Abstract
Trigger factor (TF) is the first chaperone to interact with nascent chains and facilitate their folding in bacteria. Escherichia coli TF is 432 residues in length and contains three domains with distinct structural and functional properties. The N-terminal domain of TF is important for ribosome binding, and the M-domain carries the PPIase activity. However, the function of the C-terminal domain remains unclear, and the residues or regions directly involved in substrate binding have not yet been identified. Here, a hydrophobic probe, bis-ANS, was used to characterize potential substrate-binding regions. Results showed that bis-ANS binds TF with a 1:1 stoichiometry and a Kd of 16 μM, and it can be covalently incorporated into TF by UV-light irradiation. A single bis-ANS–labeled peptide was obtained by tryptic digestion and identified by MALDI-TOF mass spectrometry as Asn391-Lys392. In silico docking analysis identified a single potential binding site for bis-ANS on the TF molecule, which is adjacent to this dipeptide and lies in the pocket formed by the C-terminal arms. The bis-ANS-labeled TF completely lost the ability to assist GAPDH or lysozyme refolding and showed increased protection toward cleavage by α-chymotrypsin, suggesting blocking of hydrophobic residues. The C-terminal truncation mutant TF389 also showed no chaperone activity and could not bind bis-ANS. These results suggest that bis-ANS binding may mimic binding of a substrate peptide and that the C-terminal region of TF plays an important role in hydrophobic binding and chaperone function.
Keywords: trigger factor, chaperone, binding site, bis-ANS, hydrophobic interaction
Trigger factor (TF) is the first protein to interact with nascent chains and facilitate their folding in bacteria (Houry 2001; Hartl and Hayer-Hartl 2002; Deuerling and Bukau 2004; Ferbitz et al. 2004; Baram et al. 2005; Maier et al. 2005). TF possesses both chaperone and peptidyl-prolyl cis-trans isomerase (PPIase) activities in vitro and in vivo (Stoller et al. 1995; Valent et al. 1995; Hesterkamp et al. 1996; Scholz et al. 1997; Huang et al. 2000; Li et al. 2001; Liu and Zhou 2004; Liu et al. 2005a), and its chaperone activity is distinct from its PPIase activity (Kramer et al. 2004a). Escherichia coli TF is 432 residues in length and consists of an N-terminal ribosomal-binding domain (1–144), a peptidyl-prolyl isomerase (PPIase) middle domain (145–247), and a C-terminal domain (248–432), whose function may be related to chaperone function (Hesterkamp et al. 1997; Houry 2001; Merz et al. 2006; Zeng et al. 2006). Recently, a number of crystal structures of TF from various sources, including the N-terminal ribosome-binding domain in complex with heterologous (Ferbitz et al. 2004) or homologous (Baram et al. 2005; Schlunzen et al. 2005) 50 S ribosome, have been solved. These structures consistently show that TF adopts an unusual extended fold resembling a “crouching dragon” with the N-terminal ribosomal-binding domain forming the “tail,” the peptidyl-prolyl isomerase domain forming the “head,” and the C-terminal domain forming the “arm” and “back” (Ferbitz et al. 2004). In a C-terminal truncated version of TF from Vibrio cholerae, the remainder of the molecule adopts a similar conformation as in full-length E. coli TF, suggesting that the absence of the C-terminal 44 amino acid does not change the overall fold of the remainder of the protein (Ludlam et al. 2004). The crystal structures of the physiological complex of the large ribosomal subunit with the N-terminal domain show that TF binds at the ribosome exit tunnel through the interaction of the highly flexible loop in its N-terminal domain with the L23, L24, and L29 proteins of the 50 S ribosome subunit to form a protective shield restricting the accessibility of nascent chains to the cellular environment (Ferbitz et al. 2004; Baram et al. 2005; Schlunzen et al. 2005). To date, only the interactions of the N-terminal binding domain of TF with the ribosome have been shown in detail, while the structural role of the rest of the TF molecule has still to be elucidated. The N-domain can partially substitute for TF in vivo (Kramer et al. 2004b). However, both the N- and C-domains are required for full expression of TF chaperone activity in vivo (Genevaux et al. 2004; Kramer et al. 2004b; Merz et al. 2006; Zeng et al. 2006) and an intact C-domain is crucial for chaperone activity in vitro (Merz et al. 2006; Zeng et al. 2006). Recently it was found that the isolated C-domain shows substantial chaperone activity in vitro and in vivo, suggesting that the C-domain of TF represents the central structural module of its chaperone activity (Merz et al. 2006). However, the residues or regions of TF that are directly involved in substrate binding have not yet been identified.
4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid (bis-ANS) (Fig. 1A) is an environment-sensitive hydrophobic fluorescent dye, and the fluorescence quantum yield of bis-ANS increases upon binding to the hydrophobic regions of proteins (Sheluho and Ackerman 2001). This feature has made the fluorescent dye popular as a probe for determining the accessibility of hydrophobic surfaces and monitoring conformational changes in proteins, including identification of peptide-binding sites in chaperones (Shi et al. 1994; Lee et al. 1997; Sharma et al. 1998a,b; Panda et al. 2001; Smoot et al. 2001). Here, we report that bis-ANS can interact with TF and can be covalently incorporated into the TF molecule by irradiation. The properties of the bis-ANS covalently labeled TF were investigated, and the bis-ANS linkage site was identified by MALDI-TOF MS. We provide experimental evidence that the bis-ANS binding site and the C-terminal residues of TF play an important role in assisting protein folding in vitro.
Figure 1.
