Abstract
RNA helicases couple the energy from ATP hydrolysis with structural changes of their RNA substrates. DEAD box helicases form the largest class of RNA helicases and share a helicase core comprising two RecA-like domains. An opening and closing of the interdomain cleft during RNA unwinding has been postulated but not shown experimentally. Single-molecule FRET experiments with the Bacillus subtilis DEAD box helicase YxiN carrying donor and acceptor fluorophores on different sides of the interdomain cleft reveal an open helicase conformation in the absence of nucleotides, or in the presence of ATP, or ADP, or RNA. In the presence of ADP and RNA, the open conformation is retained. By contrast, cooperative binding of ATP and RNA leads to a compact helicase structure, proving that the ATP- and ADP-bound states of RNA helicases display substantially different structures only when the RNA substrate is bound. These results establish a closure of the interdomain cleft in the helicase core at the beginning of the unwinding reaction, and suggest a conserved mechanism of energy conversion among DEAD box helicases across kingdoms.
Keywords: ATP-induced conformational changes, cooperativity, single-molecule FRET, RNA unwinding, YxiN
RNA helicases are ubiquitous enzymes that convert the energy of ATP hydrolysis into structural changes of RNA or RNA/protein complexes. They participate in virtually all processes involving RNA, from transcription, RNA editing, splicing, and translation to RNA decay (for a recent review see ref. 1). DEAD box helicases constitute the largest family of RNA helicases and derive their name from the conserved DEAD sequence in the Walker B motif involved in ATP hydrolysis. In contrast to the highly processive DNA helicases, which unwind thousands of base pairs before dissociating from their DNA substrate, DEAD box helicases unwind only 5–6 nucleobases (2). Recently, an unwinding mode distinct from translocating DNA helicases was suggested for DEAD box helicases (3).
A minimal DEAD box helicase consists of two RecA-like domains, the so-called helicase core, which contains nine conserved motifs required for ATP binding and hydrolysis, RNA binding, and helicase activity. In addition, many helicases contain flanking sequences of varying length that mediate additional functions such as substrate specificity. Although various structures of isolated RecA-like domains of helicases have been determined (4–9), structural information of complete DEAD box helicases (or helicase cores) is limited to eIF4A from Saccharomyces cerevisiae (PDB ID code 1fuu) (7), DeaD from Methanococcus jannaschii (mjDeaD, PDB ID code 1hv8) (10), the human splicing helicase UAP56, with (PDB ID code 1xtj) and without ADP (PDB ID code 1xti) (11), S. cerevisiae Dhh1p (PDB ID code 1s2m) (12), the Drosophila melanogaster helicase Vasa in complex with ADPNP and ssRNA (PDB ID code 2db3) (13), and human eIF4A-III as part of the exon junction complex, with ADPNP and ssRNA bound (PDB ID code 2hyi) (14, 15). Most of these structures capture the DEAD box helicases in a surprisingly wide range of open conformations in which the two RecA-like domains of the helicase core are separated and juxtaposed in different relative orientations, and contacts between the domains are largely absent. In contrast, Vasa and eIF4A-III, both in complex with ADPNP and ssRNA, display closed conformations with extensive contacts between the two RecA-like domains. Based on these structures, the formation of a closed state upon simultaneous binding of ATP and RNA was proposed (13–15). Conformational changes in the catalytic cycle of DEAD box helicases have also been predicted from biochemical data, and a number of models for RNA unwinding have been suggested that require conformational changes (reviewed in ref. 1). Hitherto, structures of open and closed conformations for one particular DEAD box helicase have not been determined. Single-molecule FRET (smFRET) experiments are ideally suited to investigate conformational changes in the catalytic cycle of RNA helicases in solution. Here, we identify different conformational states of the DEAD box RNA helicase YxiN from Bacillus subtilis that depend on the ligation state of the helicase. YxiN is involved in ribosome biogenesis (19). Specific binding of YxiN to hairpin 92 of the 23S rRNA is mediated by a C-terminal extension of the helicase core (20), which contains a classical RNA recognition motif (21).
YxiN adopts an open conformation in the absence of ligand, or in the presence of nucleotides or RNA. The open conformation is retained upon simultaneous binding of ADP and RNA. In contrast, cooperative binding of RNA and ATP promotes a closure of the cleft between the RecA-like domains in the helicase core, leading to a compact, closed conformation. Upon ATP hydrolysis, YxiN returns to the open conformation. We observe direct switching between these states in smFRET experiments. Altogether, these data can be integrated into a “switch-kink” model for YxiN-mediated RNA unwinding and for RNA unwinding by DEAD-box helicases in general.
Results
Double-Cysteine Mutants for Donor/Acceptor Labeling of YxiN Are Functional Helicases.
