Abstract
Genomic multiplication of the locus-encoding human α-synuclein (α-syn), a polypeptide with a propensity toward intracellular misfolding, results in Parkinson's disease (PD). Here we report the results from systematic screening of nearly 900 candidate genetic targets, prioritized by bioinformatic associations to existing PD genes and pathways, via RNAi knockdown. Depletion of 20 gene products reproducibly enhanced misfolding of α-syn over the course of aging in the nematode Caenorhabditis elegans. Subsequent functional analysis of seven positive targets revealed five previously unreported gene products that significantly protect against age- and dose-dependent α-syn-induced degeneration in the dopamine neurons of transgenic worms. These include two trafficking proteins, a conserved cellular scaffold-type protein that modulates G protein signaling, a protein of unknown function, and one gene reported to cause neurodegeneration in knockout mice. These data represent putative genetic susceptibility loci and potential therapeutic targets for PD, a movement disorder affecting ≈2% of the population over 65 years of age.
Keywords: Caenorhabditis elegans, neuroprotection, synuclein
In the advent of complete genomic sequences and technologies for uncovering putative protein interaction networks or whole-genome analyses, scientists have generated many “lists” of candidate genes and proteins that can be harnessed for in-depth analyses of cellular processes or disease states. In the nematode Caenorhabditis elegans these include pioneering studies defining the protein “interactome” (1), the “topology map” for global gene expression (2), and meta-analyses of predicted gene interactions (3). Application of this nematode toward human disease research has already provided insights into the function of specific gene products linked to a variety of neurological disorders (4–6). Given that the average lifespan of this nematode is only 14–17 days, it has been especially useful in its application to diseases of aging. In this study we exploited the potential predictive capacity of these C. elegans bioinformatic databases to discern genetic components and/or pathways that might represent heritable susceptibility factors for Parkinson's disease (PD).
PD involves the progressive loss of dopamine (DA) neurons from the substantia nigra, accompanied by the accumulation of proteins into inclusions termed Lewy bodies. Central to the formation of Lewy bodies is α-synuclein (α-syn), a polypeptide with a propensity toward intracellular aggregation. Genomic multiplication of the WT α-syn locus results in PD, indicating that overexpression of this protein alone can lead to the disease (7). Maintenance of DA neuron homeostasis has been hypothesized to be important for neuroprotection because an imbalance of cytosolic DA may contribute to neurotoxicity. Mechanistically, the selective loss of DA neurons in PD is very possibly due to the presence and chemical nature of DA itself. The capacity of DA for oxidation and its effect on stabilizing toxic forms of α-syn (8) represent a “perfect storm” in the context of the oxidative damage associated with the aging process, other potential environmental insults (e.g., heavy metals and pesticides), or differences in genetic predisposition.
Familial PD has been linked to specific genes, several of which function in cellular pathways involving the management of protein degradation and cellular stress (9). Although most primary insights into the molecular nature of PD have thus far come via genetic analyses of familial forms of PD, there is significant evidence that implicates a combination of environmental factors as pivotal to sporadic causality (10). Improvements in the diagnosis and treatment of PD will be contingent on increased knowledge about susceptibility factors that render populations at risk.
We previously reported the establishment of a nematode model of age-dependent α-syn-induced DA neurodegeneration that has facilitated successful identification of multiple neuroprotective factors, including those that have since been validated in other model organisms and mammals (6). Here we take advantage of the experimental attributes of C. elegans to characterize a set of neuroprotective gene products initially identified in a large-scale candidate gene screen for factors influencing misfolding of human α-syn in vivo by RNAi. These data represent a collection of functionally delineated modifiers of α-syn-dependent misfolding and neurodegeneration that enhance our understanding of the molecular basis of PD and point toward new potential targets for therapeutic intervention.
Results
Overexpression of Human α-Syn in C. elegans.
