Abstract
Conformational dynamics of proteins can be interpreted as itinerant motions as the protein traverses from one state to another on a complex network in conformational space or, more generally, in state space. Here we present a scheme to extract a multiscale state space network (SSN) from a single-molecule time series. Analysis by this method enables us to lift degeneracy—different physical states having the same value for a measured observable—as much as possible. A state or node in the network is defined not by the value of the observable at each time but by a set of subsequences of the observable over time. The length of the subsequence can tell us the extent to which the memory of the system is able to predict the next state. As an illustration, we investigate the conformational fluctutation dynamics probed by single-molecule electron transfer (ET), detected on a photon-by-photon basis. We show that the topographical features of the SSNs depend on the time scale of observation; the longer the time scale, the simpler the underlying SSN becomes, leading to a transition of the dynamics from anomalous diffusion to normal Brownian diffusion.
Keywords: single-molecule experiment, anomalous diffusion, time series analysis
Optical single-molecule spectroscopy has provided unique insights into both the distribution of molecular properties and their dynamic behavior, which are inaccessible using ensemble-averaged measurements (1–5). In principle, the complexity observed in the dynamics and kinetics of a protein originates in the underlying multidimensional energy landscape (6–12). The dynamics can be understood as the protein traversing from one state (node) to another along a complex network in conformational space or, more generally, in state space. The network properties of biological systems can provide a new perspective for addressing the nature of their hierarchical organization in multidimensional state space (10, 11, 13, 14), enabling us to ask such questions as: Is there any distinctive network topology that is characteristic for the native basin into which a protein folds? Are there any common network features that biological systems may have evolved by adapting to the changes in the environment? Motivated by questions of this nature, we address how one can extract the state space network (SSN) of multiscale biological systems explicitly from a single-molecule time series, free from a priori assumptions on the underlying physical model or rules.
Fluorescence resonance energy transfer (FRET) and electron transfer (ET) are among the mostly widely used techniques for measuring the dynamics of protein conformational fluctuations (15) and folding (16–19). For example, Yang et al. (15) used a single-molecule electron transfer experiment to reveal the complexity of protein fluctuations of the NADH:flavin oxidoreductase (Fre) complex. The fluorescence lifetimes showed that the distance between flavin adenine dinucleotide (FAD) and and a nearby tyrosine (Tyr) in a single Fre molecule fluctuates on a broad range of time scales (10−3 s to 1 s). Although the overall dynamics in the distance fluctuation are non-Brownian, they reflect normal diffusion on longer time scales. To gain further understanding of such anomalous behavior for protein conformational fluctuations, several analytical models have been proposed in terms of the generalized Langevin equation with fractional Gaussian noise (20), and the simplified discrete (21) and continuous (22) chain models. Recently, all-atoms simulations (23) were performed to extrapolate the physical origin of the anomalous FAD-Tyr distance fluctuation observed in experiment (>10−3 s) from the simulation time scales in nanoseconds.
These theoretical studies underscore the difficulties in establishing a minimal physical model for the origin of complexity in the kinetics or dynamics of biomolecules. Instead of postulating or constructing a physical model to characterize experimental results, we take a different approach to “let the system speak for itself” through the single-molecule time series. Such unbiased solutions, which are data-driven instead of model-driven, have been provided in the context of single-molecule FRET experiments (24) and emission intermittency, demonstrated in resolving quantum dot blink-ing states (25), but defining the states and the corresponding networks from a single-molecule time series remains a challenging problem. For instance, even while the system travels among different physical states, the values in the measured observable can be the same (26–28), i.e., degenerate, due to the finite resolution of the observation, noise contamination, and the limited number of measurable physical observables in the experiment. Such degeneracy can give rise to apparent long-term memory along the sequence of transitions even when the transitions among states are Markovian (29).
In this article, we present a method to extract the hierarchical SSN spanning several decades of time scales from a single-molecule time series. Within the limited information available from a scalar time series, this method lifts degeneracy as much as possible. The SSN is expected to capture the manner in which the network is organized, which may be relevant to some functions of biological systems. The crux of our approach is the combination of computational mechanics (CM) developed by Crutchfield et al. (30, 31) in information theory and Wavelet decomposition for single-molecule time series. The states are defined not only in terms of the value of the observable at each time but also the historical information of a set of the multiscale Wavelet components along the course of time evolution. Using the single-molecule ET time series of Fre/FAD complex, we demonstrate that the multiscale SSNs provide analytical expressions for the multi-time correlation function with the physical basis for the long-term conformational fluctuations.
Results and Discussion
SSN: One-Dimensional Brownian Motion on a Harmonic Well.