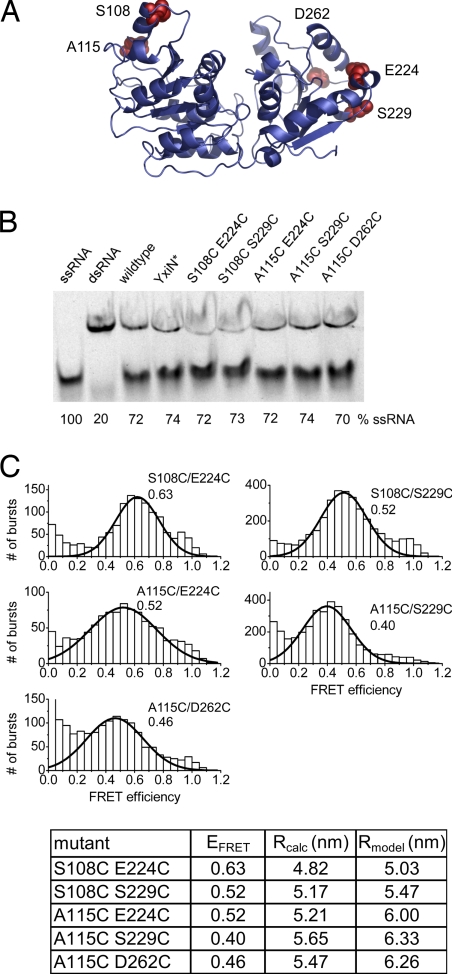
To investigate conformational changes in the catalytic cycle of the RNA helicase YxiN, we constructed double-cysteine mutants with one cysteine for fluorescent labeling on each side of the interdomain cleft. YxiN contains four intrinsic cysteines at positions 61, 243, 247, and 267. According to modification reactions of individual cysteine to serine mutants with Ellman's reagent (22), or Alexa Fluor488− (A488§−) or tetramethylrhodamine-maleimide, two of these cysteines (C243, C247) are not solvent-accessible and do not interfere with labeling reactions at site-specifically introduced cysteines. Therefore, the YxiN C61S/C267S mutant, YxiN*, was generated. Positions 108 and 115 in the N-terminal and positions 224, 229, and 262 in the C-terminal domain of the helicase core were selected for labeling (Fig. 1A), and double-cysteine mutants (S108C/E224C, S108C/S229C, A115C/E224C, A115C/S229C, and A115C/D262C) were generated. All mutants are wild type-like in terms of secondary structure, as demonstrated by their similar far-UV CD spectra (data not shown). They exhibit wild type-like RNA-stimulated ATPase activity, with KM,app values for RNA from 114 to 496 nM (wild type: 156 nM), and kcat values of 0.64–1.41 s−1 (wild type: 1.48 s−1, Table 1). Most importantly, all mutants unwind a model RNA substrate in an ATP-dependent manner (Fig. 1B) with yields comparable with wild-type YxiN, confirming that, despite the modifications, all constructs are functional RNA helicases.
Fig. 1.
Conformation of YxiN in the absence of ligands. (A) Homology model of YxiN (residues 3–367) according to the structure of M. jannaschii DeaD (mjDeaD, PDB ID code 1hv8). The two RecA-like domains of the helicase core are separated by a large interdomain cleft. The positions used for fluorophore attachment, 108 and 115 in the N-terminal, and 224, 229, and 262 in the C-terminal RecA-like domain, are highlighted. (B) RNA unwinding by wild-type YxiN and mutants. All constructs show similar unwinding yields as wild-type YxiN after 30 min of incubation with RNA substrate. (C) FRET histograms (Upper) for donor–acceptor-labeled YxiN constructs. All histograms display a unimodal distribution of FRET efficiencies, EFRET, consistent with a single conformation of YxiN in the absence of ligand. Lines show Gaussian distributions fitted to the data (see SI Table 6). The numbers denote the mean FRET efficiency. The table (Lower) summarizes the values for EFRET, the experimental distances calculated from EFRET, Rcalc, in nm, and the distances calculated from the homology model in A (Cβ–Cβ), Rmodel.
Table 1.
RNA-stimulated ATPase of YxiN constructs
| Mutant | kcat, s−1 | KM,app (RNA), nM |
|---|---|---|
| YxiN (wild type) | 1.48 (±0.22) | 156 (±74) |
| YxiN* | 0.98 (±0.10) | 170 (±61) |
| YxiN* S108C/E224C | 0.85 (±0.07) | 458 (±76) |
| YxiN* S108C/S229C | 0.64 (±0.07) | 114 (±47) |
| YxiN* A115C/E224C | 1.40 (±0.26) | 496 (±180) |
| YxiN* A115C/S229C | 1.30 (±0.25) | 324 (±149) |
| YxiN* A115C/D262C | 1.41 (±0.21) | 422 (±130) |
YxiN Conformation in the Absence of Ligands.