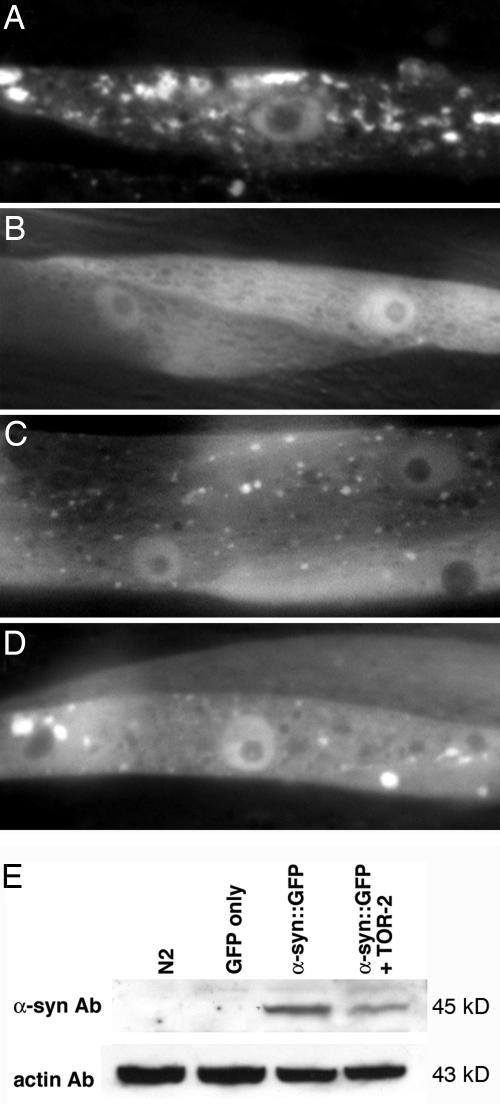
Using a fusion of human α-syn to GFP, we generated transgenic nematodes that enabled us to evaluate the consequences of α-syn overexpression and misfolding in vivo. This isogenic strain contains misfolded α-syn::GFP aggregates in body wall muscles that worsen as these animals develop and age (Fig. 1A). Coexpression of worm torsinA (TOR-2) ameliorated the formation of these fluorescent aggregates, thereby extending prior observations on torsin chaperone activity (Fig. 1B) (4, 11). Expression of an intact α-syn::GFP fusion, with and without TOR-2 coexpression, was verified by Western blotting (Fig. 1E).
Fig. 1.
RNAi knockdown of specific gene targets enhances misfolding of α-syn. (A) Isogenic worm strain expressing α-syn::GFP alone in body wall muscle cells of C. elegans. (B) The presence of TOR-2, a protein with chaperone activity, attenuates the misfolded α-syn protein. (C and D) When worms expressing α-syn::GFP and TOR-2 are exposed to candidate gene RNAi, the misfolded α-syn::GFP returns. (E) Western blot analysis of α-syn::GFP demonstrating the presence of α-syn::GFP in worms with and without TOR-2 coexpression. Actin was used as a loading control.
Worms expressing both α-syn::GFP and TOR-2 in body wall muscles represented a genetic background whereby subtle changes in α-syn misfolding could be reproducibly and effectively discerned via RNAi screening. The reasoning behind use of the body-wall muscles was 2-fold; first, these are the largest, most readily scored cell type in adult C. elegans within which to accurately judge changes in α-syn misfolding, and, second, C. elegans DA neurons are recalcitrant to RNAi (12). Moreover, we theorized that the presence of TOR-2, a protein with chaperone activity, served to maintain overexpressed α-syn at a threshold of misfolding, thereby enabling identification of genetic factors that more readily effect the formation of misfolded oligomers, or less mature α-syn aggregates, currently considered to be the more toxic species associated with degeneration (13, 14).
Hypothesis-Based RNAi Screening for Effectors of α-Syn Misfolding.
To investigate putative effectors of α-syn misfolding, we have systematically screened 868 genetic targets with the potential to influence PD by selecting for candidates that, when knocked down, enhanced age-associated aggregation of α-syn::GFP. We used the C. elegans orthologs of established familial PD genes as the foundation for constructing a candidate gene list [supporting information (SI) Table 3]. The worm genome includes orthologs of all established familial PD genes (Parkin, DJ-1, PINK1, UCHL-1, LRRK2, PARK9, and NURR1) with the exception of α-syn. Specific C. elegans bioinformatic datasets were subsequently mined to define hypothetical interrelationships between the worm PD orthologs and previously unrelated gene targets. For example, using the C. elegans topology map (2), we identified all gene products that are coexpressed with the worm PD orthologs within a radius of one. Additionally, we identified all gene products that interact with these PD orthologs, as assessed by the worm interactome (1). Also included among our RNAi targets were the worm orthologs of genes that were uncovered via screens for effectors of α-syn toxicity in Saccharomyces cerevisiae (6, 15), as well as genes encoding nematode versions of proteins identified in a proteomic analysis of rotenone-induced Lewy bodies in DA neuron cell cultures (16). We further extended our RNAi target gene set by identifying worm homologs of gene products ascribed to encompass the cellular protein degradation machinery. These included genes annotated in Wormbase as being involved in the ubiquitin–proteasome system, unfolded protein response, endoplasmic reticulum-associated degradation, and autophagy. Gene candidates derived from these pathways were assessed for homology to mammals, and nonconserved genes were excluded because it has been estimated that 47% of worm genes have no visible homology to mammals (17). SI Table 3 shows 868 candidate genes targeted for knockdown. Furthermore, we have constructed a relational interconnectivity map depicting gene targets classified in more than one category (SI Fig. 3).