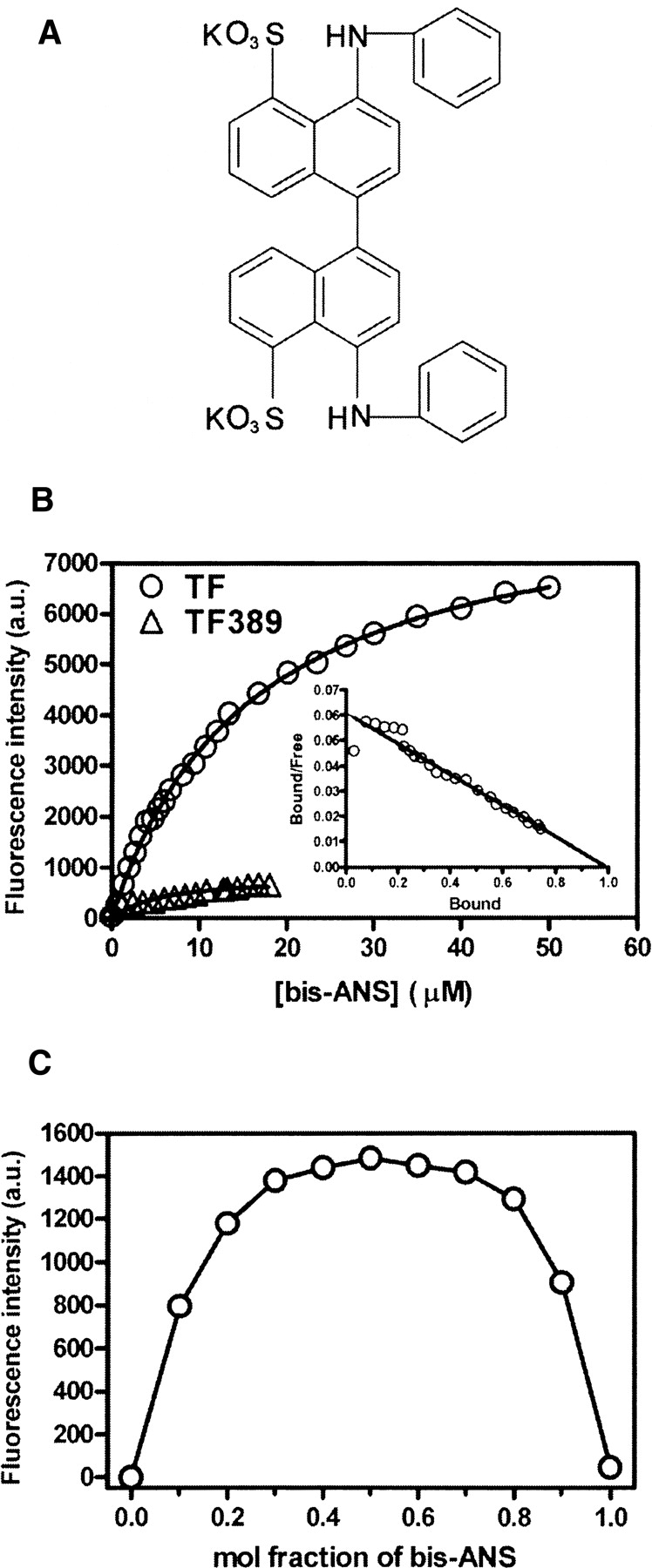
(A) Chemical structure of bis-ANS. (B) Fluorescence titration of TF and TF389 with bis-ANS. A total of 1 μM TF (○) or TF389 (▵) was titrated with 0–50 μM bis-ANS in 0.1 M phosphate buffer (pH 7.5), 2 mM EDTA. The samples were excited at 390 nm, and emission at 490 nm was measured. The inset shows the Scatchard plot for the binding of bis-ANS to TF. (C) Job plot (Huang 1982) for binding of TF to bis-ANS at a total concentration of 50 μM at 25°C.
Results
Interaction of bis-ANS with TF, but not with TF389
Bis-ANS is minimally fluorescent in aqueous buffer with an emission maximum at 525 nm. When bis-ANS bound to the hydrophobic site of TF, its fluorescence intensity increased markedly, and the emission maximum was blue-shifted to 490 nm. Figure 1B shows the titration of TF or TF389 (1 μM) with bis-ANS (0–50 μM). To obtain the dissociation constant (Kd) of bis-ANS bound to TF, fluorescence titration curves were analyzed using Equation 1 (see Materials and Methods) and a value of 16.0 (±0.4) μM was thus obtained. Scatchard analysis was also used (Equation 2; Fig. 1B, inset), which indicated a Kd of 16.5 (±0.3) μM and a stoichiometry of 1.02 (±0.08) bis-ANS per TF molecule. The stoichiometry of bis-ANS binding was also analyzed by Job plot (Fig. 1C; Huang 1982). The maximum of the curve is clearly at a mol fraction of 0.5, confirming the 1:1 stoichiometry in solution. The results indicate that only one bis-ANS molecule binds to each TF molecule, suggesting a relatively specific hydrophobic interaction between the molecules. We have shown previously that the related molecule, 1-anilinonaphthalene-8-sulfonate (ANS), binds the native state of TF with a Kd of 84 (±1) μM and a stoichiometry of 0.98 (±0.04) (Liu et al. 2005b). A further hydrophobic probe, 2-(p-toluidino)naphthalene-6-sulfonate (TNS), also showed weaker binding to TF, with a Kd of around 100 μM (data not shown). These results confirm that there is a general hydrophobic binding site in the TF molecule. Surprisingly, TF389, a TF mutant in which the C-terminal 43 residues are truncated, showed only very weak binding with bis-ANS (Fig. 1B). As the structure of this mutant is otherwise essentially the same as the full-length protein (Ludlam et al. 2004), this suggests that the truncated residues are important for bis-ANS binding to TF.
