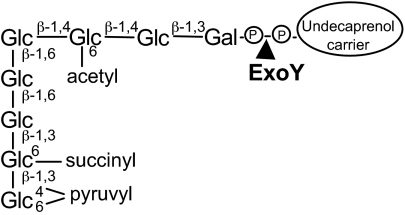Abstract
Sinorhizobium meliloti forms symbiotic, nitrogen-fixing nodules on the roots of Medicago truncatula. The bacteria invade and colonize the roots through structures called infection threads. S. meliloti unable to produce the exopolysaccharide succinoglycan are unable to establish a symbiosis because they are defective in initiating the production of infection threads and in invading the plant. Here, we use microarrays representing 16,000 M. truncatula genes to compare the differential transcriptional responses of this host plant to wild-type and succinoglycan-deficient S. meliloti at the early time point of 3 days postinoculation. This report describes an early divergence in global plant gene expression responses caused by a rhizobial defect in succinoglycan production, rather than in Nod factor production. The microarray data show that M. truncatula inoculated with wild-type, succinoglycan-producing S. meliloti more strongly express genes encoding translation components, protein degradation machinery, and some nodulins than plants inoculated with succinoglycan-deficient bacteria. This finding is consistent with wild-type-inoculated plants having received a signal, distinct from the well characterized Nod factor, to alter their metabolic activity and prepare for invasion. In contrast, M. truncatula inoculated with succinoglycan-deficient S. meliloti more strongly express an unexpectedly large number of genes in two categories: plant defense responses and unknown functions. One model consistent with our results is that appropriate symbiotically active exopolysaccharides act as signals to plant hosts to initiate infection thread formation and that, in the absence of this signal, plants terminate the infection process, perhaps via a defense response.
Keywords: nitrogen fixation, nodule, succinoglycan, microarray, legume
Nitrogen-fixing rhizobial bacteria and their host plants exchange species-specific signals that allow them to form a symbiosis based on nutrient exchange. Medicago truncatula and Medicago sativa (alfalfa) exude flavonoid compounds that are perceived by their rhizobial symbiotic partner S. meliloti (1). These flavonoids stimulate S. meliloti to produce Nod factor (NF), a lipochitooligosaccharide signal that induces multiple responses in host plants (1). These plant responses include induction of root cortical cell divisions resulting in nodule development and curling of plant root hairs that serves to trap a microcolony of S. meliloti within a tight “colonized curled root hair” (CCRH) (2). The bacteria within the CCRH then induce the formation of an infection thread, a progressive ingrowth of the root-hair cell membrane, which grows in a polarized fashion to the base of this cell (2). New infection threads are initiated at each cell layer, allowing the bacteria to penetrate deeper layers of root tissue and delivering the bacteria to the root cortex. The bacteria are engulfed within host membrane-derived compartments of the cortical cells where they differentiate into nitrogen-fixing bacteroids (2).
Analyses of S. meliloti strain 1021 performed over 20 years have revealed the critical role of acidic rhizobial exopolysaccharides in bacterial invasion (1, 2). Succinoglycan (EPS I), a polymer of octasaccharide subunits carrying succinyl, acetyl, and pyruvyl modifications (Fig. 1) is the most intensively studied of these exopolysaccharides (1, 2). Colonies of S. meliloti 1021 grown with the dye Calcofluor were found to fluoresce under UV light, facilitating the isolation of succinoglycan-deficient (exo) mutants (3). Strikingly, these exo mutants were found to be symbiotically deficient, eliciting the formation of small nodules devoid of bacteria. Studies using GFP-marked S. meliloti revealed that succinoglycan is required early in the invasion process, as mutants unable to synthesize succinoglycan are enclosed in CCRH, but do not enable infection thread formation. Acidic exopolysaccharides now are known to be required for a number of other rhizobia to establish a symbiosis (4–6).
Fig. 1.
S. meliloti succinoglycan octasaccharide. ExoY adds the first galactose to the undecaprenol carrier on which succinoglycan is synthesized.
The synthesis of succinoglycan has been studied in detail. Synthesis is initiated by the exoY gene product, which links galactose-1-phosphate to an undecaprenol-phosphate carrier (7) (Fig. 1) and is completed by the addition of seven glucoses and the noncarbohydrate substituents (7).
Succinoglycan biosynthesis now is well characterized, but the molecular mechanisms by which succinoglycan permits nodule invasion by S. meliloti are not understood, although many clues have been obtained. For example, several observations suggest succinoglycan does not act merely by forming a protective layer around the bacteria: (i) the symbiotic function of succinoglycan can be provided in trans between strains in coinoculation experiments (8) and (ii) succinoglycan produced by an exoH mutant, which lacks the succinyl group and is almost completely in the high-molecular-weight (HMW) form, is not symbiotically proficient (9, 10). The existence of plant systems that recognize structural elements of symbiotically active exopolysaccharides is implied by the ability of a particular exopolysaccharide to promote invasion by a rhizobial species on some of its plant hosts but not on other hosts (11, 12).