These candidate gene targets were knocked down by using RNAi, a method that is both rapidly and economically performed in C. elegans by feeding worms target-specific dsRNA-producing bacteria (18). In total, 13% (111/868) were lethal; however, the remaining 757 genes were analyzed for accumulation of misfolded α-syn protein. The primary RNAi screen of adult stage worms (44–48 h after eggs were laid at 25°C) revealed that 17% (125/757) of these gene targets enhanced aggregation of α-syn in worms coexpressing α-syn::GFP and TOR-2. The misfolded protein appeared over developmental time and was randomly distributed in the cytoplasm of the body-wall muscle cells (Fig. 1 C and D). RNAi was performed on 20–30 animals in duplicate for each gene. As would be expected, a significant number of genes that alter folding or protein degradation were identified (SI Table 3). Notably included within this collection of positives were worm orthologs of five familial PD genes: Parkin (K08E3.7/pdr-1), DJ-1 (B0432.2/djr1.1), PINK1 (EEED8.9/pink-1), NURR1 (C48D5.1/nhr-6), and PARK9/ATP13A2 (W08D2.5) (9, 19).
Because PD is a disease of aging, we reasoned that gene products that play a more significant functional role in the management of α-syn misfolding or clearance would exhibit a stronger effect at an earlier age. In this regard, a secondary screen of the top 125 candidates was performed in worms at the L3 larval stage of development (32–36 h after eggs are laid). This resulted in further reduction of candidates where only ≈3% (20/757) of genes enhanced misfolding of human α-syn after RNAi treatment at this earlier developmental stage (Table 1 and SI Table 3). Retained within this list of 20 more stringently selected hits were orthologs of known recessive PD genes, DJ-1 and PINK1, thereby representing internal validation of the screen. Another expected control for the screen, the C. elegans torsinA gene homolog tor-2, was also recovered. A notable gene from this dataset was T07F12.4, a serine–threonine kinase that is homologous to UNC-51, a protein similar to yeast Atg1p, required for autophagy, that also plays a role in axon elongation (20). A human ortholog of worm UNC-51, termed ULK2, was recently identified by geneticists as one of only six genes that were distinguished as significant in a genome-wide association study of single-nucleotide polymorphisms within PD patients (21). The remaining 16 (of 20) positives encompass gene products previously unassociated with either α-syn function or PD.
Table 1.
Gene identities of the 20 top candidates isolated from RNAi screening
| C. elegans gene ID | National Center for Biotechnology Information eukaryotic orthologous groups (KOGs) |
|---|---|
| B0432.2 (djr-1.1) | Putative transcriptional regulator DJ-1 |
| T05C3.5 (dnj-19) | Molecular chaperone (DnaJ superfamily) |
| C35D10.2 | RGS–GAIP interacting protein GIPC |
| C54H2.5 (sft-4) | Putative cargo transport protein ERV29 |
| EEED8.9 (pink-1) | BRPK/PTEN-induced protein kinase |
| F11H8.1 (rfl-1) | NEDD8-activating complex, catalytic component UBA3 |
| F16A11.2 | Uncharacterized conserved protein |
| F26E4.11 (hrdl-1) | E3 ubiquitin ligase |
| F32A6.3 (vps-41) | Vacuolar assembly/sorting protein VPS41 |
| F48E3.7 (acr-22) | Acetylcholine receptor |
| F55A4.1 | Synaptobrevin/VAMP-like protein SEC22 |
| F57B10.5 | Emp24/gp25L/p24 family of membrane trafficking proteins |
| F59F4.1 | Acyl-CoA oxidase |
| K11G12.4 (smf-1) | Mn2+ and Fe2+ transporters of the NRAMP family |
| M7.5 (atgr-7) | Ubiquitin-activating E1 enzyme-like protein |
| R05D11.6 | Transcription factor |
| T07F12.4 | Serine–threonine protein kinase involved in autophagy |
| T08D2.4 | Not available |
| T13A10.2 | Predicted E3 ubiquitin ligase |
| Y37A1B.13 (tor-2) | ATPase of the AAA+ superfamily |
Examination of Knockdown of α-Syn Modifiers on Polyglutamine-Induced Protein Aggregation.
Our identification of gene products that influence the misfolding of α-syn does not preclude the possibility that these proteins play a more generalized role in regulating protein misfolding or degradation. Previous screens in both C. elegans and yeast have implicated various classes of gene products that influence the misfolding or clearance of polyglutamine repeat-containing proteins (15, 22). In comparing the gene sets identified in those studies to our list of 125 less stringent modifiers of α-syn misfolding, we determined that only one positive gene was shared between these datasets, the C. elegans HSF-1 protein. HSF-1 is a critical evolutionarily conserved regulator of chaperone gene expression that would be presumed to exhibit a generalized function in mediating protein misfolding.