We briefly describe how the original CM (31) defines “states”and constructs their network from scalar time series [see the detailed descriptions in supporting information (SI) Text. For a given time series x = (x(t1), x(t2) …, x(tN)) of the physical observable x, which is continuous in value and could be the intramolecular distance reported by fluorescent probes, we first discretize it to the symbolic sequence s = (s(t1), s(t2)…, s(tN)) where s(ti) denotes the symbolized observable at time ti (see Fig. 1).An upper bound for the number of symbols may be determined by the experimental resolution. As we will see below, CM requires a statistical sampling of the subsequences in the symbolic time series s. Therefore, the choice of discretization scheme should depend not only on the experimental resolution but also on the statistical properties of the time series. A reasonable discretization is such that the topological properties of the constructed network are insensitive to the increase in the number of partitions. Second, we trace along the time series s for each time step ti to record which subsequence of length Lfuture, (sAfuture = {s(ti+1), …, s(ti+Lfuture)}), follows consecutively after the subsequence of length Lpast, (sBpast = {s(ti−Lpast+1,…, s(ti))}). Here A, B, … represent different symbolic subsequences that appear in s (see Fig. 1). The transition probability from sBpast to sAfuture [denoted by P(A|B)] can then be obtained for the time series s.
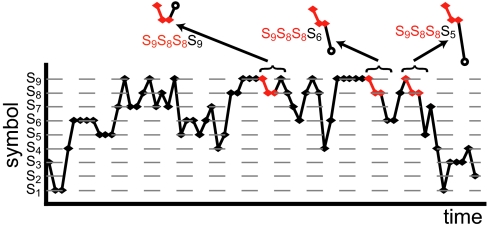
Fig. 1.
Example for a discrete-symbol time series s obtained by discretizing a time series of continuous observable. Here a particular past subsequence (s9s8s8 shown by the red dots) with Lpast = 3 can be followed by different future subsequences (the open circles) with Lfuture = 1. Going over the entire time series to build up the frequency distribution that a certain past–future sequence occurs allows us to evaluate the transition probabilities P(s′|s9s8s8) with s′ = s1, s2, … for the transition from s9 s8s8 to the next subsequence with Lfuture = 1. Transition probabilities for the other past subsequences are obtained accordingly.
Third, we define “state” (denoted by Si herein) by the set of past subsequences {sB′past, sB″past, …} with length Lpast, which make transition to the future subsequences sAfuture with the same transition probabilities (i.e., P(A|B′) = P(A|B″) = P(A|B‴) = … for all A). A directed link (i.e., transition) from a state Si to another state Sj can be drawn to represent the transition probability P(sAfuture|Si) if the subsequence sAfuture is generated from a transition from Si to Sj along the time series s. The extraction of all states and transitions among them yields a SSN associated with the time series s. The most attractive feature of the CM is that it extracts (instead of postulating) the underlying SSN from time series: the length of memory Lpast is chosen so as to make all transitions among the states Markovian; namely, the next state to visit is solely determined by the current state. The inferred SSN is, hence, regarded as a kind of hidden Markovian model extracted from the data. It has been established mathematically that such a SSN inferred from the time series, with memory effect automatically included, provides us with a minimal and optimally predictive machinery that can best reproduce the time series s in a statistical sense (31) (see also SI Text for more details).
As an example, Fig. 2 illustrates the actual SSNs constructed from the time series x(t) of a one-dimensional normal Brownian trajectory in a harmonic well. Here we discretize x using the equal probability partition with 12 symbols, i.e. each symbol has the same occurrence probability. The symbolic time series s is resampled every m time steps from which the SSN is constructed. Each node represents the state composed of the set of symbolic subsequences, which have the same transition probability as described above. The area of the node is proportional to the resident probability of the state along the time series s. The directed links connecting the nodes represent the transitions between the states. The abscissa of the circle (state) center corresponds to the average value of x over the set of symbolic subsequences assigned to that state, and the ordinate denotes the average distribution–distribution (D–D) distance Ddistrib (i), which measures on average how different from the others the transition probability of the ith state is (the detailed definition will be given later).
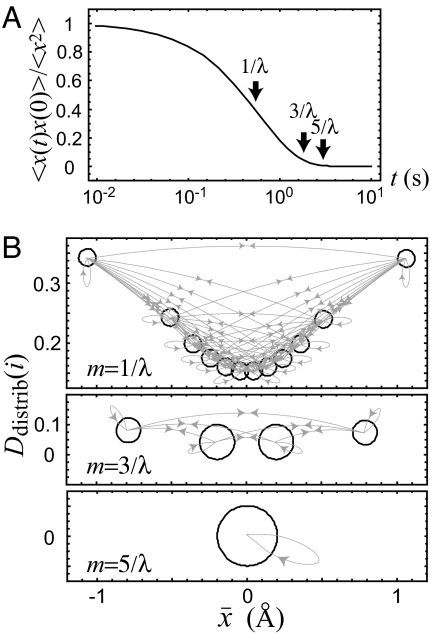
Fig. 2.