Evidence suggests that it is the low-molecular-weight (LMW) forms of succinoglycan and other exopolysaccharides that function in symbiosis: (i) the S. meliloti exoH mutant, which produces only HMW succinoglycan, cannot facilitate infection thread formation on alfalfa or M. truncatula (10) (K.M.J. and G.C.W., unpublished data); and (ii) an exoK mutant of Rhizobium NGR234, which produces only HMW exopolysaccharide, cannot invade nodules on some hosts (12). These observations argue that LMW exopolysaccharides may act as signals to the plant that enable critical events necessary for symbiosis. One possible reason for the requirement for LMW exopolysaccharide might be that only LMW forms can reach the root-hair cell membrane to deliver a signal whereas the plant cell wall prevents the access of HMW forms to the cell membrane.
The plant processes that might be affected by such exopolysaccharide-mediated signals have not yet been established. Numerous possibilities have been suggested, ranging from promoting rearrangements of the actin cytoskeleton necessary for infection thread growth to suppression of plant defense responses. Previous studies have shown that failed infections by succinoglycan-deficient S. meliloti are associated with markers of plant defense such as the deposition of callose and phenolic compounds (13). It is not yet known whether a plant defense response is causal for infection thread failure of succinoglycan-deficient S. meliloti (13). However, signs of plant defense are not always associated with failed infections and have not been observed during infection thread failure of the S. meliloti nodF/nodL double mutant on M. truncatula (14, 15).
We reasoned that differential gene expression responses might provide fundamental clues to the mechanisms underlying infection thread failure of succinoglycan-deficient bacteria, and we have used M. truncatula microarrays to compare the plant response to wild-type versus succinoglycan-deficient S. meliloti at 3 days post-inoculation (dpi), an early time point that predates infection thread failure. Our data strikingly show that inoculation with wild-type, succinoglycan-producing S. meliloti results in greater expression of a broad spectrum of translation components, protein degradation machinery, and some nodulins. Because a similar profile of functional categories is induced in plants inoculated with wild-type S. meliloti relative to uninoculated plants (16), this suggests that wild-type S. meliloti induces a metabolic shift that may prepare the plant for infection and that is absent in plants inoculated with succinoglycan-deficient S. meliloti. In contrast, inoculation with S. meliloti lacking succinoglycan results in greater expression of an unexpectedly large number of genes involved in plant defense responses and genes of unknown function relative to plants inoculated with wild-type S. meliloti. This evidence of an early defense response, before visible signs of infection thread failure, is consistent with a scenario in which the lack of a succinoglycan signal itself results in a defense response. One possibility is that, as a strategy for ensuring only an appropriate symbiont gains access, plants treat bacteria in CCRHs as pathogens unless they produce a symbiotically active exopolysaccharide, or some functionally equivalent signal, that can override the defense response.
Results
The S. meliloti exoY Mutant Cannot Form a Symbiosis, but at 3 dpi, M. truncatula Roots Inoculated with Wild-Type or exoY Mutant S. meliloti Are Not Yet Morphologically Distinguishable.
In the past, most studies of the role of succinoglycan in symbiosis have been performed on alfalfa. However, we used the alternative plant host M. truncatula in these analyses because DNA microarrays are available that allow the rapid assay of transcriptional responses. The succinoglycan-deficient S. meliloti exoY mutant cannot form a symbiosis with either alfalfa or M. truncatula (3, 11) [supporting information (SI) Fig. 4].
However, there are no morphological differences yet apparent at 3 dpi between M. truncatula roots inoculated with the wild type or the exoY mutant. At this time, both wild type and the exoY mutant are trapped within CCRHs, but infection threads have not begun to form in response to either strain (Fig. 2). Thus, no failed infection threads are yet visible in roots inoculated with the exoY mutant (Fig. 2). The root-hair curling response is fully induced at 3 dpi by exposure to S. meliloti NF, which both the wild type and the exoY mutant produce in a functional form (8). Roots inoculated with either strain are therefore expected to have similar NF gene expression responses as well as morphological responses.
Fig. 2.
Wild-type S. meliloti 1021 (a–d) and the exoY mutant (e–f) are trapped in CCRHs of M. truncatula at 3 dpi but have not yet initiated infection threads. M. truncatula roots at 3 dpi with GFP-expressing S. meliloti wild type (a–d) or exoY mutant (e–f). (a, c, and e) Brightfield images. (b, d, and f) Brightfield/GFP-fluorescence overlays. (Scale bars: 20 μm.) [In M. truncatula inoculated with strain ABS7 of S. meliloti, infection threads have been observed penetrating the root epidermis at 3 dpi (21, 49). However, for S. meliloti 1021 under our conditions, at 3 dpi, infection threads have not yet initiated.]
A very large population of root cells, including root hairs and cortical cells, exhibits gene expression changes in response to NF (17), which has probably facilitated the detection of gene expression differences between plants inoculated with wild type and NF-deficient S. meliloti mutants. In contrast, the number of root cells exhibiting gene expression changes in response to succinoglycan is predicted to be much smaller, because at most a few dozen CCRHs form per root (18). If the number of root hairs that form infection threads reflects the number of root hairs in which gene expression changes occur in response to succinoglycan, these studies require the detection of exquisitely small differences in gene expression. Nonetheless, we have determined that at 3 dpi, the root gene expression responses to S. meliloti wild type and the succinoglycan-deficient exoY mutant have already diverged considerably.