To further explore the prospect that the specific genes identified in our screen potentially act in a more generalized capacity, we used RNAi knockdown to evaluate loss of function associated with our strongest 20 α-syn modifiers in transgenic worms expressing a polyglutamine–GFP fusion protein. The results of this analysis indicate that RNAi knockdown of these targets had no significant influence on polyglutamine-dependent aggregation in vivo (SI Table 4 and SI Fig. 4). The sole exception was the TOR-2 chaperone-like protein, which served as a control in this analysis, because this protein has been shown to suppress polyglutamine aggregation in C. elegans (4). These data are consistent with a previous report that demonstrated that the toxicity mediated by overexpression of α-syn versus a mutant huntingtin fragment in yeast was regulated by nonoverlapping gene sets (15). In all, this analysis demonstrates that the strongest α-syn effector genes identified through our RNAi screening do not exert their influence via a general effect on protein misfolding, but more specifically contribute to cellular pathways associated with α-syn.
Identification of Neuroprotective Gene Products.
A distinct advantage of using C. elegans for functional investigation of gene activity is the level of accuracy that can be obtained in evaluating neurodegeneration. C. elegans has precisely eight DA neurons, with three pairs of neurons in the anterior [designated dorsal/ventral cephalic (CEP) and anterior deirid (ADE)] and one pair in the posterior [posterior deirid (PDE)] of the animal. We have established that overexpression of WT human α-syn under the control of a DA neuron-specific promoter (Pdat-1; DA transporter) results in age- and dose-dependent neurodegeneration. We generated two separate transgenic lines of animals that express α-syn at different levels, based on semiquantitative RT-PCR analysis (SI Fig. 5A). At day 7 of adulthood, 87% of animals expressing a higher level of α-syn show DA neurodegeneration (SI Fig. 5B) whereas 75% of animals expressing α-syn at lower levels display degenerative changes (data not shown). The loss of DA neurons also occurs as animals age, and no degeneration (0%) is observed in control animals (Pdat-1::GFP) lacking α-syn overexpression (SI Fig. 5B). Previously, these same animals have been used to validate the neuroprotective capacity of both worm TOR-2 and mammalian Rab1A, a GTPase involved in endoplasmic reticulum-to-Golgi transport (5, 6). Here we further extend our functional characterization of genes that resulted in enhanced α-syn misfolding when depleted by RNAi by systematically testing their prospective neuroprotective potential in vivo.
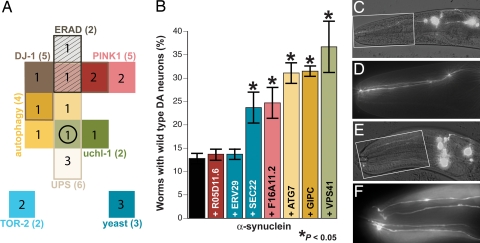
Fig. 2A depicts a classification of the 20 positives where they are displayed according to their bioinformatic associations; several of these candidates shared more than one bioinformatic relationship (SI Table 5). For example, the C. elegans ORF F32A6.3 encodes a gene (vps-41) that is coexpressed in microarrays with the worm ortholog of UCHL-1, contains a RING-finger motif common to E3 ligases, and is involved in autophagy (Fig. 2A, circle). We used inferred relationships between genes exhibiting such overlap to prioritize subsequent construction of transgenic animals to examine their ability to influence DA neuron survival.
Fig. 2.
Overexpression of candidate genes protects DA neurons from α-syn-induced degeneration. (A) Schematic representation of the distribution of the top 20 candidate genes, with bioinformatic associations used to select the gene for knockdown. Six genes identified from two or more categories are indicated with overlapping color. (B) Graph depicting the percentage of α-syn-expressing worms with WT DA neurons in 7-day-old adults when candidate genes are coexpressed. *, P < 0.05 (Student t test). (C–F) Worm DA neurons at low (C and E) and high (D and F) magnification. (C and D) At the 7-day stage, most worms expressing α-syn are missing anterior DA neurons (only two of four CEP DA neurons present). (E and F) Overexpression of C. elegans VPS41 protects from DA neurodegeneration, whereby worms display all four CEP neurons. Population analysis revealed that 36.7% of worms overexpressing VPS41 were WT compared with only 12.8% of α-syn only controls.