Energy transfer from tryptophan to bound bis-ANS in TF
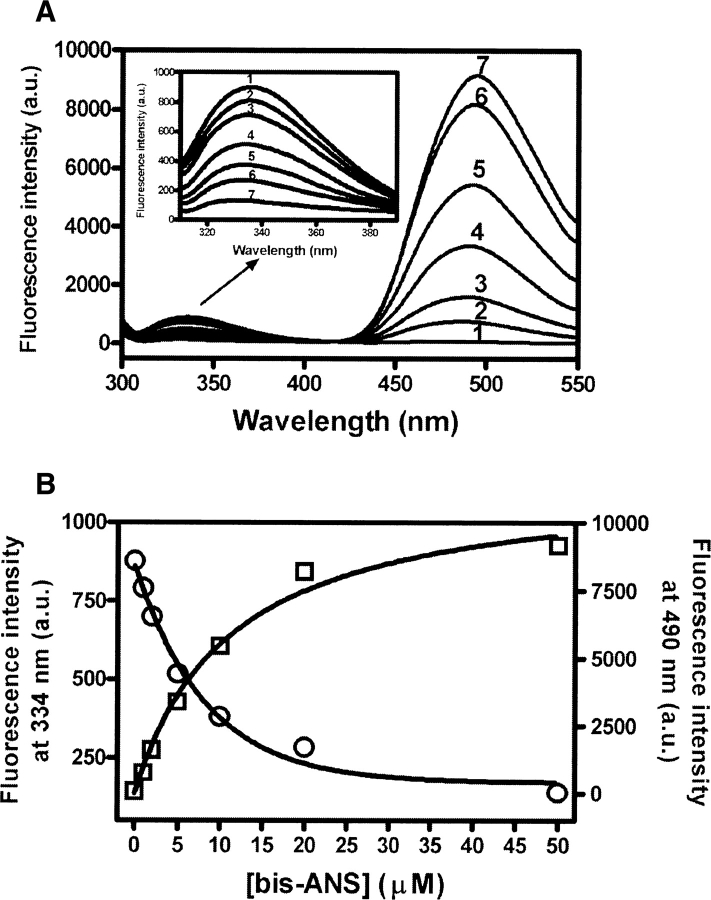
There is an overlap between the tryptophan emission spectrum of TF and the absorption spectrum of bis-ANS. The emission spectra of TF alone and of the mixtures of TF plus bis-ANS when excited at 295 nm are shown in Figure 2A. In the absence of bis-ANS, TF showed an emission maximum of 334 nm. When bis-ANS was added, there was a decrease in emission fluorescence at 334 nm, concurrent with an increase at 490 nm (Fig. 2A,B). Because the free bis-ANS shows no fluorescent emission in aqueous buffer when excited at 295 nm, the emission maximum at 490 nm represents an emission from tryptophan-excited bis-ANS. Given that TF has only a single Trp residue (Trp151) and is monomeric under the conditions of the experiment (2 μM) (Patzelt et al. 2002; Liu and Zhou 2004), this result indicates that there is energy transfer between Trp151 and the bound bis-ANS (Cantor and Schimmel 1980; Lakowicz 1983; Prasad et al. 1986b; Sheluho and Ackerman 2001). Only when the donor and acceptor are in close proximity (generally 20–50 Å), will acceptor emission be observed. The distance between bis-ANS and tryptophan in TF was estimated based on Förster theory (see Materials and Methods). Under our experimental conditions, the parameters were determined to be: ϕ = 0.118 (using an aqueous solution of tryptophan as the reference), n = 1.336, κ 2 = 2/3, JAD = 1.52 × 10−14 cm3M−1, and E = 0.21. The R0 was calculated to be 26.3 Å and R; the distance between Trp151 and bound bis-ANS in TF was 33.1 Å. As the length of the bis-ANS molecule is ∼15 Å, we can therefore predict that the bis-ANS binding site in TF lies within a 40 Å radius of Trp151.
Figure 2.
(A) Energy transfer between the tryptophan residue and bound bis-ANS in TF. The concentration of TF was 2 μM, and the bis-ANS concentrations were 0, 1, 2, 5, 10, 20, and 50 μM labeled as 1–7, respectively. The samples were excited at 295 nm, and emission spectra from 300 to 550 nm were recorded. (B) Concentration dependence of energy transfer between tryptophan and bound bis-ANS. The concentration of TF was 2 μM and the bis-ANS concentrations were 0, 1, 2, 5, 10, 20, and 50 μM. The samples were excited at 295 nm and emission was measured at 334 nm (○) and 490 nm (□).
Bis-ANS inhibits chaperone activity of TF
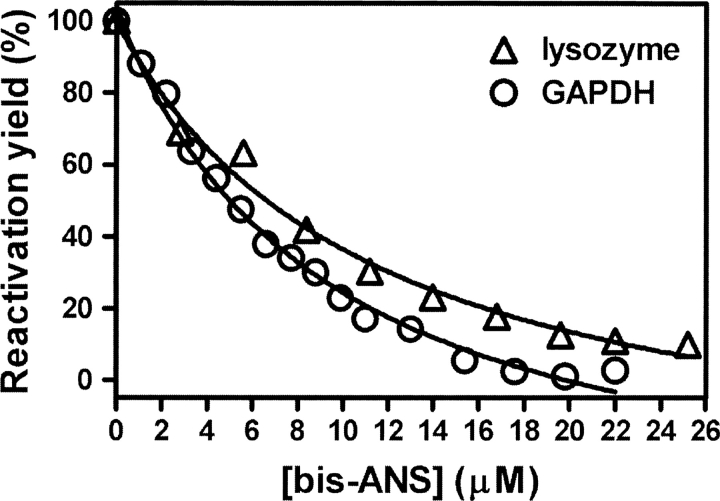
It has been shown that TF is an effective molecular chaperone in assisting reactivation of GAPDH, lysozyme, and carbonic anhydrase in vitro (Huang et al. 2000, 2002; Liu and Zhou 2004). The effect of bis-ANS on TF-assisted GAPDH and lysozyme refolding was investigated. As shown in Figure 3, the reactivation yield of GAPDH or lysozyme decreased with increasing bis-ANS concentration, indicating that TF-mediated GAPDH and lysozyme refolding is inhibited by bis-ANS. The inhibition constant Ki, which represents the apparent affinity of bis-ANS for TF, was calculated to be 9.5 (±1.3) μM and 11.4 (±2.5) μM for TF-assisted GAPDH and lysozyme refolding, respectively. The Ki values are slightly lower than the Kd (16 μM) for bis-ANS binding to TF, which may be explained by binding of bis-ANS to folding intermediates of the substrate, which could also inhibit their interaction with TF. Titration with the probe TNS also inhibited GAPDH reactivation (data not shown), but a higher concentration of probe was required, consistent with the lower affinity of TNS than bis-ANS for TF, as mentioned above.
Figure 3.
Bis-ANS inhibits chaperone activity of TF. Refolding of GdnHCl-denatured GAPDH (○) or lysozyme (▵) was initiated by 50-fold dilution (for GAPDH) or 20-fold dilution (for lysozyme) into 0.1 M phosphate buffer (pH 7.5) containing 12 μM TF (for GAPDH) or 15 μM TF (for lysozyme) and 0–25 μM bis-ANS. The final concentrations of GAPDH and lysozyme in the refolding buffer were 2.8 and 10 μM, respectively. The GAPDH reactivation mixtures were first kept at 4°C for 30 min and then for a further 3 h at 25°C before samples were taken for assay of activity (Huang et al. 2000). The lysozyme reactivation mixtures were kept at 25°C for 5 h before samples were taken for assay of activity (Huang et al. 2002). The TF-assisted reactivation yield of GAPDH or lysozyme without bis-ANS was set as 100%.