A limited number of crystal structures of DEAD box RNA helicases have been determined, most of these in the absence of ligands (7, 10–12). In all cases, the structures of the individual N- and C-terminal RecA-like domains of the helicase core are highly similar. The domains are splayed apart, with no contacts between them, and their relative orientation varies tremendously between the different structures.
To define the global conformation of YxiN in solution, smFRET experiments were performed on freely diffusing helicase molecules in a confocal microscope. YxiN mutants with one cysteine on each side of the interdomain cleft were labeled with A488 (donor) and Alexa Fluor 546 (A546, acceptor). The donor quantum yields and Förster distances are summarized in supporting information (SI) Tables 2 and 3. FRET histograms for all constructs (Fig. 1C) show a unimodal distribution of FRET efficiencies (EFRET), demonstrating that the helicase in solution adopts a single conformation already in the absence of ligands, in stark contrast to the different conformations observed in the crystal structures. The interdye distances calculated from EFRET distributions, Rcalc, (Fig. 1C) range from 4.82 to 5.65 nm and are in agreement with an open conformation of the interdomain cleft. Because of the low sequence homology between DEAD box helicases outside the conserved motifs, homology models of YxiN were generated with Geno3D by using the structures of eIF4A (PDB ID code 1fuu), mjDeaD (PDB ID code 1hv8), Dhh1p (PDB ID code 1s2m), Vasa (PDB ID code 2db3), and eIF4A-III (PDB ID code 2hyi) as templates (23) (see SI Methods) to facilitate the comparison of our experimental data with the structures of different helicases. The Cβ–Cβ distances for the cysteines used for dye coupling according to these models (SI Table 4) are virtually identical to those determined directly from the crystal structures. Strikingly, all experimental distances are shorter than expected from these models, indicating that the helicase core conformation in solution is much more compact. Most likely, the crystal structures represent an extreme opening of the cleft due to stabilization of artificial open conformations by crystal contacts. Close inspection of the low-FRET region shows that YxiN does not adopt a wide-open conformation similar to the eIF4A structure to any significant extent (SI Fig. 6 and SI Table 5).
The best agreement of the experimental distances is found with the structure of the M. jannaschii DeaD helicase (Fig. 1 A and C), with a mean distance deviation of 0.55 nm, and individual deviations ranging from 0.21 to 0.92 nm. Taking into account the length of the linkers between the cysteines and the attached dyes (0.5–1.0 nm), deviations in this range are expected.
The full width at half maximum (FWHM) of the EFRET distributions is 0.3–0.4, corresponding to an average interdye distance variation of 1.4 nm (SI Table 6). Although the width of the distribution is influenced by the segmental flexibility of the dyes attached to the protein, this broad range of experimental FRET efficiencies points toward a significant flexibility of the interdomain linker and thus of the open conformation of YxiN.
No Influence of Nucleotide and RNA Substrate on the Global Helicase Conformation.
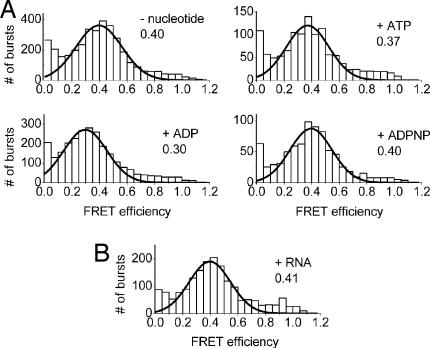
To probe possible nucleotide-induced conformational changes of YxiN, we measured the FRET efficiency of all double-labeled constructs in the presence of saturating amounts of ADP, ATP, or the nonhydrolyzable ATP analog ADPNP (Fig. 2A). According to the KD values of the YxiN/nucleotide complexes (ref. 24 and Fig. 3C), >94% saturation is achieved in these experiments. The FRET histograms are similar to the histograms for YxiN in the absence of nucleotide for each construct, irrespective of the nucleotide present. These results clearly show that the ATP- and ADP-bound conformations of YxiN in solution are virtually identical and resemble the open conformation in the absence of ligand. Hence, nucleotide binding does not induce a significant global conformational change. Similarly, FRET histograms of YxiN in the presence of RNA substrate, but without nucleotide, show no indication for a change of the donor–acceptor distances upon RNA binding (Fig. 2B). Based on the KM,app values for the RNA substrate (Table 1), the saturation of YxiN with RNA is 30–65%, and a second species with a different FRET efficiency would be clearly visible in the histogram. The results are consistent within all constructs and establish that YxiN retains the open conformation of the helicase core upon RNA binding. Neither nucleotide nor RNA alone are sufficient to convert YxiN into a compact, catalytically competent helicase.
Fig. 2.