Transgenic animals coexpressing cDNAs corresponding to seven prioritized positive targets from the RNAi screen were generated and crossed to isogenic lines of worms expressing α-syn in DA neurons (5, 6). Overexpression of α-syn alone resulted in significant degeneration, where only 12.8% of worms displayed WT DA neurons when they were assayed at the 7-day-old stage (Fig. 2 B–D). Two genes exhibited an insignificant level of DA neuroprotection; these encoded an uncharacterized transcription factor (R05D11.6) and a C. elegans ortholog of Erv29p (C54H2.5), a vesicle-associated protein involved in endoplasmic reticulum-to-Golgi transport (Fig. 2B). Strikingly, coexpression of five of seven candidate genes examined significantly rescued DA neurodegeneration with average WT worm populations from 24% to 37% (Fig. 2B). Worms were scored as WT when all six anterior DA neurons were intact (Fig. 2 E and F). Transcription of all of the gene products tested was verified via semiquantitative RT-PCR (SI Fig. 6). Three independent transgenic lines were scored per gene tested, with 30 animals analyzed in triplicate experimental trials. In considering published evidence that TOR-2, DJ-1, and PINK1 have all previously been shown to be neuroprotective as well (5, 23–26), these combined results indicate that the strategy used in our screen is highly predictive of neuroprotective genetic modifiers.
The five genes that display significant neuroprotection (P < 0.05, Student t test) in our analysis included (i) F32A6.3, the worm ortholog of VPS41, a conserved vesicular protein necessary for lysosomal biogenesis; (ii) C35D10.2, classified as GIPC, a PDZ-domain containing protein that interacts with a vesicular GTPase named RGS-GAIP involved with G protein-coupled signaling; (iii) M7.5, an ORF corresponding to an autophagy-associated regulatory gene termed ATG7; (iv) F16A11.2, a hypothetical protein of unknown function with high amino acid sequence identity to an uncharacterized human gene product; and (v) F55A4.1, the worm ortholog of Sec22p, a well characterized vesicular trafficking protein in yeast. The Blast E-values for the Homo sapiens orthologs of these C. elegans gene products indicate that they are all highly conserved (Table 2).
Table 2.
Summary of the neuroprotective genes and their human homologs
| C. elegans gene ID | Average % of worms with WT DA neurons* | NCBI KOGs | Relevance to H. sapiens |
|
|---|---|---|---|---|
| Blast E value | % length | |||
| F32A6.3 (vps-41) | 36.7 ± 5 | Vacuolar assembly/sorting protein VSP41 | 5.1e−96 | 95.7 |
| C35D10.2 | 31.5 ± 1 | RGS–GAIP interacting protein GIPC | 1.1e−49 | 75.4 |
| M7.5 (atgr-7) | 31.1 ± 1 | Ubiquitin-activating E1 enzyme-like protein | 7.4e−87 | 69.2 |
| F16A11.2 | 24.8 ± 3 | Uncharacterized conserved protein | 1.2e−207 | 99.8 |
| F55A4.1 | 23.7 ± 3 | Synaptobrevin/VAMP-like protein SEC22 | 2.3e−47 | 96.3 |
*Compared with 12.8% with α-syn alone.
Discussion
The key pathological hallmarks of PD include the development of α-syn containing protein inclusions and DA neurodegeneration. Although it remains unclear whether mature α-syn aggregates or Lewy bodies are causative for PD, evidence suggests that factors that influence the misfolding and oligomerization of this polypeptide lead to neurotoxicity (13, 14). Regardless, proteins that play a role in protecting DA neurons from the degenerative loss associated with α-syn overproduction are candidate susceptibility markers as well as potential targets for therapeutic development. Here we have combined these distinct PD-associated phenotypic readouts to discern gene products with functional consequences for PD.
Among the gene products identified via this screen, a protein that demonstrated high neuroprotective capacity was C. elegans VPS41. VPS41 is highly conserved across species and has been best characterized in S. cerevisiae, where it is involved in trafficking from the trans-Golgi to the vacuole, the yeast equivalent of the lysosome (27). Little is known about the precise function of VPS41 in mammalian systems; however, in situ hybridization predicts the VPS41 gene to be expressed in brain neurons, with strong expression localized to the DA neurons of the substantia nigra (28).
Evidence for lysosomal system dysfunction is emerging as a potential consequence of α-syn cytotoxicity. α-Syn is degraded in part by the lysosomal pathway, under the regulation of the cochaperone CHIP (29), and mutant forms of α-syn can block chaperone-mediated autophagy (30). Therefore, lysosomal failure has been proposed as a mechanism underlying the age dependence of PD (31). PARK9, a hereditary form of parkinsonism with dementia, has been recently linked to mutation of a lysosomal ATPase (19). Notably, the worm homolog of PARK9, W08D2.5, was uncovered in our original RNAi screen (one of 125 initial hits) where knockdown led to α-syn aggregation. This gene product is also neuroprotective when overexpressed in DA neurons (S.H., K.A.C., and G.A.C., unpublished observations).