At 3 dpi, 390 Genes Are Already Strongly Differentially Expressed Between Roots Inoculated with S. meliloti 1021 Wild Type or the exoY Mutant.
M. truncatula roots inoculated with either the wild type or the succinoglycan-deficient exoY mutant were harvested at 3 dpi, and differential gene expression was analyzed by using M. truncatula 16,000-gene microarrays. Of the 5,686 genes expressed sufficiently to produce signal on ≥4 of the 6 array replicates [Gene Expression Omnibus (GEO) data series GSE8509], 1,692 are differentially expressed (SI Table 4). A fold change cut-off of 2 was applied to the genes identified as significant with a false discovery rate (FDR) <0.1% (q value < 0.03) when using Significance Analysis of Microarrays (SAM) (19), which limited the analysis to 390 differentially expressed genes (116 genes expressed ≥2-fold more strongly in the wild-type samples, and 274 genes expressed ≥2-fold more strongly in the exoY samples).
Confirmations of Microarray Results.
Real-time PCR confirmed the results of the microarray for 7 of 8 messages tested when actin 11 was used as the control, and for 6 of 8 messages tested when ubiquitin was used as the control (Table 1). Ubiquitin and actin 11 were chosen because they have been previously used as controls in M. truncatula quantitative PCR (20, 21). The messages listed in Table 1 were chosen for confirmation because they gave strong signals on all six microarray replicates and gene-specific PCR primers could be selected. (SI Table 5 lists primer sequences.)
Table 1.
Real-time PCR and microarray fold change of selected differentially expressed genes
| Gene | Functional class | Medicago Gene Index identifier | Wild type/exoY |
||
|---|---|---|---|---|---|
| Microarray | Real-time PCR (actin 11 normalized) | Real-time PCR (ubiquitin normalized) | |||
| Chloroplast 30S ribosome subunit S14 | Ribosome and translation | TC105272 | 0.2 | 0.6 | 0.9 |
| Inositol polyphosphate-5-phosphatase | Signaling | TC108095 | 0.2 | 0.5 | 0.5 |
| Cinnamyl-alcohol dehydrogenase | Secondary metabolism | TC104873 | 0.3 | 0.9 | 0.5 |
| bZIP transcription factor | Transcription | TC101830 | 0.4 | 0.9 | 0.8 |
| 60S ribosome subunit L18a | Ribosome and translation | TC106500 | 3.5 | 1.1 | 0.9 |
| Ripening related protein | Pathogenesis/defense | TC106515 | 3.5 | 2.6 | 1.5 |
| Glutathione S-transferase | Pathogenesis/defense | TC100571 | 3.8 | 1.00 | 1.1 |
| Cationic peroxidase | Oxidative stress, secondary metabolism, etc. | TC94210 | 3.0 | 1.4 | 0.8 |
Microarray results confirmed for all four messages expressed more strongly in exoY-inoculated plants, relative to either control. Confirmations were more variable for the four messages expressed more strongly in wild-type-inoculated plants. Microarray results for the ripening-related protein message confirmed whether actin 11 or ubiquitin was the control. For both the 60S ribosome subunit L18a message and the cationic peroxidase message, the microarray result was confirmed when actin 11 was the control but not when ubiquitin was the control. For glutathione S-transferase message, microarray result was confirmed when ubiquitin was the control but not when actin 11 was the control
Functional Classes of the Differentially Expressed Genes.
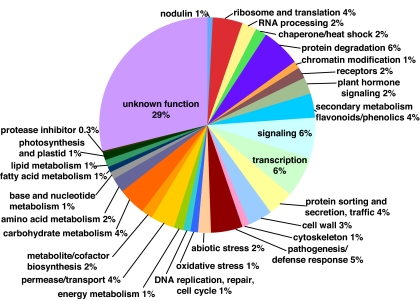
We classified all of the genes analyzed in our data set by predicted function using the Medicago Gene Index and the Gene Ontogeny for the Medicago 16K array,§,¶ but we further refined the analysis by using the entries at the National Center for Biotechnology Information (NCBI) and UniProt. The functional category distribution of all of the genes analyzed is depicted in Fig. 3. We then compared functional categories of all of the genes analyzed versus genes expressed ≥2-fold more strongly in wild-type-inoculated roots or ≥2-fold more strongly in exoY mutant-inoculated roots. Using these comparisons, we calculated the standardized difference score (z score) (Table 2; SI Tables 6 and 7) for each functional class, which provides a statistical measure of the difference between the number of genes expected to be differentially regulated and those observed to be differentially regulated (22). Statistically significant numbers of differentially expressed genes grouping into different functional categories reinforces the argument that the differences in gene expression between two treatments have not occurred by chance and therefore represent real biological differences (22).
Fig. 3.
Percentage of genes in each functional category.
Table 2.