The SSNs constructed from the time series x obeying the overdamped Langevin equation on a harmonic well, dx(t)/dt = −λx(t) + R(t). Here λ = ω2/γ is the drift coefficient that characterizes the correlation time of the stochastic variable x with γ being the friction coefficient (ω is the frequency of the harmonic well, ≈1/2ω2x2). R(t) is a Gaussian random force exerted on x with mean 〈R(t)〉 = 0 and variance 〈R(t) R(t0)〉 = 2λθδ(t − t0), where θ characterizes the magnitude of the fluctuation at a given temperature. Here, θ and λ are set to be 0.19 Å2 and 1.73 s−1, respectively, by referring to the mean force potential along the “FAD-Tyr distance”and the autocorrelation of the lifetime fluctuation of the excited state of FAD (35). We discretize x into 12 symbols with variable width Δxk such that the resident probability P(xk ≤ x(t) ≤ xk + Δxk) of kth symbol sk is the same for all k. (A) The normalized aut ocorrelation function 〈x(t)x(0)〉/〈x2〉 (≃exp(−λt)) as a function of log10 t. (B) The SSNs constructed by recording every m step from the symbolic time series s with m = 1/λ, 3/λ, and 5/λ, respectively.
It was found that past subsequences with only one symbol (Lpast = 1) are sufficient to construct the underlying SSN for the case of m = 1/λ and m = 3/λ (where 1/λ characterizes the correlation time scale of x) such that a further increase of Lpast does not change the network topology. This is due to the Markovian nature of Brownian motion that only requires the present value of the observable to “predict”the future. When the step size m ≲ 1/λ, the system cannot jump over to all the accessible regime of s and thus there are 12 states (no identical transition probability exists). The states extracted from s at such a short time scale just mimic a “trajectory” of a stochastic variable in the discretized space.
As m increases, some of the nearby states start to merge and eventually only a single state is obtained at m = 5/λ as the autocorrelation decays to almost zero, where the required Lpast is found to be zero. This manifests that it does not require any information of the current value to predict the future when the correlation is negligible. The time series recorded in every 5/λ time steps or longer is statistically equivalent to the dodecahedron's dice toss. Moreover, one can see from the Langevin case that the subsequences contained in a state are localized in the physical observable space (e.g., the x) for time scale shorter than the correlation time scale (e.g., at 1/λ). However, such localization is lost for much longer time scales (e.g., at 5/λ). A more general discussion of the connection between the changes of localization properties of the states in the physical observable space as a function of time scale and its relation to the state transition probability similarity is given in SI Text.
One can expect that CM is able to extract the time scale on which the system loses memory in the observable. It also reveals how the system smears out the fine structure of the state space in terms of the time scale-dependent SSN. Such a “model-free”approach is crucial in capturing the complexity in the kinetics and dynamics observed in single-molecule experiments. However, there are several practical drawbacks in the standard form of CM, especially for systems with hierarchical time and space scales.
First, the number of possible past subsequences sApast grows exponentially with Lpast and the sampling of sApast becomes worse rapidly due to the finite length of the time series. So it is difficult to properly resolve the SSN when long-term memories exist. Although the CM discussed above using skipping time steps works well for the Markovian Brownian dynamics, it skips and so neglects the information between consecutive sampling steps that may contains important non-Markovian properties in real single-molecule time series. Second, CM relies on the concept of stationarity for the underlying processes. This implies that the statistical properties of the system changes slowly within the length of the time series from which the SSN is constructed. However, this is not necessarily the case for real systems where the existence of hierarchical time and space scales provides a diverse dynamical properties over different scales. Therefore, a decomposition of the observable time series into a set of hierarchical, stationary (and nonstationary) processes with different time scales is highly desirable for the prescription of CM.
Most importantly, after one extracts the underlying dynamics for each characteristic time scale associated with the long memory process, there may exist “mutual correlation”or “nonadiabatic coupling”across different hierarchies of different time scales. Hence, the incorporation of the mutual correlation across the decomposed hierarchies is important for establishing the correct SSN hidden in the time series for multiscale complex systems.
Below we propose a scheme of multiscale CM based on the discrete wavelet decomposition, which can not only overcome the existing difficulties in the current form of CM but also resolve the cumbersome degeneracy problem in single-molecule measurements as much as possible. Here, we apply our method to the delay-time time series of the Fre/FAD complex (15) in the ET experiment. We note, however, that our method is general and should be applicable to any time series.
Hierarchical SSN: Anomalous Conformational Fluctuation in Fre/FAD Complex.
