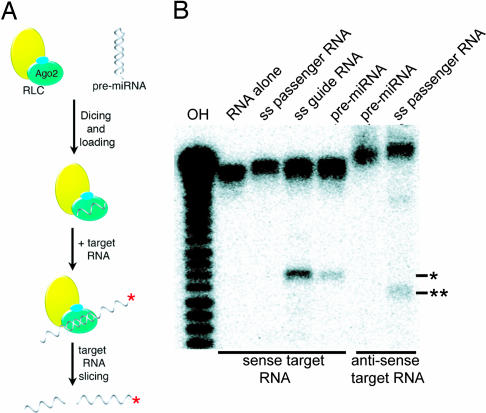
Upon finding that the reconstituted RLC retains the dicing and slicing nucleolytic activities of its respective Dicer and Ago2 subunits, we wondered whether the reconstituted complex is competent for RISC loading. The process of RISC loading involves several RNA-processing steps, including pre-miRNA recognition, dicing, guide RNA selection, Ago2 loading, and passenger strand removal (Fig. 3A). To test for RISC-loading activity, the reconstituted RLC was subjected to a two-step in vitro assay. In the first step, ≈200 nM RLC was incubated with 200 nM Dmpre-let7 RNA hairpin. During this first incubation, the complex is expected to process the pre-let7A hairpin and load the mature let7 guide RNA into Ago2. The pre-let7A duplex intermediate is thermodynamically asymmetric so that only one strand in the duplex should be loaded into the guide position of Ago2 (19, 20). In the second step of the assay, radiolabeled target RNA (37-nt A, 10 nM) was added, and slicing activity was measured as described above.
Fig. 3.
RISC-loading activity of the reconstituted RLC. (A) Overview of the RISC-loading process, involving pre-miRNA recognition, dicing, passenger strand removal, and guide strand RNA loading onto Ago2, as well as target RNA slicing. The red asterisk indicates the 5′ end 32P-labeled RNA strand. (B) RISC-processing assays. Lanes indicate the addition of unlabeled single-stranded 21-nt guide RNA (ss guide RNA), single-stranded 21-nt passenger RNA (ss passenger RNA), or Dmpre-let7 RNA (pre-miRNA) to RLC and 5′ end 32P-labeled sense strand target RNA, 37 nt-A (first four lanes), or to 5′ end 32P-labeled antisense target RNA, 37-nt B (last two lanes). Asterisks indicate the slicing product expected for the sense (∗) and antisense (∗∗) targets.
Results from the RISC-loading assay reveal that the reconstituted RLC is capable of RISC loading. Upon incubation with a target RNA, the pre-let7A-loaded complex displayed discrete slicing activity (Fig. 3B). To test for correct guide strand selection, we examined the slicing activity of an antisense target RNA (complementary to the passenger strand). Slicing of an antisense target RNA is diagnostic of Ago2 loading the incorrect RNA (passenger or let7*) strand. The pre-let7A-loaded RLC did not slice the antisense target, indicating that the recombinant RLC is capable of selecting the correct guide strand. As a positive control, RLC loaded with a single-stranded passenger RNA did slice the antisense target RNA. These results demonstrate that the reconstituted RLC can correctly process and load a pre-miRNA to generate an active RISC and show that the three proteins, Dicer, TRBP and Ago2, are sufficient for the RISC loading.
The RLC Disassembles After RISC Loading.
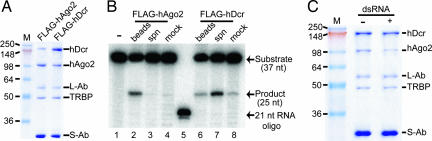
There is conflicting evidence regarding the fate of the RLC after Ago2 is loaded with a guide RNA. One report suggests that the complex remains intact and that the presence of Dicer and TRBP stimulates multiple rounds of substrate turnover by Ago2 (12). A second report shows that the human Ago2 disassociates from Dicer after Ago2 is loaded with a guide RNA (13). A third report observed an intact human RLC that was Ago2-loaded (9). In Drosophila extracts, Dicer-2 and R2D2, a fly ortholog of TRBP, stay associated with Ago2, which is subsequently incorporated into a large 80S particle called “holo-RISC” (21). To determine the fate of the reconstituted human RLC after Ago2 loading, we performed an RLC disassembly assay based on a previous study (13).