Our identification of C. elegans ATG7 as a neuroprotective gene product is further suggestive of a significant role for autophagy and lysosomal function in restoring homeostatic balance to DA neurons in response to excess α-syn. ATG7 is an E1-like enzyme required for the initiation of autophagosome formation. Added validation for these worm data comes from mammals where it was shown that loss of the Atg7 gene in mice results in neurodegeneration and that this protein may function to prevent neuronal impairment and axonal degeneration (32, 33).
Among the factors that mediate DA neuron homeostasis is the interplay of DA production, transport, and receptor signaling. In the “classical” view of DA, D2 autoreceptors modulate a putative presynaptic feedback mechanism resulting in a net neuroprotective effect (34). As the complexities of D2 signaling continue to be unraveled, it is critical to consider that this model does not take into account the largely unknown impact of α-syn misfolding and overabundance associated with PD.
Here we describe evidence indicating that GIPC (GAIP-interacting protein, C terminus), a conserved cellular scaffold-type protein, has the capacity to function in a neuroprotective manner against α-syn-induced neurodegeneration. GIPC has been shown to interact with mammalian D2 and D3 receptors in heterologous cell cultures, where its expression appears to mediate endosomal trafficking and receptor stability (35). GIPC was originally identified in a screen for proteins that bind to GAIP (Gα-interacting protein) (36), a member of the large family called regulators of G protein signaling (RGS), yet GAIP is the only RGS protein that binds GIPC. Overexpression of GAIP has been shown to stimulate protein degradation via Gαi-mediated induction of autophagy in human intestinal cells (37). It is interesting to speculate that GIPC serves to modulate a presynaptic protein-coupled pathway that can somehow combat the effects of α-syn misfolding and accumulation, perhaps by a DA or DA receptor-regulated manner.
Our data demonstrating that the C. elegans F55A4.1 gene product, an ortholog of Sec22p, is neuroprotective accentuates the importance of vesicular trafficking between the endoplasmic reticulum and Golgi as an integral process affected by α-syn. We previously hypothesized that α-syn-dependent blockage of vesicular trafficking could lead to the limitation of available monoamine vesicular transporters (i.e., VMAT2) (6). This would theoretically result in an excess pool of cytosolic DA and contribute to selective DA neurodegeneration. Indeed, α-syn overexpression may exacerbate this process, leading to increased cytosolic catecholamine concentration (38, 39). Thus, cellular proteins, like Sec22p or those in the Rab GTPase family (i.e., Rab1) that enhance vesicular trafficking and the removal of DA from the cytosol, likely contribute to neuroprotection by relieving this α-syn-mediated blockade (6). Although hypotheses focused on the intrinsic contributions of DA to cytotoxicity are appealing, it is important to remember that other neuronal subtypes are also susceptible in PD and that disruption of basic cellular functions has implications beyond the DA system.
The candidate gene approach may limit the ability to make generalized conclusions about all possible gene families, pathways, or nonbiased gene sets that could potentially be revealed by genome-wide screening. By design, the genes preselected for analysis in our RNAi screen will not reveal all possible effectors of α-syn misfolding and neuroprotection in C. elegans. They are further limited by factors such as lethality and redundancy. Nevertheless, this focused strategy did not restrict the detection of unexpected effectors, as evidenced by the identification of the F16A11.6 gene product, which has not been previously linked to neuroprotection. This protein, which contains an RNÁ-binding motif, has >99% identity to an uncharacterized human gene product that is associated with neuronal RNA-rich granules where it may be involved in transport (40). This is intriguing considering the recent discovery that miRNAs in DA neurons may play a role in neurodegenerative process (41).
We contend that our a priori elimination of targets without significant homology to mammals, as well as prioritizing targets with putative relationships to known PD genes via our “guilt by association” bioinformatics selection strategy, significantly enhanced our ability to identify functionally relevant effectors. For example, we determined that genes coexpressed with known PD genes that are also components of cellular pathways implicated in PD have a far greater likelihood of significantly effecting α-syn misfolding (17% vs. 3% strong positives for the entire gene set) (SI Fig. 7A). Furthermore, we discovered that 11% (3/28) of genes coexpressed with both DJ-1 and PINK1 were significantly enriched within our top 20 hits, as compared with the other top candidates that represented a 3% (17/757) hit rate (P < 0.05, Fisher exact test) (SI Fig. 7B). In the era of completed genome sequences, this directed approach is applicable to other disease gene studies in model organisms and serves to accelerate the pace of gene discovery. With continued expansion in large-scale genomic/proteomic studies, established means for rapid functional validation of gene candidates will be important and necessary for reconciling disparate datasets, such as those obtained from the initial genome-wide analyses on PD (42).