Nucleotide or RNA binding does not influence the conformation of YxiN. (A) FRET histograms for YxiN* A115C/S229C (A488/A546), in the absence of ligands, and in the presence of ADP, ATP, and the nonhydrolyzable ATP analog ADPNP. (B) FRET histogram for YxiN* A115C/S229C (A488/A546) in the presence of 200 nM RNA substrate. RNA binding does not induce a conformational change of YxiN. Nucleotides or RNA do not affect the FRET efficiencies and thus do not induce a global conformational change. Lines show Gaussian distributions fitted to the data (SI Table 6). The numbers denote the mean FRET efficiency.
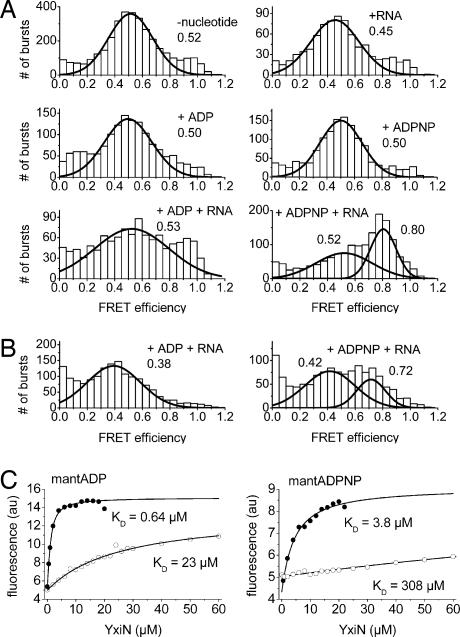
Fig. 3.
Cooperative binding of RNA and ADPNP induces a closure of the interdomain cleft. Neither ADP or ADPNP nor RNA binding show any effect on the helicase conformation, whereas binding of RNA and ADPNP decreases the FRET efficiency from ≈0.5 in the apoprotein to ≈0.8, corresponding to a change in interdye distance from 5.16 to 4.13 nm. No such effect is detected upon binding of ADP and RNA. Consequently, the structures of the helicase in the ATP and the ADP form are significantly different when RNA is bound. (A) FRET histograms for YxiN* S108C/S229C in the absence of ligands, in the presence of ADP or ADPNP, of RNA, or of ADP and RNA or ADPNP and RNA.(B) FRET histograms for YxiN* A115C/S229C in the presence of ADP and RNA or ADPNP and RNA. (C) Cooperative binding of ADPNP and RNA. mantADP (1 μM) (Left) or mantADPNP (1 μM) (Right) were titrated with YxiN (wild type) in the presence (filled symbols) and absence (open symbols) of 154-mer RNA. The KD values determined are 23 (±5.9) μM (mantADP/YxiN) and 308 (±16) μM (ADPNP) in the absence, and 0.64 (±0.05) μM (mantADP) and 3.8 (±0.6) μM (ADPNP) in the presence of RNA, corresponding to a 35-fold increase in mantADP affinity and a 80-fold increase in mantADPNP affinity when RNA is bound.
Cooperative Binding of RNA and ATP Induces a Closure of the Interdomain Cleft.
In contrast, smFRET experiments in the presence of both ADPNP/ATP and RNA substrate, show the appearance of a high-FRET species for all YxiN constructs. The data illustrating the closure of the interdomain cleft in response to ADPNP and RNA binding for the YxiN* S108C/S229C and A115/S229C mutants are shown in Fig. 3, and the distances determined for the closed conformation for all constructs are summarized in Fig. 4. In contrast, no such conformational change occurs in the presence of ADP and RNA (Fig. 3), but the open conformation is retained.
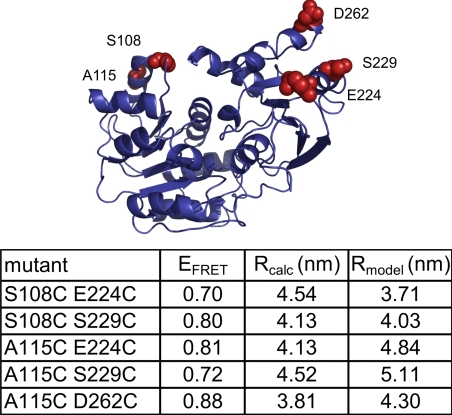
Fig. 4.
The closed conformation of the RNA helicase YxiN. Homology model of YxiN (1–354) based on the crystal structure of the DEAD box helicase Vasa in the presence of RNA and ADPNP (PDB ID code 2db3). The table below summarizes FRET efficiencies of the closed conformation from smFRET experiments for all constructs, the interdye distance calculated from EFRET, Rcalc, and the distance expected from the homology model (Cβ–Cβ), Rmodel, in nm.