Standardized difference scores (z scores) for genes in each functional class that are significantly differentially expressed in wild-type-inoculated or exoY mutant-inoculated roots
| Functional classes |
z score (observed vs. expected) |
|
|---|---|---|
| Wild-type-inoculated | exoY-inoculated | |
| Pathogenesis/defense | 0.76 | 1.75 (21 vs. 14.5) |
| Unknown function | −3.08 (19 vs. 37.8) | 2.44 (112 vs. 89.3) |
| Ribosome/translation | 8.20 (25 vs. 5.7) | −2.07 (6 vs. 13.4) |
| Protein degradation | 2.01 (13 vs. 7.5) | −0.43 |
| Photosynthesis | 1.99 (4 vs. 1.6) | 0.18 |
| Nodulin, nodulin-like | 1.69 (3 vs. 1.2) | −0.49 |
| Signaling | −1.72 (3 vs. 7.7) | −0.06 |
Positive z scores indicate categories represented at a higher than expected frequency, and negative z scores indicate categories represented at a lower than expected frequency. z scores >1.65 or <−1.65 are significant and shown in bold (90% confidence). See SI Table 6 for values used to calculate z scores.
In wild-type-inoculated roots, the functional categories overrepresented among more highly expressed genes include ribosomal components/translation factors and protein degradation machinery, suggesting active changes in metabolism occur in wild-type-inoculated roots (23). In exoY mutant-inoculated roots, only two functional categories are overrepresented among more highly expressed genes: plant defense genes and genes of unknown function.
Functional Classes Overrepresented Among Genes Expressed More Strongly in Wild-Type-Inoculated Roots.
Ribosomal components and translation factors are highly overrepresented among genes expressed more strongly in wild-type-inoculated roots. It has been reported that genes encoding translation apparatus components are induced in roots by wild-type S. meliloti between 1 and 3 dpi relative to uninoculated roots (16). These findings are consistent with wild-type-inoculated plants undergoing changes in metabolism necessary to create a special environment for symbiosis, whereas this stimulation is absent in exoY mutant-inoculated plants.
Cellular components involved in protein degradation are overrepresented among genes expressed more strongly in wild-type-inoculated roots. The central importance of regulated proteolysis in plant development has been long recognized (23). El Yahyaoui et al. (24) observed differential regulation of components of the protein degradation machinery during nodule development; however, our study reports a significant global regulation of this class associated with nodulation. The expression of a significant number of protein degradation components in wild-type-inoculated roots is intriguing and may suggest that a global developmental change in roots occurs before infection thread formation.
Photosynthesis and plastid component-encoding genes are overrepresented among genes expressed more strongly in wild-type-inoculated roots. Under our growth conditions, all roots experienced slight greening, indicative of chlorophyll production caused by light exposure (25). Nevertheless, the overrepresentation of this category under these conditions was unexpected.
Nodulins are a diverse class of factors induced in host roots during nodule development. M. truncatula nodulins are overrepresented among genes expressed more strongly in wild-type-inoculated roots. However, the overrepresentation consists of only three genes (1.2 genes expected) (Table 2; SI Table 6). In fact, in this study, the vast majority of analyzed nodulins are either not differentially expressed or are differentially expressed by <2-fold (SI Table 4), suggesting that many early responses of plant roots to exoY mutant S. meliloti are normal or nearly normal. For example, early nodulin 12 (ENOD12) (TC101825) has been shown previously to be induced 8.9-fold in plants inoculated with the wild type at 3 dpi (16). However, ENOD12 expression was only 1.4-fold greater in roots inoculated with the wild type than those inoculated with the exoY mutant (SI Table 4).
Intriguingly, one of the nodulin genes that is expressed more strongly in wild-type-inoculated roots is the LysM domain-containing receptor-like kinase 6 (LYK6) (TC104234), which is quite similar to the LYK3 gene encoding the NF entry receptor (15). This finding suggests the interesting possibility that exposure to succinoglycan might stimulate expression of the NF perception machinery and thereby further sensitize the interior of the infection thread to NF. Because NF perception is required for infection thread extension (15), this fits with a model in which NF and succinoglycan work together to facilitate infection thread penetration.
Plant Defense Genes and Genes of Unknown Function Are Expressed More Strongly in exoY Mutant-Inoculated Roots.
Expression of plant defense genes in an unexpectedly large number is the most striking feature of the genes expressed more strongly in exoY mutant-inoculated plants (Table 3). Of 301 defense genes analyzed, 21 are expressed more strongly in exoY mutant-inoculated roots.
Table 3.