Taken together, the results of this study indicate that further characterization of the genes identified in our RNAi screen will yield additional insights into mediators of α-syn-induced cytotoxicity. An emerging model underlying PD involves dysfunction within a variety of intersecting pathways that maintain homeostasis via a compensatory balance between intracellular protein trafficking and degradation systems, as well as other signaling mechanisms triggered by stress. The manner by which α-syn impacts these mechanisms remains poorly defined, and factors that influence the stability, production, and clearance of this protein likely represent effectors of disease onset and progression. Identification of critical cellular mediators within these processes will enhance development of biomarkers and therapeutic agents to halt this disease.
Methods
Nematode Strains.
Nematodes were maintained by using standard procedures (43). To make transgenic lines, each expression plasmid was injected into WT N2 (Bristol) worms at 50 μg/ml. For the RNAi screen, UA52 [baInl12; Punc-54::gfp, rol-6 (su1006)], UA49 [baInl2; Punc-54::a-syn::gfp, rol-6 (su1006)], and UA50 [baInl13; Punc-54::a-syn::gfp, Punc-54::tor-2, rol-6 (su1006)] were integrated as previously described (5) and outcrossed at least three times to N2 worms. The polyglutamine aggregation analysis was performed by using integrated isogenic strain UA6 [UA6 (baIn6)] coexpressing Q82::GFP and TOR-2 (4). Details on strains and crosses are available in SI Materials and Methods.
Plasmid Constructs.
Plasmids were constructed by using Gateway Technology (Invitrogen); details are available in SI Materials and Methods.
Preparation of Worm Protein Extracts and Western Blotting.
Worm protein extracts were prepared and Western blotting was performed as described previously (5). Details are available in SI Materials and Methods.
RNAi Screen and Analysis of α-Syn Misfolding or Polyglutamine Aggregation.
RNAi feeding was performed as described (18) with modifications listed in SI Materials and Methods. Bacterial clones leading to enhanced α-syn misfolding were tested in two trials, and the clones resulting in significant aggregation (80% of worms with increased quantity and size of α-syn aggregates) were scored as positive. For each trial, 20 worms were transferred onto a 2% agarose pad, immobilized with 2 mM levamisole, and analyzed. The identities of the top 20 positive hits from the RNAi screen were sequence-verified. For polyglutamine aggregation analysis, 20 worms at the L3 stage were scored for aggregate number in two separate trials.
Candidate Gene Analysis for Neuroprotection.
Synchronized embryos expressing GFP and DsRed2 were transferred onto NGM plates and grown at 20°C for 7 days. For each trial, 30 worms were transferred to a 2% agarose pad, immobilized with 2 mM levamisole, and scored. Worms were considered rescued when all four CEP and both ADE neurons were intact and had no visible signs of degeneration. Each stable line was analyzed three times (90 worms per transgenic line). Three separate transgenic lines were analyzed per gene (270 animals per gene).
RNA Isolation and Semiquantitative RT-PCR.
Worms were harvested and snap-frozen in liquid nitrogen. After total RNA and cDNA preparation, semiquantitative RT-PCR was performed as previously described (44). Protocols and primer sequences are available in SI Materials and Methods.
Imaging and Statistics.
Imaging and statistics details are available in SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We acknowledge the cooperative spirit of all members of the K.A.C. and G.A.C. laboratory. Special thanks go to Michelle Norris (RNAi screening), Lynn Boyd (E3 ligase cDNAs), Cody Locke (RNA isolation), and A. J. Burdette (RT-PCR), as well as David Standaert, Erich Schwarz, and Laura Berkowitz for helpful discussions. This work was supported by the Bachmann–Strauss Dystonia & Parkinson Foundation, the United Parkinson Foundation, the American Parkinson Disease Association, the Parkinson's Disease Association of Alabama, the Michael J. Fox Foundation for Parkinson's Research, and an Undergraduate Research Science Program Grant from the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: G.A.C. and K.A.C. serve as scientific advisors to QRxPharma, Ltd., from whom they receive monetary compensation and a sponsored research agreement.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711018105/DC1.