In the disassembly assay, the RLC was immobilized on an α-FLAG antibody resin through noncovalent interactions with FLAG-tagged Dicer (Fig. 4A). The bound complex was then incubated with a pre-let7A hairpin or single-stranded guide RNA to allow loading of Ago2. After incubation, the resin-bound material was pelleted by centrifugation, and the supernatant fluid was tested for slicing activity. In our hands, the reconstituted RLC was destabilized after Ago2 loading. The material released from the resin after incubation with RNA had clear slicing activity when FLAG-tagged hDicer was used (Fig. 4B, lane 7). Consistently, the slicing activity was mainly associated with the resin when FLAG-tagged hAgo was used (Fig. 4B, lane 2). Furthermore, SDS/PAGE analysis of boiled samples showed that >60% of Ago2 dissociated from the resin upon Ago2 loading (Fig. 4C and SI Fig. 5). This result supports some previous work with immunoprecipitated RLC samples and suggests that Dicer and TRBP do not participate directly in target RNA processing.
Fig. 4.
RNA-dependent disassembly of the human RLC complex. (A) Coomassie-stained SDS/PAGE gel showing reconstitution of the RLC complex by using FLAG-tagged Argonaute 2 (FLAG-hAgo2) or FLAG-tagged Dicer (FLAG-hDcr). Samples were assembled by incubation of RLC components as described in Fig. 1 and were isolated by using Agarose bead-coupled anti-FLAG antibodies. RLC components (hDcr, hAgo2, and TRBP) and antibodies (L-Ab and S-Ab) are labeled. (B) Phosphorimage of a denaturing polyacrylamide gel showing slicing activity of bead-bound (beads) and supernatant (spn) samples from A after incubation with 5′ end 32P-labeled 37-nt A target (substrate) RNA and unlabeled Dmpre-let hairpin RNA. Target and Dmpre-let RNAs incubated in buffer alone (mock) provided controls for nonspecific degradation. Mobilities of RNA substrate, 25-nt slicing product, and a 21-nt RNA marker oligo are indicated on the right. (C) Coomassie-stained SDS/PAGE gel analysis of FLAG-hDcr/hAgo2/TRBP samples prepared as in A after incubation in buffer alone (−) or buffer containing duplex RNA substrate 37-nt A and B (+). The buffer contains no Mg2+ but is supplemented with 5 mM EDTA.
Discussion
RNAi and related gene-silencing processes are invariably carried out by large and complex ribonucleoproteins (RNPs). To date, >15 Ago-associated proteins have been identified in humans (reviewed in ref. 22), raising the possibility that a variety of RISCs function with distinct biochemical activities in living cells. Although efforts to identify the different proteins involved in RNAi have been extensive, studies that characterize the exact biochemical function of any particular protein or complex, as well as structural studies of such samples, remain limited because most investigations have relied on either crude cell extracts or small amounts of purified endogenous complexes. Here we have shown that it is technically feasible to reassemble the minimal human RLC in vitro from recombinant components. The reconstituted complex has the ability to correctly recognize and process a pre-miRNA, load the correct guide miRNA into Ago2, and form a catalytically active RISC.
Interestingly, however, there is at least one subtle difference between our reconstituted RLC and the reported biochemical properties of the endogenous complex. In our hands, the reconstituted RLC does not display significant coupling between Dicer activity and the formation of the active slicer (SI Fig. 6). This result contrasts with intact RLC samples prepared from HEK293 cells, in which pre-miRNA hairpins (that are cleaved by Dicer) have been shown to generate more slicer activity than the corresponding prediced miRNA duplexes (12, 13). For this reason, applications of RNAi by using exogenous microRNAs typically employ hairpin substrates that are processed by Dicer to yield the mature miRNA of interest. An important challenge is to determine the factors or modifications that are present in RLC samples derived from human cell lines that endow the complex with this activity. It also would be interesting to see how other known Ago2-associated proteins, including PACT, MOV10, TRNC7, and RNA helicase A, influence RISC and RISC-loading activities. The results presented here provide the groundwork for such studies, allow the future dissection of RLC biochemical activity, and pave the way for more rigorous structural investigations.
Materials and Methods
Synthetic RNA Oligos.
The following RNAs (synthesized by Dharmacon) are the RNA oligos used in this study: Dmpre-let7, 5′-AAUGAGGUAGUAGGUUGUAUAGUAGUAAUUACACAUCAUACUAUACAAUGUGCUAGCUUUCU-3′; 37-nt A, 5′-UGAGGUAGUAGGUUGUAUAGUUUGAAAGUUCACGAUU-3′, and its complementary partner 37-nt B, 5′-UCGUGAACUUUCAAACUAUACAACCUACUACCUCAUU-3′; 21-nt guide RNA, 5′-UGAGGUAGUAGGUUGUAUAGU-3′; and 21-nt passenger RNA, 5′-UAUACAAUGUGCUAGCUUUCU-3′.
Purification of Dicer, Ago2, and TRBP.