The protein structure of the Fre/FAD complex and the position of the tyrosin residue Tyr35, relative to the FAD substrate are shown in Fig. 4A. Fig. 3 illustrates the multiscale CM scheme based on the discrete wavelet decomposition of the delay-time time series. The delay time of the fluorescence photons are recorded with respect to the excitation pulse as a function of the chronological arrival time of the detected photon (15). Discrete wavelet decomposition (33) produces a family of hierarchically organized decompositions from a scalar time series τ = (τ1,…,τN),
where A(j) = (A1(j), …, AN(j) and D(j) = (D1(j), …, DN(j) are called as j-level approximation and detail, respectively. In the case of dyadic decomposition, which is applied in this paper, A(j) approximates τ with a time resolution of 2j time steps by discarding fluctuations with time scale smaller than 2j time steps, and D(j) captures the fluctuation of τ over the time scale of 2j time steps. The time series can be reconstructed by adding back all fluctuations with timescales smaller than or equal to those of the approximation. Moreover, approximations of different time scales are related by A(j) = A(j+1) + D(j+1) with j ≥ 0. In this paper, the Haar wavelet is adopted for its simple interpretation: Ai(j) and Di(j) of the Haar wavelet are the mean and the mean fluctuation over a bin of 2j time steps, respectively (see SI Text for details).
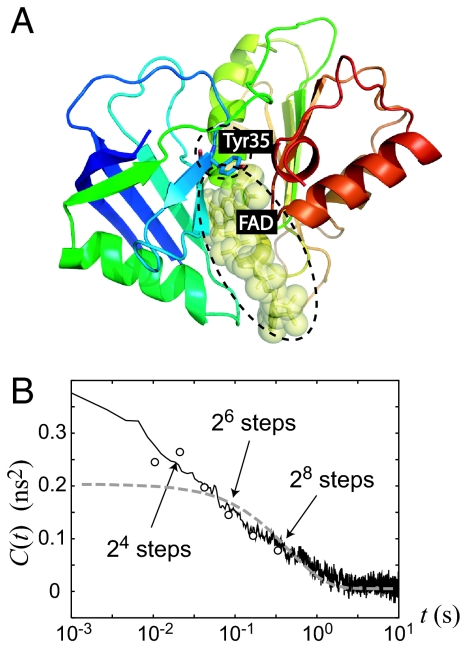
Fig. 4.
Visualization of the chromophore in protein and its fluctuation correlation. (A) The protein structure of Fre/FAD complex. The tyrosine residue Tyr35 and the FAD substrate, which are responsible for the fluorescence quenching, are shown in the dash circles. (B) The autocorrelation function of fluorescence lifetime fluctuation 〈δγ−1 (t)δγ−1 (0)〉 evaluated by Eq. 6 with the transition time of 2n (n = 3, 4, …, 8) (denoted by the circles) in linear-logarithmic plot. For comparison, the previous numerical result from a photon-by-photon-based calculation is also depicted (the solid line) together with the normal Brownian diffusion model (gray dashed line) represented by the overdamped Langevin equation on a harmonic potential well presented in Fig. 2. θ and λ are set to be 0.19 Å2 and 1.73 s−1, respectively, with β = 1.4 Å−1 (15). The arrows indicate the time scales at which 〈δγ−1 (t)δγ−1 (0)〉 is evaluated according to the multiscale SSNs shown in Fig. 5. The results indicate that the SSN indeed captures the system's multiscale dynamics.
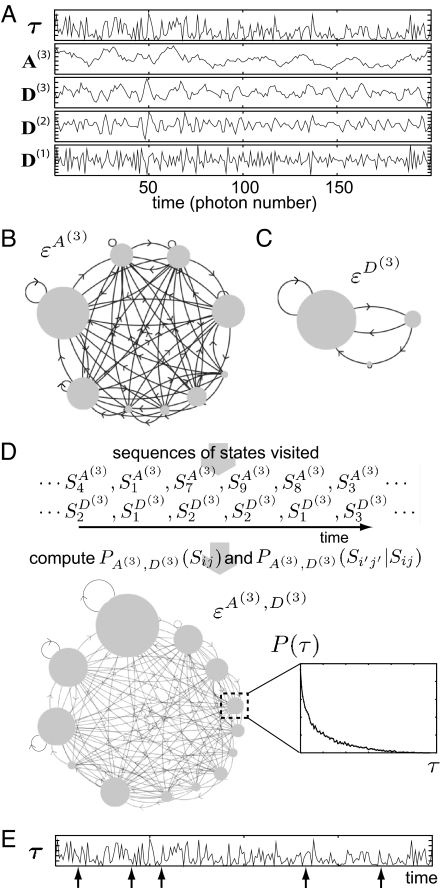
Fig. 3.
Construction of multiscale SSNs. (A) A portion of the delay-time time series of ET single-molecule measurement for the Fre/FAD complex [15] and its wavelet decomposition with n = 3 (c.f. Eq. 1). (B and C) The sub-SSNs of A(3) and D(3). Note that a jump from one state to another in both εA(3) and εD(3) corresponds to 23 time steps in the time series. A directed arrow is drawn between two states (gray circles) if the transition probability among the states is not zero. The size of the circle is proportional to the resident probability of the state, and the number of inferred states for εA(3) and εD(3) and NA(3) and ND(3) are nine and three, respectively. (D) A multiscale SSN combining the sub-SSNs in B and C (see text for details). Note that the number of state in the combined SSN εA(3),D(3) is lesser than NA(3) × ND(3), implying that εA(3) and εD(3) are mutually correlated. (E) Some data points in the delay-time time series, indicated by arrows, where the system visited at the state in the dash box of D. The ensemble of such data points provides a unique delay-time probability density function P(τ) for each state, as shown in D.