Differentially expressed defense and pathogenesis-related genes
| Predicted function | Medicago Gene Index identifier | Fold change wild type-inoculated/exoY-inoculated |
|---|---|---|
| Endo-1,4-β-glucanase | TC109893 | 0.34 |
| β-1,3-glucanase 3 | TC98780 | 0.48 |
| Pathogenesis-related protein 4A | TC94004 | 0.45 |
| Avr9/Cf-9 rapidly elicited protein 231 | TC109373 | 0.48 |
| Syringolide-induced protein 19–1-5 | TC95498 | 0.47 |
| Similar unknown Arabidopsis thaliana protein | TC107534 | 0.38 |
| Disease resistance protein-like MsR1 | TC100870 | 0.45 |
| Probable glutathione S-transferase | TC106945 | 0.46 |
| Probable glutathione S-transferase | TC95231 | 0.46 |
| Glutathione synthetase GSHS1 | TC108090 | 0.48 |
| Blight resistance protein SH20-like | TC96813 | 0.27 |
| Leucine-rich repeat protein | TC108077 | 0.46 |
| Leginsulin | TC94252 | 0.49 |
| Defender against cell death 1 | TC110386 | 0.37 |
| SAG101 | TC108405 | 0.49 |
| Ripening-related protein | TC106496 | 0.44 |
| ADR6 protein | TC100948 | 0.44 |
| ER6 protein universal stress | TC107043 | 0.50 |
| Ubiquitin/ribosomal protein S27a protein | TC93921 | 0.49 |
| Brassinosteroid-regulated protein BRU1 | TC105679 | 0.39 |
| Putative extensin | TC108103 | 0.47 |
| EDGP precursor | TC94310 | 2.11 |
| Glutathione S-transferase 15 | TC100571 | 3.78 |
| Similar to Cf2/Cf5 disease resistance protein | TC103114 | 2.05 |
| Albumin 1 precursor | TC94218 | 2.38 |
| Lipoxygenase | TC100162 | 2.72 |
| Ripening-related protein | TC106515 | 3.52 |
| Ubiquitin/ribosomal S27a fusion protein | TC93958 | 2.50 |
| Extensin-like protein | TC106576 | 2.14 |
Defense genes expressed more strongly in exoY-inoculated roots include a putative blight resistance protein SH20 (TC96813), an endo-1,4-β-glucanase (TC109893) similar to plant cellulases that protect against fungal attack, and defender against cell death 1 (TC110386), which is similar to genes involved in suppression of apoptosis.§
Examples of defense genes expressed during normal nodulation have been noted before (16, 24, 26, 27). Interestingly, Lohar et al. (16) found that defense-related genes are underrepresented among genes induced at 2–3 dpi by wild-type S. meliloti but were significantly overrepresented at the very early time point of 1 h post-inoculation. NF itself might even activate this very early defense response. An intriguing possibility is that exoY mutant-inoculated roots are arrested in the development of the symbiosis such that they fail to suppress a defense response that occurs very early post-inoculation. Succinoglycan is a possible candidate for suppression of such a defense response.
Genes of unknown function were all classed together for our purposes. Although a heterogeneous group, these genes are overrepresented among those expressed more strongly in exoY-inoculated roots, which is consistent with the fact that broad-spectrum gene expression differences between exoY mutant-inoculated and wild-type-inoculated roots may not have been highlighted and studied before. In contrast, this class is underrepresented among genes expressed more strongly in wild-type-inoculated roots, which is not surprising given that a large number of M. truncatula genes induced during a successful symbiosis with the wild type have been identified in previous studies and characterized extensively.
Other Genes of Interest.
Several of the most intriguing differentially expressed genes are predicted to be involved in signal perception and transmission. Two putative receptors expressed more strongly in wild-type-inoculated roots are a β-glucan receptor (TC104170) and a leucine-rich-repeat (LRR) receptor (TC103114). Both of these genes are candidates for a succinoglycan receptor. If succinoglycan were to up-regulate its own receptor, this could increase the supply of receptor available for insertion in the new membrane material at the growing tip of the infection thread. Four different putative receptor genes are expressed more strongly in exoY mutant-inoculated plants (SI Tables 4 and 8). One encodes a CHRK1-like receptor (TC104154), which helps maintain cell specificity and regulates cytokinin levels in Nicotiana tabacum plants (28). Another encodes a PERK1-like receptor kinase, which is induced by wounding and pathogen attack in Brassica napus (29).
Interesting signaling pathway components are expressed more strongly in exoY mutant-inoculated plants, including a predicted inositol polyphosphate-5-phosphatase (TC108095), which is involved in phospholipid signaling, and two calmodulins (TC106874 and TC99865), which are involved in calcium signaling.§ Some of these receptors and signaling components have been selected for inclusion in an RNAi knockdown screen in collaboration with J. Stephen Gantt's laboratory (University of Minnesota) (www.medicago.org/rnai).
Several genes predicted to modulate transcription also are differentially expressed. One, expressed more strongly in exoY mutant-inoculated roots, is predicted to encode a WRKY transcription factor (TC105848),§ which is particularly interesting because WRKY transcription factors regulate plant defense responses (30).
Vesicular trafficking and modulation of the actin cytoskeleton are critical for cellular tip growth, as occurs in root hairs (31). An actin-related protein 8A (TC99917) gene predicted to be involved in polymerization of actin filaments is expressed more strongly in wild-type-inoculated roots (32). Conversely, a predicted profilin (TC99903), a type of protein that prevents actin polymerization, is expressed more strongly in exoY mutant-inoculated roots (33). Perhaps factors that promote actin polymerization are up-regulated in wild-type-inoculated roots that are soon to form infection threads, whereas factors that prevent actin polymerization are expressed more strongly in exoY mutant-inoculated roots. Several genes predicted to encode Rab-like GTPases and ADP-ribosylation factors also are differentially regulated. The precise function of particular Rab-like proteins and ADP-ribosylation factors in tip growth of legume root hairs is not yet known, but this is sure to be an exciting area of discovery (31).
Further Comparisons with Other Studies.