References
- 1.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SK, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 3.Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell GA, et al. Suppression of polyglutamine-induced protein aggregation in Caenorhabditis elegans by torsin proteins. Hum Mol Genet. 2003;12:307–319. doi: 10.1093/hmg/ddg027. [DOI] [PubMed] [Google Scholar]
- 5.Cao S, Gelwix CC, Caldwell KA, Caldwell GA. Torsin-mediated neuroprotection from cellular stresses to dopaminergic neurons of C. elegans. J Neurosci. 2005;25:3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singleton AB, et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 8.Conway KA, et al. Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early-onset Parkinson's disease: Implications for pathogenesis and therapy. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 10.Tanner CM. Is the cause of Parkinson's disease environmental or hereditary? Evidence from twin studies. Adv Neurol. 2003;91:133–142. [PubMed] [Google Scholar]
- 11.McLean PJ, et al. TorsinA and heat shock proteins act as molecular chaperones: Suppression of alpha-synuclein aggregation. J Neurochem. 2002;83:846–854. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- 12.Asikainen S, Vartiainen S, Lakso M, Nass R, Wong G. Selective sensitivity of Caenorhabditis elegans neurons to RNA interference. NeuroReport. 2005;16:1995–1999. doi: 10.1097/00001756-200512190-00005. [DOI] [PubMed] [Google Scholar]
- 13.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 14.Lee HJ, Khoshaghideh F, Patel S, Lee SJ. Clearance of alpha-synuclein oligomeric intermediates via the lysosomal degradation pathway. J Neurosci. 2004;24:1888–1896. doi: 10.1523/JNEUROSCI.3809-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, et al. Analysis of alpha-synuclein-associated proteins by quantitative proteomics. J Biol Chem. 2004;279:39155–39164. doi: 10.1074/jbc.M405456200. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz EM. Genomic classification of protein-coding gene families. WormBook. 2005 doi: 10.1895/wormbook.1.29.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez A, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki N, et al. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: Possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res. 2000;85:1–12. doi: 10.1016/s0169-328x(00)00218-7. [DOI] [PubMed] [Google Scholar]
- 21.Fung HC, et al. Genome-wide genotyping in Parkinson's disease and neurologically normal controls: First stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 22.Nollen EA, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menzies FM, Yenisetti SC, Min KT. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr Biol. 2005;15:1578–1582. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 24.Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J Biol Chem. 2005;280:43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 25.Petit A, et al. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 26.Pridgeon JW, Olzmann JA, Chin L-S, Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rehling P, Darsow T, Katzmann DJ, Emr SD. Formation of AP-3 transport intermediates requires Vps41 function. Nat Cell Biol. 1999;1:346–353. doi: 10.1038/14037. [DOI] [PubMed] [Google Scholar]
- 28.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 29.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 30.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 31.Chu Y, Kordower JH. Age-associated increases of alpha-synuclein in monkeys and humans are associated with nigrostriatal dopamine depletion: Is this the target for Parkinson's disease? Neurobiol Dis. 2007;25:134–149. doi: 10.1016/j.nbd.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;444:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 33.Komatsu M, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozzi Y, Borrelli E. Dopamine in neurotoxicity and neuroprotection: What do D2 receptors have to do with it? Trends Neurosci. 2006;29:167–174. doi: 10.1016/j.tins.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Jeanneteau F, Diaz J, Sokoloff P, Griffon N. Interactions of GIPC with dopamine D2, D3 but not D4 receptors define a novel mode of regulation of G protein-coupled receptors. Mol Biol Cell. 2004;15:696–705. doi: 10.1091/mbc.E03-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Vries L, Lou X, Zhao G, Zheng B, Farquhar MG. GAIP is membrane-anchored by palmitoylation and interacts with the activated (GTP-bound) form of Gαi subunits. Proc Natl Acad Sci USA. 1998;95:12340–12345. [Google Scholar]
- 37.Ogier-Denis E, Petiot A, Bauvy C, Codogno P. Control of the expression and activity of the Galpha-interacting protein (GAIP) in human intestinal cells. J Biol Chem. 1997;272:24599–24603. doi: 10.1074/jbc.272.39.24599. [DOI] [PubMed] [Google Scholar]
- 38.Mosharov EV, et al. Alpha-synuclein overexpression increases cytosolic catecholamine concentration. J Neurosci. 2006;26:9304–9311. doi: 10.1523/JNEUROSCI.0519-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caudle WM, et al. Reduced vesicular storage of dopamine causes progressive nigrostriatal neurodegeneration. J Neurosci. 2007;27:8138–8148. doi: 10.1523/JNEUROSCI.0319-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 41.Kim J, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Tur J. Parkinson's disease genetics: A complex disease comes to the clinic. Lancet Neurol. 2006;5:896–897. doi: 10.1016/S1474-4422(06)70580-4. [DOI] [PubMed] [Google Scholar]
- 43.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locke CJ, Williams SN, Schwarz EM, Caldwell GA, Caldwell KA. Genetic interactions among cortical malformation genes that influence susceptibility to convulsions in C. elegans. Brain Res. 2006;1120:23–34. doi: 10.1016/j.brainres.2006.08.067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.