Photoincorporation of bis-ANS into TF
To further investigate the properties of bis-ANS-labeled TF and identify the linkage site of bis-ANS on the TF molecule, TF covalently labeled with bis-ANS was prepared by UV-light irradiation and analyzed by native-PAGE and mass spectrometry. After UV-light irradiation, the unbound bis-ANS was removed using a Sephadex G-25 column and the stoichiometry of bound bis-ANS on TF was determined to be 0.98 (±0.01), using the absorbance values at 385 and 280 nm, and the respective extinction coefficients for bis-ANS and TF (see Materials and Methods). As shown in Figure 4A, the bis-ANS-labeled TF can emit fluorescence when illuminated at 360 nm on the polyacrylamide gel. This property indicates that bis-ANS was covalently photoincorporated into the TF molecule. The mass spectrum of a 1:1 mixture of TF and bis-ANS–labeled TF is shown in Figure 4B, indicating that the molecular weight of TF and bis-ANS-labeled TF are 49,894 ± 25 Da and 50,509 ± 25 Da, respectively. The difference in molecular mass between labeled and unlabeled TF (615 ± 35 Da) corresponds to the molecular mass of a single bis-ANS molecule (597 Da), confirming the conclusion from the stoichiometry calculation, and consistent with the Scatchard and Job plot analysis, which showed that there is only one bis-ANS molecule bound to each TF molecule prior to UV incorporation.
Figure 4.
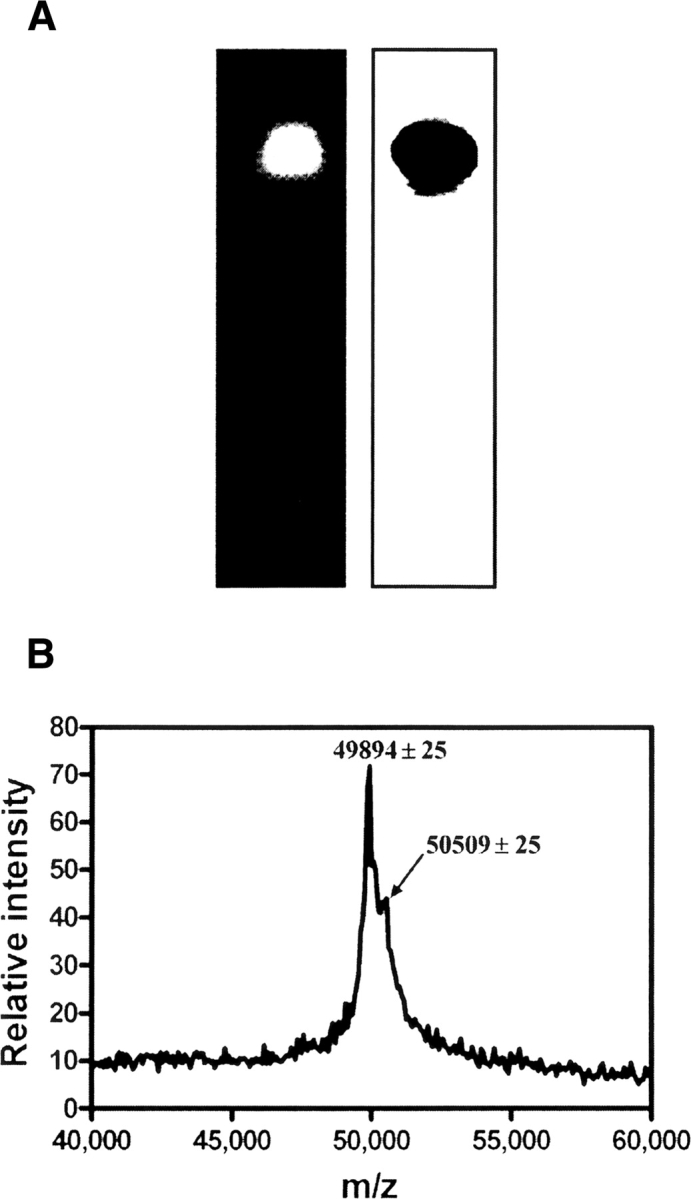
(A) Identification of bis-ANS labeled TF. Bis-ANS was photoincorporated into TF (see Materials and Methods) and detected by 15% native-PAGE. (Left) Labeling products were visualized under a UV-light box. (Right) Labeling products were stained with Coomassie Brilliant Blue. (B) MALDI-TOF mass spectrum of a 1:1 mixture of bis-ANS labeled TF and unlabeled TF.
Bis-ANS covalently links to the C-terminal of TF at Asn391-Lys392
To identify the peptide region to which bis-ANS covalently links, the bis-ANS-labeled TF was thoroughly digested by trypsin, and the digested peptides were separated on a C18 reverse-phase column using a water-acetonitrile gradient with 0.1% TFA. The eluate was monitored at 220 nm for general peptides and 385 nm for bis-ANS-labeled peptide (Fig. 5A). A single peak was identified with absorption at both 220 and 385 nm, showing that this was the only peptide labeled with bis-ANS. The bis-ANS-labeled peptide was further analyzed by MALDI-TOF MS (Fig. 5B). A single peak at 855 ± 2 was found. Subtracting the molecular mass of bis-ANS (597 Da) from the peak yields a difference of 259 ± 2 Da, giving the molecular mass of the peptide bound to bis-ANS. We assessed database information about TF hydrolyzed by trypsin and found that in all likelihood, Asn391-Lys392 is the dipeptide that covalently links to bis-ANS. Although the mode of covalent linkage between bis-ANS and peptides is not understood, this nevertheless provides us with information about the location of the bis-ANS binding site on the TF structure.
Figure 5.