The structures of the DEAD box RNA helicases Vasa and eIF4A-III in complex with RNA and ADPNP (refs. 13–15 and Fig. 4) show a closed conformation of the helicase core, with an extensive interface between the two RecA-like domains. The donor–acceptor distances calculated from the FRET histograms of double-labeled YxiN in the presence of ADPNP and RNA agree with the values expected from Vasa and eIF4A-III for all constructs studied here. The average differences between experimental and expected distances are low, with 0.56 nm (eIF4A-III, range 0.18–0.87 nm) or 0.55 nm (Vasa, range 0.11–0.71 nm), which confirms that YxiN adopts a similar compact conformation as Vasa and eIF4A-III in the presence of ADPNP and RNA substrate. Additionally, the FWHM of the EFRET distribution is reduced to 0.1–0.2 in the closed conformation, corresponding to an average interdye distance variation of 0.87 nm (SI Table 6), consistent with an increased rigidity compared with the open conformation.
For a few helicases, cooperative binding of ATP or ADPNP and RNA substrate, but not of ADP and RNA substrate, has been demonstrated (3, 16, 18). We have shown before that mantADP is a suitable fluorescent probe for nucleotide binding to YxiN (24) and we have now investigated the effect of RNA substrate on the interaction of YxiN with nucleotides in fluorescence equilibrium titrations (Fig. 3C). For the mantADPNP/YxiN complex in the absence of RNA, the KD value is 308 (±16) μM. In the presence of RNA, this value decreases 80-fold to 3.8 (±0.6) μM, revealing a clear coupling between mantADPNP and RNA binding. Although mantADP binding is also affected by RNA, the increase in affinity is only 35-fold, with a KD value of 23 (±6) μM in the absence and 0.64 (±0.05) μM in the presence of RNA. As a consequence of these different extents of cooperativity, the affinities for ATP and ADP are different in the absence of RNA [346 (±29) and 55 (±4) μM, respectively (24)] but very similar in the presence of RNA (≈4.3 and ≈1.6 μM). In turn, these results prove that RNA binds 2- to 3-fold more tightly to the ATP form of YxiN than to the ADP form. A similar factor is estimated from comparison of KM,app values for RNA from ATPase assays at saturating and subsaturating ATP concentrations (data not shown).
These results demonstrate that the ATP- and ADP-bound states of RNA helicases display substantially different structures only in the presence of RNA substrate. Although the cooperative binding of ATP and RNA promotes the closure of the interdomain cleft, binding of ADP and RNA is not sufficient for this conformational change.
Direct Transitions Between Open and Closed Conformations.
A strict coupling of the simultaneous binding of ATP and RNA substrate with the conformational change of the YxiN helicase ensures a tight regulation of ATP hydrolysis and RNA helicase activity. Consequently, a coordination of the switching between the two conformations with ATP hydrolysis is expected. The rate constants for ATP hydrolysis by the YxiN constructs at saturating RNA concentrations are 0.64–1.41 s−1 (Table 1), translating into one hydrolysis event every 0.71–1.56 s on average. The observation time in the confocal microscope is typically <5 ms, with only a few events >10 ms. Therefore, the probability to observe such a switching event on this time scale is <1%. Nevertheless we recorded FRET traces with a jump from low to high FRET, corresponding to a closure of the interdomain cleft or with a sudden drop from high to low FRET, reflecting an opening of the interdomain cleft (SI Fig. 7). Although confocal microscopy does not allow kinetic experiments on this time scale, our experiments open avenues to investigate the conformational changes of RNA helicases during their catalytic cycle in real time by increasing the observation time, e.g., by immobilizing the double-labeled helicases on a surface.
In summary, we have shown that ATP and RNA bind cooperatively to the DEAD box helicase YxiN and promote a closure of the interdomain cleft in the helicase core. ATP hydrolysis induces a reopening of the interdomain cleft and reduces the affinity for RNA. The similar closed conformations of YxiN, Vasa, and eIF4A-III suggest a conserved mechanism of energy conversion among DEAD box helicases.
Discussion
We have shown here that the helicase core of the RNA helicase YxiN adopts a flexible, open conformation in solution, with a wide cleft between the two RecA-like domains. In the absence of RNA, this open conformation is retained irrespective of nucleotides bound. Only the cooperative binding of ATP and RNA induces a concerted conformational change, leading to a well defined closed conformation of the helicase core. In single-molecule experiments, the direct switching of the helicase between open and closed conformations during the unwinding cycle is observed.
Flexible proteins such as DEAD box helicases are notoriously difficult to crystallize and are prone to artifacts, which may explain why only a limited number of full-length helicase structures have been determined. SmFRET is an ideal technique to characterize the conformation of such a flexible enzyme in solution. The measured EFRET distributions are rather broad, characteristic of high interdomain flexibility, but do not cover such a broad range as the crystal structures suggest. All interdye distances from FRET experiments are shorter than expected from the structures of open helicase conformations, demonstrating that the crystal structures do not reflect the open conformation of DEAD box helicases in solution. Rather, the opening of the interdomain cleft is exaggerated, probably because of capture of rare conformations that maximize crystal contacts.