A small number of genes have been previously determined, by Northern blot analysis or real-time PCR, to be differentially expressed between wild-type- and exo mutant-inoculated roots (34–37). Our data set, which represents roots harvested at 3 dpi, is earlier than the time points at which expression was assayed for most of the genes previously determined to be differentially expressed (34–36). MtLEC4, encoding a putative lectin (TC94544), is expressed in wild-type-inoculated roots but not in exo mutant-inoculated roots at 4 weeks post-inoculation (wpi) (§, 34). In our study, no MtLEC4 signal was detected by microarray (GEO GSE8509) or by Northern blot analysis in plants inoculated with either the wild type or the exoY mutant (data not shown). Another gene, MtN1 (TC86337), encoding a putative antimicrobial peptide, is expressed in wild-type-inoculated roots but not in exoY mutant-inoculated roots at 3 wpi, although expression of MtN1 has been detected as early as 4 dpi in wild-type-inoculated roots (35, 36). However, in our data set, no MtN1 signal was detected (GEO GSE8509). The studies of differential gene expression responses of M. truncatula roots at 3–4 wpi, after wild-type-inoculated roots have successfully formed a symbiosis and exoY mutant-inoculated roots have become nitrogen-starved, may have assayed changes that are a downstream consequence of differential invasion events.
Only one gene has a different expression ratio at a comparable time point in our study and a previous study. The MtMMPL1 gene (TC95584), encoding a member of the matrix metalloendoprotease family, previously was found to be expressed more strongly in wild-type-inoculated roots than in exo mutant-inoculated roots at 3 dpi (37), whereas in our study, no significant difference in expression was detected between wild-type-inoculated and exoY mutant-inoculated roots.
Discussion
We have found that M. truncatula roots inoculated with wild-type, succinoglycan-producing S. meliloti express ribosomal components/translation factors and protein degradation machinery more strongly than those inoculated with the exoY mutant, suggesting active changes in metabolism in these roots. In contrast, roots inoculated with a succinoglycan-deficient exoY mutant of S. meliloti express an unexpectedly large number of plant defense genes more strongly than roots inoculated with the wild type. Differential gene expression between roots inoculated with these strains is evident at 3 dpi, before morphological differences in infection threads are visible. Our results suggest that M. truncatula plants sense the presence of succinoglycan early in the infection process and make profound metabolic adjustments that prepare the roots for invasion by S. meliloti. Because NF perception is required for infection thread extension (15, 38), this fits with a model in which NF and succinoglycan work together to facilitate infection thread penetration. In the absence of succinoglycan, these metabolic changes do not occur and plant defense genes are expressed. Plant defense responses have been implicated in the termination of excess infection threads during a normal invasion (39), and it is possible that plant defense is involved in termination of infection by succinoglycan-deficient S. meliloti as well. One possible model is that the plants treat succinoglycan-deficient S. meliloti as although it were an invading pathogen and mount a defense response to inhibit this invasion.
Analysis of M. truncatula invasion phenotypes and gene expression responses to S. meliloti exo mutants that make only HMW or structurally incomplete succinoglycan will likely yield a more complete picture of how exopolysaccharides might signal plant hosts to permit rhizobial invasion. Expression time courses of selected defense genes may clarify the kinetics of defense response induction and suppression by S. meliloti. Analysis of S. meliloti mutants may yield the identity of these defense triggers and clarify the role of succinoglycan in defense suppression.
How general are these findings? Do exopolysaccharides act similarly in other rhizobial symbioses, or even more broadly, might they act similarly as modulators in other bacterial/eukaryotic host interactions? Addressing this issue may be complicated because of the difficulties functional redundancy presents to genetic studies. Rhizobia are now known to synthesize structurally different, but functionally equivalent, acidic exopolysaccharides that play an essential role in nodule invasion by enabling infection thread formation. In retrospect, the choice of strain 1021 for the original genetic studies of succinoglycan was serendipitous because succinoglycan is the only symbiotic exopolysaccharide made in a functional form by Sm1021 (40, 41). We now know that EPS II, which also can promote nodule invasion, is not produced by Sm1021 because of the presence of a insertion sequence in the positive regulator expR (40), whereas the exopolysaccharide K antigen is produced but in a symbiotically inactive form (41). Had a strain that produces a second functional exopolysaccharide been chosen for study, the requirement for symbiotically active exopolysaccharides in invasion would have been missed because of this functional redundancy. Perhaps functional redundancy accounts for the failure to detect a requirement for exopolysaccharide function for symbiosis in some rhizobium/legume pairs (42). More broadly, because many bacteria make multiple surface polysaccharides, the existence of functional redundancy could have obscured key roles for polysaccharides in genetic analyses of other symbiotic or pathogenic interactions between a bacterium and a eukaryotic host.
Materials and Methods
Bacterial Strains.
S. meliloti 1021, the exoY210:Tn5 mutant, the pHC60 plasmid, and growth conditions have been described (3, 10, 43).
Plant Material.
M. truncatula cv. Jemalong A17 seedlings were prepared as described (44, 45), with modifications: plates were supplemented with 1 mM of the ethylene synthesis inhibitor α-aminoisobutyric acid to facilitate nodulation (45); growth was at 25°C, in a 16:8 h light:dark cycle, with roots shielded. Roots were inoculated with 25 ml of OD600 = 0.05 S. meliloti. Root hairs were imaged by using a Zeiss Apotome microscope and Zeiss Axiovision Software.
RNA Isolation and cDNA Production.