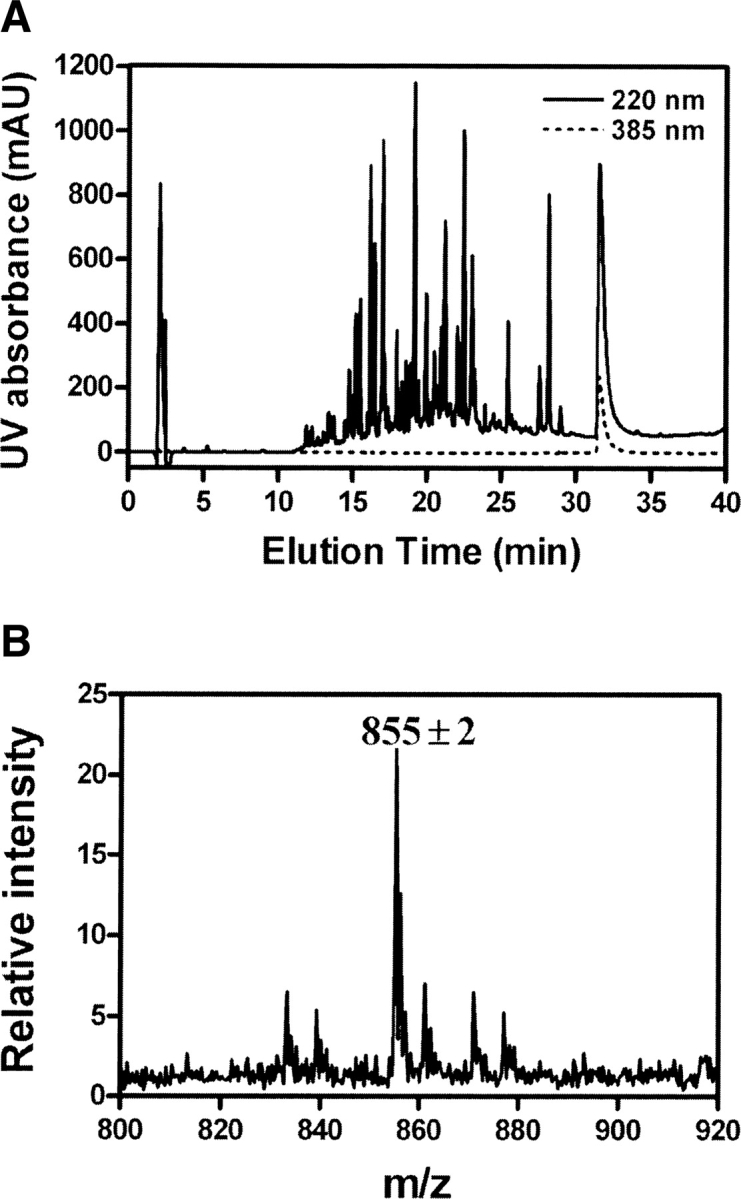
(A) Separation of bis-ANS-labeled peptide by HPLC. Trypsin-digested bis-ANS-labeled TF was separated on a C18 reverse-phase column using a gradient of 0.1% TFA in water and 0.1% TFA in acetonitrile. The eluate was monitored at 220 nm (solid line) for general peptides and 385 nm (dashed line) for bis-ANS-labeled peptides. (B) MALDI-TOF mass spectrum of bis-ANS-labeled peptide. The MS spectrum of the labeled peptide (m/z 855.30) was obtained from the MALDI-TOF MS analysis of 100 pmol-labeled peptide.
Hydrophobic residues in TF are protected by bis-ANS binding
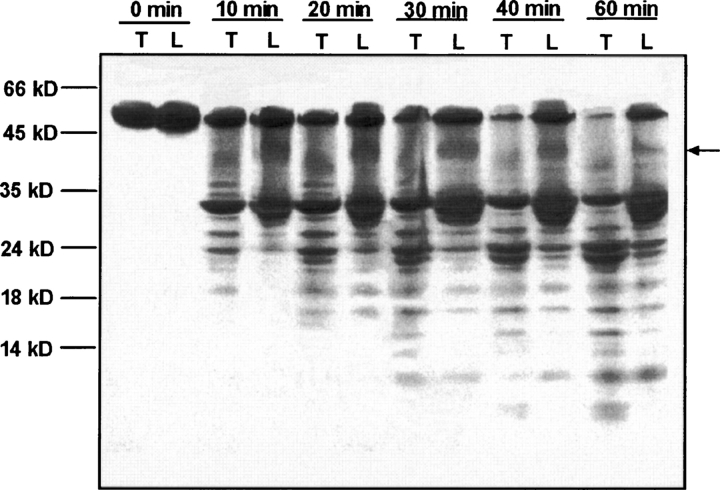
Limited proteolysis of TF and bis-ANS-labeled TF by α-chymotrypsin was compared (Fig. 6). The hydrolysis rate of bis-ANS-labeled TF was slower than that of unlabeled TF under the same conditions, and the hydrolysis pattern was significantly different. There was a high molecular weight band showing a very slow digestion rate that appeared only in the bis-ANS-labeled TF (indicated by the arrow in Fig. 6). α-chymotrypsin specifically hydrolyzes aromatic and hydrophobic residues in a protein molecule. The results here show that the TF sites accessible to α-chymotrypsin were changed upon bis-ANS binding. This change could have occurred because some of the α-chymotrypsin-accessible sites were directly blocked by the bis-ANS molecule or could be due to a conformational change in the molecule induced by bis-ANS binding, resulting in reduced accessibility of these residues. A comparison of the far-UV circular dichroism spectra of unlabeled and bis-ANS-labeled TF shows no detectible difference (data not shown), indicating that incorporation of bis-ANS induces no significant structural change in TF. This, therefore, suggests that protection of α-chymotrypsin hydrolysis sites by bis-ANS labeling principally reflects blocking of access to hydrophobic (or aromatic) residues.
Figure 6.
Limited digestion of TF and bis-ANS-labeled TF by α-chymotrypsin. A total of 10 μg of protein was digested by 0.05 μg of α-chymotrypsin for different times at 25°C; the digestion reactions were stopped by the addition of 2 mM PMSF, and the mixture was resolved by 18% SDS-PAGE. The digestion times and proteins were as labeled. (T) TF; (L) bis-ANS-labeled TF.