Fig. 3A exemplifies the discrete wavelet decomposition with n = 3 using the delay-time time series (denoted here by τ). The sub-SSNs, denoted by εA(n) and εD(j), are constructed from the time series of A(n) and from that of D(j) (j ≤ n) by the same algorithm used in Fig. 2, respectively. Fig. 3 B and C present εA(3) and εD(3) extracted from the time series of A(3) and D(3), both with the time steps 23. Due to the nature of A(n) (the binned average), the constructed sub-SSN εA(n) averages out the information contained in each bin. This suppresses the noise from photon statistics but on the other hand suffers from information loss inside the bins (24). Therefore, a combined SSN should be constructed by incorporating the SSNs of fluctuations inside the bin (the details) and the correlations among them into εA(n). For instance, the incorporation of εD(n) and εD(n − 1) into εA(n) gives the SSN that describes dynamics with time scale 2n by taking account of fluctuations down to the bin size of 2n − 1.
Fig. 3D demonstrates how εA(3) and εD(3) can form the combined SSN εA(3),D(3). By tracing A(3) and D(3) time series, one can identify which state the system visited at each time step in εA(3) and εD(3), respectively. The possible states of the combined SSN εA(3),D(3) are given by the product set {SiA(3),SjD(3)} (≡ Sij), where SiA(3) and SjD(3) denote the ith and jth state in εA(3), and εD(3) (where i = 1, …, NA(3) and j = 1, …, ND(3)). The resident probability of the Sij denoted by PA(3),D(3)(Sij) can then be computed as follows:
where N(Sij) is the number of simultaneous occurrence at the states SiA(3) and SjD(3) along the time series, and N is the number of data points of the series. In general PA(3),D(3) (Sij) ≠ PA(3) (SiA(3)) PD(3) (SjD(3)) because the two time series A(3) and D(3) are statistically correlated. On the other hand, the transition probability from Sij to Si′j′ can also be obtained by
where N(Si′j′, Sij) is the number of visiting Si′j′ at 23 time steps passed after visiting Sij. In general, a transition from one state to another in εA(n),D(n) takes 2n time steps as in εA(n).
The combined SSN εA(n),D(n) corresponds to a “splitting” of the states SiA(n) of the approximation to Sij with 1 ≤ j ≤ ND(n) by incorporating the fluctuation inside the bins. Similarly, other sub-SSNs (εD(2), εD(1)) can be incorporated into εA(3),D(3) one by one, depending on how fine the fluctuations one wishes to see.
Moreover, because the original τ is decomposed into a vector time series with approximation and details as components, degeneracy is expected to be further lifted by this multiscale CM compared with the original CM in terms of scalar time series τ. The stationarity of the approximation and details can be inspected by evaluating their autocorrelations. The autocorrelation of D(j) decays rapidly on a timescale of 2j time steps with small oscillations for longer time. Therefore, the D(j)'s are ‘approximately’ stationary with time scale of 2j. On the other hand, the autocorrelation of A(j) remains approximately constant for 2j steps and shows similar behavior to those of τ for time scales longer than 2j. This indicates that A(j) capture all the nonstationarity of τ with time scales longer than 2j (see also SI Text for more details). Hence, Eq. 1 enables us to naturally decompose the original time series into a set of hierarchical stationary processes (the details) at different time scales and their nonstationary counterpart (the approximation).
Lifetime Spectrum and the Average Interdye Distance Associated with a State in the Multiscale SSN.
Once the multiscale SSN is extracted up to the desired level by combining the sub-SSNs, one can then build up an unique delay-time distribution for each state in the network as shown in the inset of Fig. 3D. The delay-time probability density function P(τ) is related to the spectrum of lifetime α(γ−1) by (see SI Text for details)
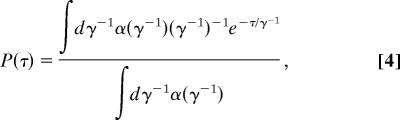 |
with ∫ dτP(τ) = 1. The conformational information of a state can be obtained from α(γ−1) and γ−1(t) = [γ0 + kET(t)]−1 ≈ kET−1(t) [γ0 is the fluorescence decay rate in the ab sence of quencher(s) and kET the ET rate] as follows: The average rate (inverse of lifetime) of the Ith combined state, γ̄I, can be calculated easily from its lifetime spectrum αI(γ−1) and delay-time probability density function PI(τ) as
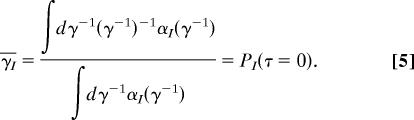 |
The averaged donor(D)–acceptor(A) distance R associated with the Ith state, R̄I, can, then, be evaluated by R̄i ≈ R0 − β̄−1 log γ̄I under the assumption of kET(t) ∼ exp[−βR(t)] with β = 1.4 Å−1 for proteins (34).