Roots from ≈50 plants per experiment were harvested at 3 dpi as described (16). RNA was DNased with RNase-free DNase (Qiagen) or DNA-free DNase (Ambion). DNased RNA was pooled from five separate experiments for each S. meliloti strain treatment. A 66-μg aliquot of each RNA pool was used for cDNA synthesis with random primers by using the 3DNA Array 50 Expression Array Detection Kit (Genisphere).
Microarray Processing and Data Analysis.
M. truncatula genome-wide glass slide microarrays were produced by using the 16,000 70-mer Medicago Array-Ready Oligonucleotide Set (Operon GS-1700–02, Version 1.0). Each of three technical replicate microarrays was hybridized with Cy3-labeled cDNA from S. meliloti wild-type-inoculated plants and Cy5-labeled cDNA from exoY mutant-inoculated plants. Another three microarrays were hybridized with cDNAs labeled in the reverse dye direction (dye swap). Hybridization procedures for the 3DNA Array Expression Array Detection Kit (Genisphere) have been described (16).
Microarray slides were imaged and normalized as described with an Axon scanner and Genepix software (16). Normalization was by local linear regression analysis (LOWESS) and by four-way analysis of variants (ANOVA) (46, 47). Only spots represented on at least four of six slides were included in the analysis.
Reverse Transcription and Real-Time PCR.
Transcriptor Reverse Transcriptase (Roche) reactions were performed on 180 ng of each RNA pool. SYBR green core real-time PCR (ABI) reactions were performed on diluted reverse-transcription reactions in an ABI 7500 Fast Real-Time PCR System. SI Table 5 lists real-time PCR primer sequences and conditions. Ubiquitin (GenBank accession no. AJ245511) (20) and actin 11 (TC106785) (21) were control messages. Other real-time PCR primers were selected by using the Primer 3 software at http://frodo.wi.mit.edu (48).
Functional Classifications of Analyzed Genes.
Classifications of the 5,686 genes analyzed in our data set are based on the Medicago Gene Index and the Gene Ontogeny for the Medicago 16K array mapped to MtGI Release 8.0 and links to NCBI (www.ncbi.nlm.nih.gov) and UniProt (www.pir.uniprot.org).§,¶ z scores were calculated by using the values in SI Table 6 using methods in ref. 22.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Kevin A. T. Silverstein (University of Minnesota) for file updates; Gregory May (Noble Foundation), Chris Town and Foo Cheung [The Institute for Genomic Research (TIGR)], Operon Technologies, and the Medicago community for their roles in development of the Medicago Array-Ready Oligonucleotide Set (GS-1700–02); Deborah Samac (University of Minnesota) and Stephen Bell (Massachusetts Institute of Technology) for real-time PCR access; Alan Grossman (Massachusetts Institute of Technology) for Axon Software access; Dennis Kim (Massachusetts Institute of Technology) for microscope access; and members of the G.C.W. and K.A.V. laboratories for input. This work was supported by National Institutes of Health Grant GM31030 (to G.C.W.), the National Science Foundation Plant Genome Project Grant 0110206, and University of Minnesota College of Biological Sciences support (to K.A.V.). G.C.W. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8509).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709338105/DC1.
The Gene Index Databases, Computational Biology and Functional Genomics Laboratory, Dana Farber Cancer Institute, Boston, MA 02115.
The Institute for Genomic Research (TIGR), Rockville, MD 20850.
References
- 1.Gage DJ. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leigh JA, Signer ER, Walker GC. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolfe B, Carlson R, Ridge R, Dazzo F, Mateos P, Pankhurst C. Aust J Plant Physiol. 1996;23:285–303. [Google Scholar]
- 5.Djordjevic SP, Chen H, Batley M, Redmond JW, Rolfe BG. J Bacteriol. 1987;169:53–60. doi: 10.1128/jb.169.1.53-60.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borthakur D, Barber CE, Lamb JW, Daniels MJ, Downie JA, Johnston AWB. Mol Gen Genet. 1986;203:320–323. [Google Scholar]
- 7.Reuber TL, Walker GC. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 8.Klein S, Hirsch AM, Smith CA, Signer ER. Mol Plant Microbe Interact. 1988;1:94–100. doi: 10.1094/mpmi-1-094. [DOI] [PubMed] [Google Scholar]
- 9.Leigh JA, Reed JW, Hanks JF, Hirsch AM, Walker GC. Cell. 1987;51:579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- 10.Cheng HP, Walker GC. J Bacteriol. 1998;180:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glazebrook J, Walker GC. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 12.Staehelin C, Forsberg LS, D'Haeze W, Gao MY, Carlson RW, Xie ZP, Pellock BJ, Jones KM, Walker GC, Streit WR, Broughton WJ. J Bacteriol. 2006;188:6168–6178. doi: 10.1128/JB.00365-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niehaus K, Kapp D, Puhler A. Planta. 1993;190:415–425. [Google Scholar]