Bis-ANS-labeled TF loses the ability to assist GAPDH or lysozyme reactivation
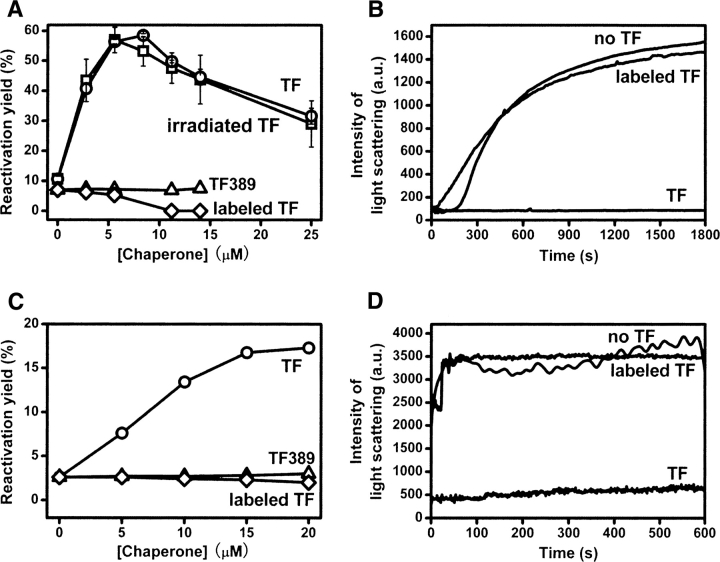
The ability of unlabeled and bis-ANS-labeled TF to assist GAPDH or lysozyme reactivation were compared. With increasing TF concentration, the reactivation yields of GAPDH or lysozyme showed a clear increase and the degree of aggregation was inhibited significantly (Fig. 7A–D). However, bis-ANS-labeled TF completely lost the ability to assist GAPDH or lysozyme reactivation and to prevent aggregation (Fig. 7A–D). This is consistent with the loss of TF chaperone activity when titrated with free bis-ANS (Fig. 3). As a further control, UV-irradiated TF (in the absence of bis-ANS) showed the same ability in assisting refolding and preventing aggregation of GAPDH as untreated TF, eliminating the possibility that this loss of chaperone activity in bis-ANS-labeled TF is due to irradiation damage of the TF molecule.
Figure 7.
(A) Effect of TF or bis-ANS-labeled TF on GAPDH reactivation in phosphate buffer (conditions as described in the legend to Fig. 3). GAPDH refolding in the presence of TF (○), UV-irradiated TF (□), TF389 (▵), and bis-ANS-labeled TF (◊). (B) Suppression of aggregation of GAPDH by TF or bis-ANS-labeled TF. Aggregation of GAPDH upon dilution was monitored at 25°C by 90° light scattering at 488 nm. The concentration of GAPDH was 2.8 μM. The time course of the change in light scattering due to GAPDH aggregation in the presence of 0 μM TF, 11.2 μM TF, or 11.2 μM bis-ANS-labeled TF were measured as described (Huang et al. 2000). (C) Effect of TF or bis-ANS-labeled TF on lysozyme reactivation in phosphate buffer (conditions as described in the legend to Fig. 3). Lysozyme refolding in the presence of TF (○), TF389 (▵), and bis-ANS-labeled TF (◊). (D) Suppression of aggregation of lysozyme by TF or bis-ANS-labeled TF. Aggregation of lysozyme upon dilution was monitored at 25°C by 90° light scattering at 488 nm. The concentration of lysozyme was 10 μM. The time course of the change in light scattering due to lysozyme aggregation in the presence of 0 μM TF, 15 μM TF, or 15 μM bis-ANS-labeled TF were measured as described (Huang et al. 2002).
Bis-ANS is a strong hydrophobic probe. TF binds to both bis-ANS and the folding intermediates of GAPDH or lysozyme by hydrophobic interactions. These results suggest that the substrate-binding site of TF overlaps with the bis-ANS binding site, so that when bis-ANS is bound, the substrate binding sites are occupied and TF can no longer bind to the folding intermediates of GAPDH or lysozyme. Thus, TF loses its ability to assist in reactivation and prevention of aggregation of these proteins.
We also compared the ability of TF389 to assist GAPDH or lysozyme refolding under the same conditions. Similarly, TF389 completely lost the ability to aid GAPDH or lysozyme reactivation (Fig. 7A–C). This finding is consistent with the bis-ANS binding results (Fig. 1B), which showed that the C-terminal residues play an important role in forming the binding site that accommodates the hydrophobic molecule bis-ANS. It appears that the C-terminal residues of TF are crucial for hydrophobic substrate binding.
Bis-ANS binds in the pocket formed by the C-terminal arms of TF
Molecular docking is a computational process of searching for a ligand that is able to fit both geometrically and energetically into the binding site of a protein. To investigate the possible binding sites for bis-ANS on the TF molecule, we docked the bis-ANS structure onto two crystal structures of TF: full-length E. coli TF (Ferbitz et al. 2004) and the C-terminal truncated Vibrio cholerae TF (TF378) (Fig. 8A,B; Ludlam et al. 2004). The docking program found only one suitable binding site for the bis-ANS molecule on TF, consistent with the 1:1 binding stoichiometry observed experimentally. The location of this binding site in the full-length structure is in the pocket formed by the C-terminal arms, surrounded by hydrophobic residues Val 313 (in α-helix 9), Leu 351 (in α-helix 11), Val 401 (in α-helix 14), and Leu 403 (in α-helix 14) in the full-length structure (Fig. 8A,C). According to the docked structure, bis-ANS interacts with hydrophobic peptides 343–353 (RRVVVGLLLGE), 370–373 (LIEE), 384–386 (VIE), and 401–415 (VALEEQAVEAVLAKA) (Fig. 8C), which forms a hydrophobic pocket (Fig. 8D). The putative binding site identified by docking is adjacent to the dipeptide identified by mass spec analysis of the bis-ANS linkage site (Fig. 8A,C) and the docked bis-ANS molecule lies around 38 Å from Trp151 at the nearest point (Fig. 8A). Allowing for slight conformational differences between crystal and solution structures and/or a minor conformational change on hydrophobic substrate binding, this distance agrees well with the 40 Å radius predicted by the observation of Förster energy transfer (Fig. 2). Interestingly, when the same docking method was used, no docking site for bis-ANS could be found in the structure of the C-terminally truncated TF (Fig. 8B). From comparison of the structures, it appears that C-terminal truncation results in loss of the bis-ANS binding cavity (Fig. 8A,B). This is also consistent with the lack of bis-ANS binding for the TF389 mutant observed experimentally (Fig. 1B), and further emphasizes that the residues in the C-terminal region of TF are important for hydrophobic binding.
Figure 8.