In 1998, a nucleotide- and RNA-induced conformational cycle was suggested for the DEAD box helicase eIF4A based on different limited proteolysis patterns in the absence and presence of ADP, ADPNP, or RNA (16, 17). Despite numerous efforts (11, 12, 25, 26), the nature of these conformational changes and the trigger for their interconversion has remained elusive. The proposed closure of the interdomain cleft in helicases in response to ATP binding has been difficult to prove because no ATP-bound DEAD box helicase crystal structure has been reported, and there is no DEAD box helicase for which structures of two (or more) different conformations have been determined. Our single-molecule studies reveal open and closed conformations of the same helicase and demonstrate that neither ADP nor ATP binding alone induce a conformational change in the helicase core. Similarly, the helicase does not respond to RNA binding alone but retains its open conformation. Only when ATP/ADPNP and RNA are bound simultaneously does the closed conformation become populated. The closed conformation is well defined and in agreement with the helicase core conformation in the structures of Vasa (13) and eIF4A-III (14, 15). The structural conservation of the DEAD box helicase core and the identical arrangement of all nine conserved helicase motifs on this scaffold suggest that the closed conformation is conserved among DEAD box helicases across the kingdoms of life.
In crystal structures of open helicase conformations, residues from both RecA-like domains that are involved in ATP hydrolysis are far apart. The ATPase site becomes fully assembled only when the closed conformation is induced (13–15). The switching from an inactive open conformation to the active closed conformation only when both ATP and RNA are present ensures a tight coupling between ATP hydrolysis and structural changes in the RNA substrate. Motif IV is the only motif that interacts with both ADPNP and RNA in the closed conformation (13), which renders this motif a likely candidate to act as a sensor for the two ligand-binding sites. In addition, various “uncoupled” mutants that retain ATPase and RNA binding but lack RNA unwinding activity have been described. These mutants may be deficient in undergoing the conformational change in the presence of RNA and ATP, thus preventing helicase activity.
The structures of Vasa and eIF4A-III (13–15) define the bipartite RNA-binding site constituted by residues on the surface of both RecA-like domains. Tight RNA binding therefore requires contacts with both domains, which is impossible in the open conformation. Chemical and enzymatic footprinting of RNA bound to DbpA revealed that the conformation of helicase-bound RNA is not affected by ADP binding but changes significantly when ADPNP is bound (27). Consistent with this change in footprinting patterns in the presence of ADPNP, the RNA bound to eIF4A-III and Vasa exhibits a kinked backbone, and it was suggested that the kink is stabilized only when ATP binding favors closure of the interdomain cleft that leads to formation of the extended RNA-binding surface (13–15). The kink in the RNA backbone is not compatible with double-stranded RNA (13), and switching between the two helicase conformations thus leads to local RNA unwinding. This distortion of the local RNA structure also explains other structural changes in the RNA substrate promoted by DEAD box helicases, such as displacement of proteins (28).
A destabilization model has been proposed for the mechanism of RNA helicases (1, 29), according to which either ATP binding or ATP hydrolysis induce a closure of the interdomain cleft and thereby change the affinity of the helicase for its RNA substrate. The smFRET experiments allowed us to capture the conformations of the DEAD box helicase YxiN before ATP hydrolysis (in the presence of ADPNP and RNA) and after hydrolysis (in the presence of ADP and RNA), and the open and closed conformations are now unambiguously assigned to the individual ligation states: The interdomain cleft is closed in the ATP state and open in the ADP state.
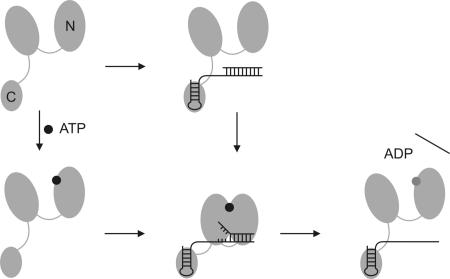
These results can be reconciled in a modified destabilization or “switch-kink” model for DEAD box helicase-mediated RNA remodeling (Fig. 5). In the absence of ligand, or in the presence of either ATP or RNA, the helicase adopts an inactive, open conformation. In the presence of RNA, the helicase acts as a nucleotide-dependent switch: ATP, but not ADP, binding leads to a closure of the interdomain cleft. In the closed conformer, the ATPase site and the bipartite RNA binding site are assembled. At the same time, the unwinding reaction is initiated by kinking the RNA backbone, which separates the adjacent base pairs. When ATP hydrolysis generates the ADP state, the helicase returns to the open form. As a consequence, the ATPase- and RNA-binding sites are disrupted, and the affinity for the RNA is reduced. RNA and ADP release allow for further catalytic cycles.
Fig. 5.