The Autocorrelation of Lifetime Fluctuation.
What kinds of physical quantities can be extracted from such multiscale SSNs? For instance, the autocorrelation function of lifetime fluctuation C(t) = δγ−1(t)δγ−1(0), where δγ−1(t) = γ−1(t) − γ−1 is readily elucidated from the inferred SSNs: The autocorrelation function C(t = 2n) is represented from the multiscale SSN with transition step of 2n as
where P2(n)(SJ, SI) = P2n(SJ|SI)P(SI) is the joint probability of visiting SI followed by SJ after 2n steps; the higher-order correlation functions can be also derived straightforwardly (see SI Text for details). Fig. 4B shows that the multiscale SSN can naturally produce the autocorrelation function which agrees well with the photon-by-photon based calculation (15, 35). The physical origin of anomaly presented in the autocorrelation function C(t) of the fluorescence lifetime fluctuation was conjectured as follows (15): the conformational states corresponding to local minima on the multidimensional energy landscape have vastly different trapping times because the energy barrier heights for the interconversion among local minima are expected to be broadly distributed. Such a broad distribution of trapping time at a particular D–A distance R should give rise to a rugged “transient” potential for short time scales, resulting in subdiffusion and the stretched exponential in C(t). However, for longer time scales, the apparent potential becomes a smooth harmonic mean force potential and converges to a single state. In the following, we will show that our multiscale SSN naturally reveals such time-dependent topographical features of the underlying network in the state space.
Time Scale-Dependent Topographical Features of the SSN.
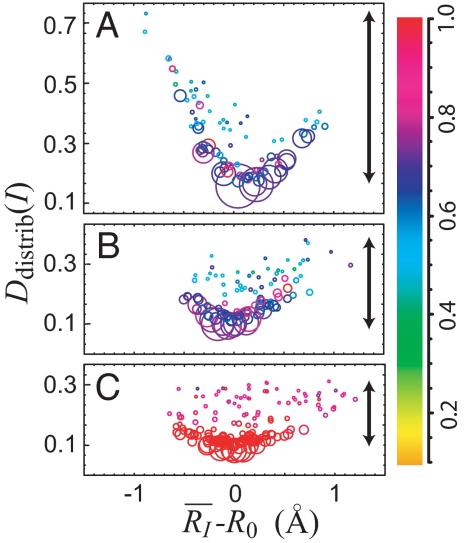
Fig. 5 illustrates how the SSN topography depends on the time scale by projecting the network onto a two-dimensional plane composed of the average FAD-Tyr distance RI − R0 and the average D–D distance of the state from the others, where a state is represented by a circle as in Fig. 2. Here, three combined SSNs are shown with transition time steps of 24, 26, and 28, approximately corresponding to 30, 120, and 500 ms, respectively. The average D–D distance of the Ith state to all other states in the network is defined by Ddistrib(I) = ΣJ=1NS P(SJ)dH(I, J), where P(SJ) and NS denote the resident probability of the Jth state, SJ, and the total number of states, respectively. dH(I, J) is the distance between two distributions in terms of the Hellinger distance (36) [∫−∞∞ (PI(η)1/2 − PJ(η)1/2)2dη]1/2, where PI(η) and PJ(η) are the transition probability P(ηfuture|SI) and P(ηfuture|SJ). The smaller the value of Ddistrib(I), the closer the Ith state is located to the center of the network. Furthermore, the variance of Ddistrib(I) over the set of states in the network (see the black arrows in Fig. 5) measures how diverse the transition probabilities of the states is. One can see in Fig. 5 that, as the time scale increases from 24 to 28 steps, the variance of Ddistrib(I) decreases and the network becomes more compact. This indicates the trend for the states to merge together for longer time scales.
Fig. 5.
The dependence of the topographical feature of SSNs on the time scales: εA(4),D(4),D(3) with 24 steps (≈ 30 ms) (A), εA(6),D(6),D(5) with 26 steps (≈ 120 ms) (B), and εA(8),D(8),D(7) with 2 8 steps (≈ 500 ms) (C). The abscissa and ordinate denote the average interdye distance R̄I − R0 evaluated by −β log γ̄I and the average D–D distance Ddistrib(I) associated with the Ith state in each SSN, respectively. The area of the circle is proportional to the resident probability of the state and the arrow indicates the variance of Ddistrib(I). States of distinct Ddistrib but exhibiting the same (R̄I − R0) can now be resolved, lifting the degeneracy (cf. Fig. 2 where there is no degeneracy). See text for more detail.