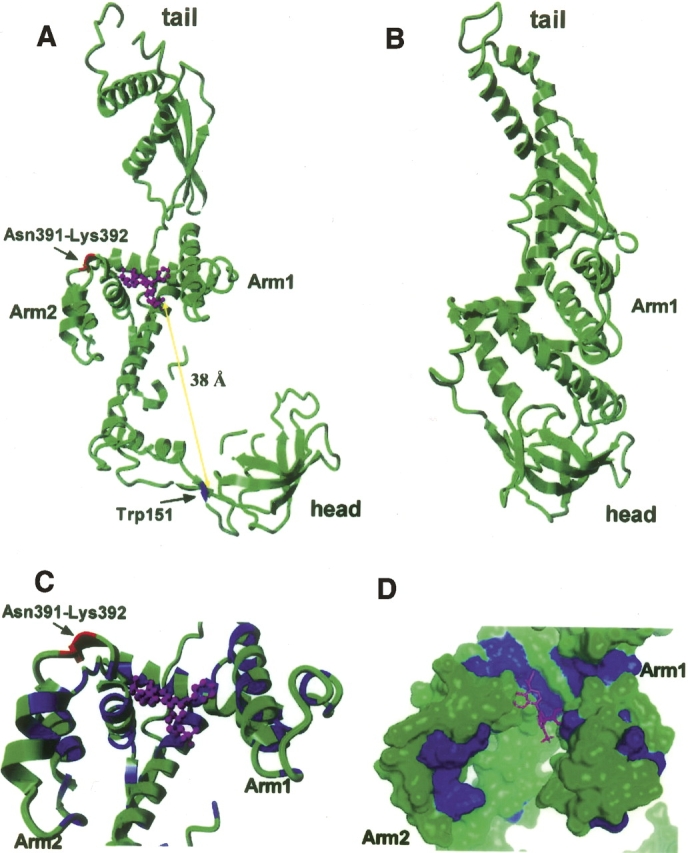
Docking of bis-ANS into the TF crystal structure. (A) TF PDB data 1w26 (Ferbitz et al. 2004) and the programs Hex 4.5 and Yasara were used to dock bis-ANS into the TF crystal structure. bis-ANS is shown in magenta, Asn391-Lys392 in red, and Trp151 in blue. The distance between Trp151 and the putative bis-ANS binding site is consistent with the observation of excitation energy transfer (Fig. 2A). (B) Crystal structure of TF378 (Ludlam et al. 2004), in which no docking site for bis-ANS could be found. (C) Details of the bis-ANS binding region; bis-ANS is shown in magenta, Asn391-Lys392 in red, hydrophobic residues are blue, and hydrophilic residues are green. (D) Molecular surface of TF; bis-ANS is shown in magenta, hydrophobic residues are shown in blue, and hydrophilic residues in green.
Discussion
Based on the crystal structure of the TF N-domain in complex with the large ribosomal subunit, a model of chaperone function of TF in vivo has been suggested: TF hunches over the polypeptide exit of the ribosomal tunnel and positions a hydrophobic area formed by its tail, back, and arms directly above the emerging nascent chain, creating a protected folding space where nascent polypeptides may be shielded from proteases and aggregation (Ferbitz et al. 2004). This protected environment provided by TF may be particularly important for large multidomain proteins to fold productively into their native states (Hoffmann et al. 2006). However, to date, little is understood about which part of the TF molecule is directly involved in binding to target proteins. Here, we use bis-ANS to mimic a hydrophobic substrate in order to search for the regions on the TF molecule that might be involved in substrate binding.
The environment-sensitive hydrophobic fluorescent dye bis-ANS has been used to provide information about hydrophobic surfaces and to detect conformational changes in proteins (Prasad et al. 1986a,b; Sharma et al. 1998a,b). In particular, bis-ANS has been used as a probe to identify hydrophobic peptide binding sites important for chaperone function (Seale et al. 1995, 1998; Lee et al. 1997; Sheluho and Ackerman 2001). Generally, bis-ANS is not a specific probe: A protein may have several exposed hydrophobic regions and could potentially bind several bis-ANS molecules. However, bis-ANS showed specific 1:1 binding with the TF molecule, allowing us to investigate the properties of TF covalently labeled with bis-ANS, and to determine the location of the bis-ANS binding site. The results obtained provide new insight into the structural basis of the chaperone function of TF.
Bis-ANS binds to TF with 1:1 stoichiometry and a Kd of 16 μM, and it can be covalently incorporated into TF by UV-light irradiation (Figs. 1, 4). Binding of bis-ANS, whether or not covalently bound, resulted in loss of the ability to assist GAPDH or lysozyme refolding (Figs. 3, 7). Incorporation of bis-ANS also resulted in partial protection against digestion by α-chymotrypsin (Fig. 6), a protease that recognizes aromatic and/or hydrophobic residues. The results of in silico docking suggest that bis-ANS interacts with hydrophobic residues Val 313, Leu 351, Val 401, and Leu 403, which are located between α-helix 9, α-helix 11 (in arm 1), and α-helix 14 (in arm 2) (Fig. 8C). The putative bis-ANS binding pocket lies ∼40 Å from Trp151 (Fig. 8A), consistent with observation of Förster energy transfer (Fig. 2). Further, the site is adjacent to the dipeptide Asn391-Lys392 (Fig. 8A), which is the bis-ANS linkage point we identified by MALDI-TOF MS. According to the crystal structure of TF, the N-, and C-domains form a protected folding environment or “cradle” for folding intermediates (Ferbitz et al. 2004). Recent results suggest that an intact C-domain is crucial for the chaperone activity of TF (Merz et al. 2006; Zeng et al. 2006) and that the C-domain in fact represents the central structural module for this activity (Merz et al. 2006). The bis-ANS binding site is located in the second arm of the C-domain of TF and near the entrance of the “cradle” (Fig. 8A). Our results suggest that when bis-ANS is bound to the C-domain of TF, the hydrophobic surface of the C-domain is blocked and the hydrophobic pocket is occupied, so the C-domain of TF is no longer able to bind folding intermediates. As a result, bis-ANS-labeled TF loses the ability to assist GAPDH or lysozyme refolding. These findings clearly show that the hydrophobic bis-ANS binding site of the C-domain is a key element for substrate binding of TF. The importance of hydrophobic interaction in TF-assisted protein folding has also been found by Tomic and coworkers (Tomic et al. 2006). They suggest that TF is not a general shield for nascent chains and show that the protease protection for ribosome-bound proteins appears to depend on a hydrophobic interaction between TF and nascent polypeptides.