“Switch-kink model” for DEAD box helicase-mediated RNA unwinding. The two RecA-like domains of the helicase core are depicted as large gray ovals (N, N-terminal domain), and the C-terminal domain mediating RNA binding is depicted as a small oval (C, C-terminal RNA-binding domain). The hairpin of the RNA substrate binds to the RNA recognition motif in the C-terminal domain, and ATP binds to the N-terminal RecA domain in the helicase core. Upon binding of RNA and ATP, YxiN adopts a closed conformation. The conformational change of the helicase core assembles the ATPase site and the RNA-binding site, establishing contacts with the adjacent double-helical region of the RNA. Introduction of a kink into the RNA backbone leads to local unwinding (“switch-kink” model). Upon ATP hydrolysis, YxiN returns to the open conformation. The affinity for RNA substrate is reduced, and the RNA is released. Kinking the RNA is not sufficient for unwinding, but an additional “power stroke” upon ATP hydrolysis is required to complete the catalytic cycle.
This “switch-kink” model rationalizes the RNA-stimulated ATPase activity and the cooperative binding of RNA and ATP or ADPNP but not (to the same extent) of RNA and ADP (16, 18, 30, 31) (Fig. 3C). In addition, it illustrates why DEAD box helicases do not act as highly processive motor proteins but as nonprocessive switches. The RNA kink is observed with the nonhydrolyzable ADPNP, suggesting that ATP binding is sufficient to induce this structural change in the RNA. However, even in large excess over RNA substrate, YxiN does not efficiently catalyze RNA unwinding in the presence of ADPNP (ref. 20 and SI Fig. 8). Hence, an additional “power stroke” upon ATP hydrolysis appears to be required to complete the unwinding reaction.
ATP binding and ADP release are rapid compared with the steady-state ATPase rate of YxiN and DbpA (data not shown and ref. 32), leaving either ATP hydrolysis, phosphate release, or an associated conformational change as the rate-limiting step. It is likely that the large conformational change from the open to the closed helicase constitutes this rate-limiting step in the overall helicase cycle. Our experiments open avenues to track this conformational change in the catalytic cycle in real time and to correlate its kinetics with unwinding activity to answer this question.
Materials and Methods
Cloning, Mutagenesis, Protein Production, and Purification.
YxiN wild type was purified as described (24). The YxiN mutant C61S/C267S (YxiN*) was generated via stepwise site-directed mutagenesis (QuikChange; Stratagene) and used as a template for generating double-cysteine mutants for donor–acceptor labeling (S108C/E224C, S108C/S229C, A115C/E224C, A115C/S229C, and A115C/D262C). Sequences were confirmed, and proteins were produced and purified as wild-type YxiN.
Fluorescence Measurements.
Fluorescence measurements were performed at 20°C by using a Fluoromax-3 fluorimeter. mantADP (1 μM) or mantADPNP (1 μM) in 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 2 mM 2-mercaptoethanol were titrated with YxiN in the absence and presence of saturating concentrations of 154-mer RNA substrate (200 nM), and KD values were determined as described (24). Amplitudes in the absence and presence of RNA were fixed during the fit.
Steady-State ATPase Assay.
ATP hydrolysis was monitored in a coupled enzymatic assay at 37°C via the decrease in A340 because of oxidation of NADH to NAD+ (33) as described (24).
RNA Substrates and Unwinding Assay.
A 154-mer comprising nucleotides 2,481–2,634 of the B. subtilis 23S rRNA was generated by T7 polymerase in vitro transcription as described (24). In all experiments, a C2538G/C2606G mutant (lacking an internal T7 promotor sequence) was used.
For RNA-unwinding assays, a minimal helicase substrate consisting of a synthetic, fluorescently labeled 9-mer and a 32-mer was used. Unwinding reactions were performed as described (24).
Fluorescent Labeling.
Proteins were labeled with a mixture of A488- (donor) and A546-maleimide (acceptor), respectively, for 1 h at 25°C in the presence of 1 mM Tris(2-carboxyethyl)phosphine. The ratio of the dyes was varied from 1:0.5:5 (protein/donor/acceptor) to 1:5:10 to maximize the formation of donor/acceptor-labeled protein. Labeling efficiencies were determined from absorbance ratios at 495 nm (A488, corrected for A546 contributions) or 555 nm (A546) and 280 nm (protein, corrected for dye contributions).
Determination of Quantum Yields and Förster Distances.
Quantum yields of the donor A488 attached to positions 108, 115, 224, 229, or 262 of YxiN were determined as described (34) relative to fluorescein in 0.1 M NaOH (φ = 0.92) (35).
Förster distances were calculated according to Eq. 1 (36) from normalized absorbance spectra of acceptor-only labeled protein, εA(λ), normalized fluorescence spectra of the donor-only labeled protein, FD(λ), and the quantum yield of the donor, φD. The refractive index n of water is 1.33, and N is the Avogadro constant. Rapid rotational averaging of the dyes attached to the protein was confirmed in anisotropy decays of singly labeled YxiN (see SI Methods), and the orientation factor κ2 was therefore set as 2/3.