The radius of the circles reflects the stability of the corresponding states. One can see that, as |R̄I − R0| → 0, the states in the networks tend to be more stabilized, implying that R0 corresponds to the equilibrium FAD-Tyr distance of the mean force potential with respect to R. The more striking feature is that the number of states can be more than one at a given value of the FAD-Tyr distance R, and there exists a broad distribution in the size of the states especially around R0 at the short time scale of 24 steps. The latter is in big contrast to Fig. 2, where only a single state presents at a given value of x (no degeneracy) because of the one dimensional nature of the system. This clearly indicates the degeneracy lifting properties that the multiscale SSN can differentiate states having almost the same value in the observable and therefore reflects the multidimensional nature of the underlying landscape. Furthermore, compared with longer time steps like 28, some “isolated” states with larger circles (weighted more) exist in regions that are far away from R0 at 24 steps (note the time scale of m = 1/λ ≈ 580 ms in Fig. 2 is expected to be close to 28 steps here). This is a manifestation of frustration on the multidimensional energy landscape resulting in a vast number of different trapping times at short time scale as inferred in ref. 15.
Further insight into the nature of the conformational dynamics can be acquired by considering how different states are connected in the SSN. The simplest measure of connectivity among the nodes is the degree of node kI, that is, the number of transitions or links from the Ith node to the others. In Fig. 5, the color of the states (the circles) indicates the value of the normalized degree kI/kmax where kmax = max{k1, …, kNS} is the maximum degree among all states in the chosen SSN. As indicated by the color bar in the figure, more links or transitions from a state is denoted by color towards the red end of the spectrum. The saturating red color signifies that the state connects or communicates to almost all of the states in the network. As the time scale increases, say, from 24 to 28, the nodes tend to acquire more connections on average, indicated by the shift of color to the red end. This reflects the fact that the system, given more time, can explore more thoroughly the remote regions on the energy landscape.
On the other hand, one can see from the degree dependence on the stability of states in the multiscale SSNs (SI Fig. 9) that the greater the degree of the node, the larger the node size. This can be regarded as the first experimental manifestation so far observed in the network of multidimensional conformational space of biomolecules (10, 11); that is, the state tends to be more stabilized when there exist more transition paths from the state. Moreover, a large diversity of degrees for a given state size (or stability) is observed for short time scales (e.g., 24 and 26), which provided us with the evidence of heterogeneity in the state connectivity. Its implication and the degree dependence of the stability of states will be discussd in more detail in SI Text.
As a summary, at a typical time scale of “subdiffusion,” e.g., 24 steps as shown in Fig. 5A, the underlying network exhibits strong diversity in the transition and morphological features of the state space, which should arise from the frustration of the multidimensional energy landscape. However, on the time scale of 28 steps, which can be regarded as a turning point from the subdiffusion regime to the Brownian diffusion regime, the topographical feature of the underlying network becomes relatively compact, leading to the consolidation of all states so that the number of links from each state become uniformly close to maximum.
Conclusions
In this article, we have presented a method to extract the multiscale network in state space from a single-molecule time series, with the ability to lift the degeneracy inherent to finite scalar time series. In contrast to models that are postulated for the underlying physical mechanism, the multiscale SSN can objectively provide us with rules about the underlying dynamics that one can learn “directly” from the experimental single-molecule time series. The network topography depends on the time scale of observation; in general, the longer the observation, the less complex the underlying network appears.
Our method also provides a means to introduce several concepts of complex networks into single-molecule studies, which have been developed extensively in different fields sharing similar organization such as biology, technology, or sociology (13). For instance, modules or communities and “small-world”concepts in biological networks are expected to be relevant to specific functions and hierarchical organization of the systems. This multiscale SSN can also examine the time scale on which the concept of a Markovian process is valid. Most importantly, it provides a natural way of investigating how multiscale systems evolve in time with mutual interference across the hierarchical dynamics in different time scales.
As for the future works, a rigorous connection of the concepts in the multiscale SSNs and those in the context of dynamical theory can further enhance our understanding of the multiscale dynamics of complex systems. Moreover, we expect that, by monitoring the change of multiscale SSN that is locally constructed from a set of finite consecutive periods along the course of time (37), it will be possible to shed light on how the system adapts to time-dependent external stimuli under thermal fluctuation.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Satoshi Takahashi and Mikito Toda for valuable comments, X. S. Xie and G. Luo for providing time series of the ET single-molecule experiment of Fre/FAD complex, and Kazuto Sei for helping us to draw Fig. 3. Parts of this work were supported by JSPS, JST/CREST, Priority Areas “Systems Genomics,” “Molecular Theory for Real Systems” and the 21st century COE (Center of Excellence) of Earth and Planetary Sciences, Kobe University, MEXT (to T.K.) and by U.S. National Science Foundation (to H.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707378105/DC1.