- 14.Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome JC, Denarie J, Truchet G. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 16.Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KA, VandenBosch KA. Plant Physiol. 2006;140:221–234. doi: 10.1104/pp.105.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V. Mol Plant Microbe Interact. 2001;14:737–748. doi: 10.1094/MPMI.2001.14.6.737. [DOI] [PubMed] [Google Scholar]
- 18.Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kalo P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. Plant Cell. 2007;19:1221–1234. doi: 10.1105/tpc.106.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzer P, Bonanomi A, Beyer K, Vogeli-Lange R, Aeschbacher RA, Lange J, Wiemken A, Kim D, Cook DR, Boller T. Mol Plant Microbe Interact. 2000;13:763–777. doi: 10.1094/MPMI.2000.13.7.763. [DOI] [PubMed] [Google Scholar]
- 21.Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, Vandenbosch KA. Plant Physiol. 2004;136:3682–3691. doi: 10.1104/pp.104.045575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan JA, Shirasu K, Deng XW. Nat Rev Genet. 2003;4:948–958. doi: 10.1038/nrg1228. [DOI] [PubMed] [Google Scholar]
- 24.El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al. Plant Physiol. 2004;136:3159–3176. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A. Plant Cell Physiol. 2004;45:1798–1808. doi: 10.1093/pcp/pch205. [DOI] [PubMed] [Google Scholar]
- 26.Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Puhler A, Perlick AM, Kuster H. Mol Plant Microbe Interact. 2004;17:1063–1077. doi: 10.1094/MPMI.2004.17.10.1063. [DOI] [PubMed] [Google Scholar]
- 27.Tesfaye M, Samac DA, Vance CP. Mol Plant Microbe Interact. 2006;19:330–341. doi: 10.1094/MPMI-19-0330. [DOI] [PubMed] [Google Scholar]
- 28.Lee JH, Takei K, Sakakibara H, Sun Cho H, Kim DM, Kim YS, Min SR, Kim WT, Sohn DY, Lim YP, Pai HS. Plant Mol Biol. 2003;53:877–890. doi: 10.1023/B:PLAN.0000023668.34205.a8. [DOI] [PubMed] [Google Scholar]
- 29.Silva NF, Goring DR. Plant Mol Biol. 2002;50:667–685. doi: 10.1023/a:1019951120788. [DOI] [PubMed] [Google Scholar]
- 30.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. Trends Plants Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 31.Samaj J, Muller J, Beck M, Bohm N, Menzel D. Trends Plants Sci. 2006;11:594–600. doi: 10.1016/j.tplants.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 32.McKinney EC, Kandasamy MK, Meagher RB. Plant Physiol. 2002;128:997–1007. doi: 10.1104/pp.010906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snowman BN, Kovar DR, Shevchenko G, Franklin-Tong VE, Staiger CJ. Plant Cell. 2002;14:2613–2626. doi: 10.1105/tpc.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitra RM, Shaw SL, Long SR. Proc Natl Acad Sci USA. 2004;101:10217–10222. doi: 10.1073/pnas.0402186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamas P, Niebel Fde C, Lescure N, Cullimore J. Mol Plant Microbe Interact. 1996;9:233–242. doi: 10.1094/mpmi-9-0233. [DOI] [PubMed] [Google Scholar]
- 36.Gamas P, de Billy F, Truchet G. Mol Plant Microbe Interact. 1998;11:393–403. doi: 10.1094/MPMI.1998.11.5.393. [DOI] [PubMed] [Google Scholar]
- 37.Combier JP, Vernie T, de Billy F, El Yahyaoui F, Mathis R, Gamas P. Plant Physiol. 2007;144:703–716. doi: 10.1104/pp.106.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timmers AC, Auriac MC, Truchet G. Development. 1999;126:3617–3628. doi: 10.1242/dev.126.16.3617. [DOI] [PubMed] [Google Scholar]
- 39.Vasse J, de Billy F, Truchet G. Plant J. 1993;4:555–566. [Google Scholar]
- 40.Pellock BJ, Teplitski M, Boinay RP, Bauer WD, Walker GC. J Bacteriol. 2002;184:5067–5076. doi: 10.1128/JB.184.18.5067-5076.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharypova LA, Chataigne G, Fraysse N, Becker A, Poinsot V. Glycobiology. 2006;16:1181–1193. doi: 10.1093/glycob/cwl042. [DOI] [PubMed] [Google Scholar]
- 42.Hotter GS, Scott DB. J Bacteriol. 1991;173:851–859. doi: 10.1128/jb.173.2.851-859.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glazebrook J, Walker GC. Methods Enzymol. 1991;204:398–418. doi: 10.1016/0076-6879(91)04021-f. [DOI] [PubMed] [Google Scholar]
- 44.Penmetsa RV, Cook DR. Plant Physiol. 2000;123:1387–1398. doi: 10.1104/pp.123.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engstrom EM, Ehrhardt DW, Mitra RM, Long SR. Plant Physiol. 2002;128:1390–1401. doi: 10.1104/pp.010691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang YH, Buckley MJ, Dudoit S, Speed TP. Comparison of Methods for Image Analysis on cDNA Microarray Data. Berkeley: Statistics Dept, Univ of California; 2000. [Google Scholar]
- 47.Kerr MK, Martin M, Churchill GA. J Comput Biol. 2000;7:819–837. doi: 10.1089/10665270050514954. [DOI] [PubMed] [Google Scholar]
- 48.Rozen S, Skaletsky H. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editors. Totowa, NJ: Humana; 2000. pp. 365–386. [Google Scholar]
- 49.Penmetsa RV, Cook DR. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.