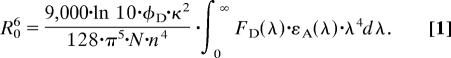 |
Single-Molecule FRET Experiments.
SmFRET experiments were performed by using a home-built confocal microscope (SI Methods). Only fluorescence bursts above a threshold of 50 photons in total were considered in the analysis. Measured background-corrected fluorescence intensities (ID, IA) were corrected for cross-talk (α, donor cross-talk in acceptor channel, β, acceptor cross-talk in donor channel), different quantum yields and detection efficiencies of donor and acceptor fluorescence (γ), and direct excitation of the acceptor (δ) and converted into FRET efficiencies according to Eq. 2.
 |
Measurements were performed at room temperature (25°C) in 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2 with 40 pM YxiN (concentration of donor fluorophore), 5 mM nucleotide, and 200 nM 154-mer RNA.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Ramona Heissmann, Ines Hertel, and Andreas Schmidt for technical assistance; Florian Spreitler for performing initial smFRET experiments and for contributions to the data correction procedure; and Markus Rudolph for critical reading of the manuscript. This work was supported by the VolkswagenStiftung and the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705488105/DC1.
References
- 1.Cordin O, Banroques J, Tanner NK, Linder P. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Rogers GW, Jr, Richter NJ, Merrick WC. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 3.Yang Q, Jankowsky E. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 4.Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. J Mol Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 5.Caruthers JM, Hu Y, McKay DB. Acta Crystallogr F. 2006;62:1191–1195. doi: 10.1107/S1744309106044642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurimoto K, Muto Y, Obayashi N, Terada T, Shirouzu M, Yabuki T, Aoki M, Seki E, Matsuda T, Kigawa T, et al. J Struct Biol. 2005;150:58–68. doi: 10.1016/j.jsb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Caruthers JM, Johnson ER, McKay DB. Proc Natl Acad Sci USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benz J, Trachsel H, Baumann U. Structure (London) 1999;7:671–679. doi: 10.1016/s0969-2126(99)80088-4. [DOI] [PubMed] [Google Scholar]
- 9.Johnson ER, McKay DB. RNA. 1999;5:1526–1534. doi: 10.1017/s1355838299991410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Story RM, Li H, Abelson JN. Proc Natl Acad Sci USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Proc Natl Acad Sci USA. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Z, Coller J, Parker R, Song H. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 14.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 15.Bono F, Ebert J, Lorentzen E, Conti E. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Lorsch JR, Herschlag D. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 17.Lorsch JR, Herschlag D. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 18.Polach KJ, Uhlenbeck OC. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 19.Kossen K, Uhlenbeck OC. Nucleic Acids Res. 1999;27:3811–3820. doi: 10.1093/nar/27.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kossen K, Karginov FV, Uhlenbeck OC. J Mol Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. RNA. 2006 [Google Scholar]
- 22.Ellman GL. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 23.Combet C, Jambon M, Deleage G, Geourjon C. Bioinformatics. 2002;18:213–214. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- 24.Karow AR, Theissen B, Klostermeier D. FEBS J. 2007;274:463–473. doi: 10.1111/j.1742-4658.2006.05593.x. [DOI] [PubMed] [Google Scholar]
- 25.Talavera MA, Matthews EE, Eliason WK, Sagi I, Wang J, Henn A., De La Cruz EM. J Mol Biol. 2006;355:697–707. doi: 10.1016/j.jmb.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 26.Henn A, Shi SP, Zarivach R, Ben-Zeev E, Sagi I. J Biol Chem. 2002;277:46559–46565. doi: 10.1074/jbc.M207438200. [DOI] [PubMed] [Google Scholar]
- 27.Karginov FV, Uhlenbeck OC. Nucleic Acids Res. 2004;32:3028–3032. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 29.Rogers GW, Jr, Lima WF, Merrick WC. J Biol Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 30.Tsu CA, Uhlenbeck OC. Biochemistry. 1998;37:16989–16996. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 31.Iost I, Dreyfus M, Linder P. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 32.Talavera MA, De La Cruz EM. Biochemistry. 2005;44:959–970. doi: 10.1021/bi048253i. [DOI] [PubMed] [Google Scholar]
- 33.Adam H. Methoden der Enzymatischen Analyse. Weinheim, Germany: Bergmeyer; 1962. pp. 573–577. [Google Scholar]
- 34.Parker CA, Rees WT. Analyst. 1960;85:587–600. [Google Scholar]
- 35.Magde D, Wong R, Seybold PG. Photochem Photobiol. 2002;75:327–334. doi: 10.1562/0031-8655(2002)075<0327:fqyatr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Förster T. Disc Faraday Soc. 1959;27:7–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.