N-terminally His6-tagged human Dicer, Ago2, and TRBP were purified separately from Sf9 cells infected with baculovirus bearing the gene of the desired protein. For each protein, 1 × 109 cells grown in 1 liter of Excel 420 media were harvested 72 h after infection. Cells were pelleted gently by centrifugation, resuspended in 30 ml of ice-cold lysis buffer [300 mM NaCl, 0.5% Triton X-100, 5% glycerol, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), 10 mM imidazole, and 50 mM Na2HPO4 (pH 8.0)] containing an EDTA-free protease inhibitor mixture (Roche), and lysed with eight strokes of the B pestle of a 40-ml glass homogenizer (Fisher). Insoluble material was pelleted by 20,000 × g centrifugation for 15 min, and the supernatant solution was applied to 5 ml (packed) of Ni-NTA resin (Qiagen) in a 50-ml Falcon tube. After 20 min of gentle rocking, the resin was pelleted by brief centrifugation and washed by resuspending in 45 ml of wash buffer [300 mM NaCl, 5% glycerol, 0.5 mM TCEP, 20 mM imidazole, and 50 mM Na2HPO4 (pH 8.0)]. The resuspended resin was pelleted again and subjected to four more rounds of washing. Protein was eluted from the washed resin with 10 ml of elution buffer [300 mM NaCl, 5% glycerol, 0.5 mM TCEP, 300 mM imidazole, and 50 mM Na2HPO4 (pH 8.0)] and then was dialyzed against 1 liter of wash buffer overnight. The N-terminal His6-tag was removed by including 1 mg of tobacco etch virus (TEV) protease in the dialysis bag. Dialyzed protein was then applied to a 5-ml Ni-nitrilotriacetic acid (NTA) column. The unbound material was collected, concentrated to 1 ml, and applied to a Superdex 200 16/60 column (Amersham Pharmacia) equilibrated in gel filtration buffer [100 mM KCl, 5% glycerol, 1 mM DTT, and 20 mM Hepes (pH 7.5)]. Fractions containing nonaggregated protein were pooled, concentrated to 5–10 mg/ml, and used in subsequent reconstitution experiments. All purification steps were carried out at 4°C. Protein concentrations were determined by the Bradford assay (Bio-Rad).
RLC Reconstitution.
First, 705 μg (3 nmol) of Dicer, 600 μg (6 nmol) of Ago2, and 550 μg (11 nmol, assuming a dimer) of TRBP were mixed in 250 μl (final volume) of gel filtration buffer. The protein solution was incubated on ice for 10 min and then was applied to a Superose 6 10/30 column (Amersham Pharmacia) equilibrated in gel filtration buffer. Fractions were analyzed by SDS/PAGE, and those fractions containing the RLC were pooled and concentrated to 1.5 mg/ml. Aliquots were stored frozen at −80°C.
MS.
Mass spectra collected for the intact RLC were recorded on a Q-TOF2 (Micromass) mass spectrometer modified for high mass detection (23). The 1 μg/μl RLC complex was exchanged into 100 mM ammonium acetate buffer (pH 7.5) by using MicroBioSpin 6 columns (Bio-Rad), and 2-μl aliquots were introduced by gold-coated nanoflow capillaries prepared in-house. The conditions within the mass spectrometer were adjusted to preserve noncovalent interactions. The mass spectrometer was operated at a capillary voltage of 1,600 V and at a sampling cone voltage of 100 V. Tandem MS (MS/MS) was performed on a QSTAR XL (MDS Sciex) mass spectrometer also modified for high mass isolation and detection. MS/MS spectra of the RLC complex were obtained by isolation of the complex at m/z 8,300 and an acceleration with a 150-V collision energy.
RNA Filter-Binding Assay.
Protein- and RNA-binding assays were performed by using a previously described protocol with minor modifications (24). A constant concentration of 5′ end-labeled Dmpre-let7 hairpin RNA (<2 pM) was incubated in a 50-μl reaction volume containing 20 mM Hepes (pH 7.5), 60 mM KCl, 5 mM EDTA, 1 mM DTT, 0.01% Igepal-680, and 0.1 mg/ml tRNA. After renaturation by heating/cooling in a buffer containing 10 mM Tris·HCl (pH 7.5), 1.5 mM Mg2+, and 50 mM NaCl, the RNA was incubated with protein for at least 1.5 h before application to filters. For each experiment, two filters were used: a BA-85 nitrocellulose filter to retain protein–RNA complexes and a Hybond-N plus nylon membrane to retain free RNA. These filters were soaked in a buffer containing 20 mM Hepes (pH 7.5) for 1 h before use. A 40-μl aliquot from each reaction was applied to the filters, and then vacuum was applied to draw the samples through. The filters were not washed with additional buffer after the sample was drawn through; after brief air drying, the free and bound RNA was quantified by phosphorimaging the filters.