References
- 1.Xie XS, Trautman JK. Annu Rev Phys Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 2.Moerner WE, Fromm DP. Rev Sci Inst. 2003;74:3597–3619. [Google Scholar]
- 3.Barkai E, Jung Y, Silbey R. Annu Rev Phys Chem. 2004;55:457–507. doi: 10.1146/annurev.physchem.55.111803.143246. [DOI] [PubMed] [Google Scholar]
- 4.Lippitz M, Kulzer F, Orrit M. ChemPhysChem. 2005;6:770–789. doi: 10.1002/cphc.200400560. [DOI] [PubMed] [Google Scholar]
- 5.Michalet X, Weiss S, Jager M. Chem Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 7.Stillinger FH. Science. 1995;267:1935–1939. doi: 10.1126/science.267.5206.1935. [DOI] [PubMed] [Google Scholar]
- 8.Wales DJ. Energy Landscapes. Cambridge, UK: Cambridge Univ Press; 2003. [Google Scholar]
- 9.Krivov SV, Karplus M. Proc Natl Acad Sci USA. 2004;101:14766–14770. doi: 10.1073/pnas.0406234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao F, Caflisch A. J Mol Biol. 2004;342:299–306. doi: 10.1016/j.jmb.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 11.Gfeller D, Rios PDL, Caflisch A, Rao F. Proc Natl Acad Sci USA. 2007;104:1817–1822. doi: 10.1073/pnas.0608099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ball KD, Berry RS, Kunz RE, Li F-Y, Proykova A, Wales DJ. Science. 1996;271:963. [Google Scholar]
- 13.Albert R, Barabási A-L. Rev Mod Phys. 2002;74:47–97. [Google Scholar]
- 14.Gallos LK, Song C, Havlin S, Makse HA. Proc Natl Acad Sci USA. 2007;104:7746–7751. doi: 10.1073/pnas.0700250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Luo G, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun L, Xie XS. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 16.Talaga DS, Lau WL, Roder H, Tang JY, Jia YW, DeGrado WF, Hochstrasser RM. Proc Natl Acad Sci USA. 2000;97:13021–13026. doi: 10.1073/pnas.97.24.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuler B, Lipman EA, Eaton EA. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 18.Rhoades E, Gussakovsky E, Haran G. Proc Natl Acad Sci USA. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita M, Kamagata K, Maeda M, Goto Y, Komatsuzaki T, Takahashi S. Proc Natl Acad Sci USA. 2007;104:10453–10458. doi: 10.1073/pnas.0700267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min W, Luo G, Cherayil BJ, Kou SC, Xie XS. Phys Rev Lett. 2005;94:198302. doi: 10.1103/PhysRevLett.94.198302. [DOI] [PubMed] [Google Scholar]
- 21.Tang J, Marcus RA. Phys Rev E. 2006;73:022102. doi: 10.1103/PhysRevE.73.022102. [DOI] [PubMed] [Google Scholar]
- 22.Debnath P, Min W, Xie XS, Cherayil BJ. J Chem Phys. 2005;123:204903. doi: 10.1063/1.2109809. [DOI] [PubMed] [Google Scholar]
- 23.Luo G, Andricioaei I, Xie XS, Karplus M. J Phys Chem B. 2006;110:9363–9367. doi: 10.1021/jp057497p. [DOI] [PubMed] [Google Scholar]
- 24.Watkins LP, Yang H. Biophys J. 2004;86:4015–4029. doi: 10.1529/biophysj.103.037739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watkins LP, Yang H. J Phys Chem B. 2005;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- 26.Edman L, Rigler R. Proc Natl Acad Sci USA. 2000;97:8266–8271. doi: 10.1073/pnas.130589397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witkoskie JB, Cao J. J Chem Phys. 2004;121:6361–6372. doi: 10.1063/1.1785783. [DOI] [PubMed] [Google Scholar]
- 28.Flomenbom O, Klafter J, Szabo A. Biophys J. 2005;88:3780–3783. doi: 10.1529/biophysj.104.055905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwanzig R. Nonequilibrium Statistical Mechanics. New York: Oxford Univ Press; 2001. [Google Scholar]
- 30.Crutchfield JP, Young K. Phys Rev Lett. 1989;63:105. doi: 10.1103/PhysRevLett.63.105. [DOI] [PubMed] [Google Scholar]
- 31.Shalizi CR, Crutchfield JP. J Stat Phys. 2001;104:819–881. [Google Scholar]
- 32.Brandes U, Kenis P, Raab J, Schneider V, Wagner D. J Theor Politics. 1999;11:75–106. [Google Scholar]
- 33.Daubechies I. Ten Lectures on Wavelets. New York: Soc Indust Appl Math; 1992. [Google Scholar]
- 34.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Xie XS. J Chem Phys. 2002;117:10965–10979. [Google Scholar]
- 36.Krzanowski WJ. J App Stat. 2003;30:743–750. [Google Scholar]
- 37.Nerukh D, Karvounis G, Glen RC. J Chem Phys. 2002;117:9611–9617. doi: 10.1063/1.1780152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.