Dicing Assays.
Dicing assays were carried out by using either Dmpre-let7 hairpin RNA or 37-nt A and B RNAs as indicated in the figure legends. For hairpin RNA, ≈10 nM 5′ end 32P-labeled RNA was incubated in reaction buffer [100 mM NaCl, 40 mM Hepes (pH 7.5), 1 mM DTT, and 3 mM MgCl2] at 65°C for 10 min and then snap-cooled by immediately placing on ice before every assay. For forming duplex 37-nt A and B, 5′ end 32P-labeled 37-nt A and unlabeled 37-nt B RNA oligos were denatured in the above buffer and then slowly cooled to room temperature. Dicing assays were carried out in reaction buffer with radiolabeled pre-let7A RNA at 37°C for 1 h in the presence of 200 nM RLC or 200 nM Dicer alone. RNA products were resolved by denaturing PAGE (16%) and visualized by phosphorimaging.
Slicing Assays.
Slicing assays were performed at 30°C for 90 min in 10 μl of reaction volume containing 60 nM protein, 60 nM siRNA (either 21-nt guide or passenger RNA), 1 unit/μl RNasin (Promega), 20 mM Tris·HCl (pH 6.5), 50 mM KCl, 5% glycerol, 1.5 mM MgCl2, and 6 nM 5′ end-radiolabeled target 37-nt A RNA oligo (5,000 cpm). The reaction was stopped by adding 12 μl of loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, 0.1% xylene cyanol, and 0.1% bromophenol blue). After heating at 75°C for 10 min, the samples were analyzed by electrophoresis on a 16% polyacrylamide/7 M urea gel run in TBE buffer. The gel was dried, and the products were detected by PhosphorImager (GE Healthcare). ATP was not included in any activity assays.
RLC Disassembly Assays.
In vitro RLC disassembly was analyzed after three steps: RLC assembly, substrate RNA (Dmpre-let7, dmlet7) processing, and target RNA slicing. For in vitro RLC formation, 5 μg of FLAG-tagged protein (either hDicer or hAgo2) was bound to 10 μl of α-FLAG beads by incubation for 80 min on ice in TBS buffer [50 mM Tris·HCl (pH 7.5) and 0.15 M NaCl]. After washes with 4 × 500 μl of TBS buffer, 10 μg each of untagged proteins (TRBP and hAGo2 for FLAG-hDicer or TRBP and hDicer for FLAG-hAgo2) was added to the beads and incubated on ice for 60 min. After washes with 5 × 500 μl of WB buffer [20 mM Tris·HCl (pH 6.5), 50 mM NaCl, 1% glycerol, and 1.5 mM MgCl2], material from half of the beads was subjected to SDS/PAGE. Material from the remaining beads was tested for RNA processing activity by first adding and removing 10 μl of slicing buffer [1 unit/μl of RNasin, 20 mM Tris·HCl (pH 6.5), 50 mM KCl, 5% glycerol, and 1.5 mM MgCl2] to be reserved as a mock reaction sample. Another 10 μl of slicing buffer containing 4 pmol of Dmpre-let-7 hairpin RNA was added and incubated for 30 min at 30°C. The beads and supernatant (saved) were separated, and the beads were washed with 4 × 500 μl of WB buffer. Slicing activity was then tested by adding 10 μl of slicing buffer containing 4 nM 5′-32P-labeled target 37-nt A RNA oligo to the beads and incubating for 30 min at 30°C. To the mock (negative control) or supernatant (spn), 4 nM 5′-32P-labeled target 37-nt A RNA oligo was added and incubated for 30 min at 30°C. Reactions were stopped by adding 12 μl of loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, 0.1% xylene cyanol, and 0.1% bromophenol blue). After heating at 75°C for 10 min, the samples were analyzed by electrophoresis through a 16% polyacrylamide/7 M urea gel run in TBE buffer. The gel was dried, and the products were detected by PhosphorImager (GE Healthcare).
Supplementary Material
ACKNOWLEDGMENTS.
We thank members of the J.A.D. Laboratory for helpful discussions and suggestions. This work was supported by National Institutes of Health Grant GM073794 (to J.A.D.) and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710869105/DC1.
References
- 1.Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 4.Meister G, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 5.Mourelatos Z, et al. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomari Y, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 7.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 8.Caudy AA, et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Gen Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahbaz N, et al. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosentino GP, et al. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 17.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J Am Chem Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 18.Rivas FV, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 20.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 21.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 22.Peters L, Meister G. Argonaute proteins: Mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 24.Batey RT, Sagar MB, Doudna JA. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J Mol Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- 25.Benesch JL, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Tandem MS reveals the quaternary organization of macromolecular assemblies. Chem Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.