The modular structure of TF and the functional contributions of its individual domains have been investigated extensively both in vivo and in vitro (Hesterkamp and Bukau 1996; Hesterkamp et al. 1997; Zarnt et al. 1997; Kramer et al. 2004b; Merz et al. 2006; Zeng et al. 2006). The C-domain is important for chaperone activity, while the N-domain is required for ribosome binding, and the M-domain carries PPIase activity. The synergetic interaction of N- and C-domain is needed for efficient chaperone function of TF in vitro. When either the N- or C-domain is truncated, the fragments NM or MC lose chaperone activity toward chemically denatured GAPDH and cannot be cross-linked with GAPDH intermediates (Kramer et al. 2004b). The NC fragment is the only domain combination that displays chaperone activity toward chemically denatured GAPDH (Kramer et al. 2004b) and protects nascent polypeptides from proteolysis (Hoffmann et al. 2006). We previously found that partial truncation of the C terminus results in a dramatic loss of function as a chaperone toward GAPDH in vitro (Zeng et al. 2006). These findings suggest that the C-domain may make a major contribution to the chaperone activity of TF. Here, we compared the bis-ANS binding ability and the chaperone function of TF and the mutant TF389, in which the C-terminal 43 residues are truncated. The results clearly show that TF389 is unable to bind bis-ANS, and the ability to assist GAPDH or lysozyme refolding is also completely lost. This finding coincides with the results of Merz et al. (2006) that show the truncation of the C-terminal 53 amino acid residues of TF caused the complete loss of chaperone activity in vitro and severely impaired its chaperone function in vivo. This provides further experimental evidence that the C-terminal residues, 390–432, are involved in forming a hydrophobic-binding region on TF and that they are important for its molecular chaperone function. From the crystal structure, residues 390–432 represent the main part of arm2 and cooperate with arm1 and the N-domain to form the hydrophobic cavity (Fig. 8A). When the C-terminal residues are truncated, this hydrophobic cavity is disrupted (Fig. 8B). Consistent with this, no suitable binding site for bis-ANS could be identified on the C-terminally truncated TF molecule by molecular docking and no binding was detected experimentally for this mutant. From the structure of the complex between the N-terminal binding domain of TF from Deinococcus radiodurans with the large ribosomal subunit from the same organism, Schluenzen et al. (2005) conclude that the cradle formed between TF and the ribosome may not be between the N-terminal binding domain and the arms since ribosomal L24 occupies this space, but instead such a cavity may be located more toward the central body and arms of TF, implying the importance of the C-terminal domain of TF in peptide substrate binding, consistent with the results we present here.
Conclusions
The hydrophobic probe bis-ANS was used to characterize the hydrophobic binding region of E. coli trigger factor. Experimental and molecular docking results show that there is only one bis-ANS binding site located on the TF molecule, which is located in the C-terminal region. Bis-ANS-labeled TF, and a C-terminal truncation mutant (TF389), which is otherwise essentially the same in structure to full-length TF, completely lose the ability to assist GAPDH or lysozyme refolding. These results suggest that the bis-ANS binding site is a key element for hydrophobic substrate binding, and that the C-terminal region of TF may play an important role in its in vivo chaperone function.
Materials and Methods
Materials
Bis-ANS was purchased from Sigma. The stock solutions of bis-ANS were prepared in 100% methanol, and the concentration was determined by absorbance at 385 nm using an extinction coefficient of 16,790 M−1cm−1 (Sharma et al. 1998b). DL-glyceraldehyde-3-phosphate (GAP), DL-Dithiothreitol (DTT), β-NAD, hen egg-white lysozyme, GSSG, GSH, trypsin, chymotrypsin, and Micrococcus lysodeikticus cell walls were purchased from Sigma. Guanidine hydrochloride (GdnHCl) was a product of ICN Biomedicals. All other chemicals were local products of analytical grade.
Plasmid pQE60 containing the wild-type tig gene that encodes E. coli TF was donated by Professor G. Fischer (Max Planck Research Unit for Enzymology of Protein Folding, Halle, Germany). The C-terminal truncation mutant, TF389, in which the C-terminal 43 residues are deleted, was prepared as described (Zeng et al. 2006). Full-length TF and TF389 were expressed in E. coli JM109 and purified according to the methods of Stoller et al. (1995) and Zeng et al. (2006), respectively. Final protein preparations were typically >90% homogeneous as judged by SDS-PAGE. Absorbance coefficients at 280 nm of 15,930 M−1cm−1 for TF and 16,645 M−1cm−1 for TF389 were used for protein concentration determination.
Purification of rabbit muscle GAPDH was performed as previously described (Liang et al. 1990). An absorbance coefficient of ɛ280nm = 144,000 M−1cm−1 was used for enzyme concentration determination.
Fluorescence measurements
Fluorescence measurements were made on a Hitachi F-4500 spectrofluorimeter. Proteins (1 μM) were mixed with bis-ANS of various concentrations in buffer containing 0.1 M sodium phosphate and 2 mM EDTA (pH 7.5). The samples were excited at 390 nm, and the emission was measured at 490 nm or recorded from 400 to 550 nm. The excitation and emission slits were set to 5 nm. All measurements were made at 25°C. To obtain the dissociation constant (Kd), fluorescence titration curves were analyzed using the equation:
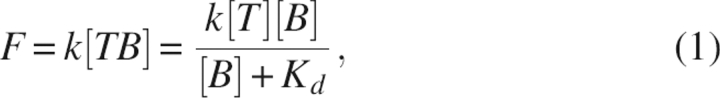 |
where F is the fluorescence intensity of the TF·bis-ANS complex, [TB] is the concentration of the TF·bis-ANS complex, k is a proportionality constant, [T] is the concentration of TF, and [B] is the concentration of free bis-ANS at equilibrium (Lookene et al. 2003). The data were also analyzed by Scatchard plot:
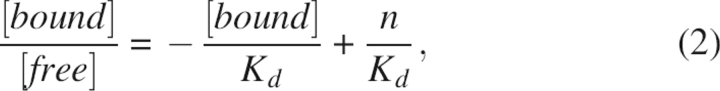 |
where [bound] and [free] is the concentration of bound and free bis-ANS at equilibrium, and n is the number of bis-ANS molecules bound to TF.
The stoichiometry of TF binding to bis-ANS was also analyzed by Job plot as follows: equimolar solutions (50 μM) of TF and bis-ANS were prepared in 0.1 M phosphate buffer (pH 7.5), 2 mM EDTA, and mixed in various ratios. The total concentration of TF and bis-ANS was kept constant at 50 μM and only the TF/bis-ANS ratio was varied. The samples were excited at 390 nm, and emission at 490 nm was measured. The excitation and emission slits were set to 2.5 nm. The data were analyzed by the Job method (Huang 1982). The fluorescence intensity was plotted as a function of the mol fraction of bis-ANS.
For energy-transfer experiments, an excitation wavelength of 295 nm was used to selectively excite tryptophan residues. Emission spectra were recorded between 300 and 550 nm. The distance between bis-ANS and tryptophan in TF was determined based on Förster theory, as follows:
 |
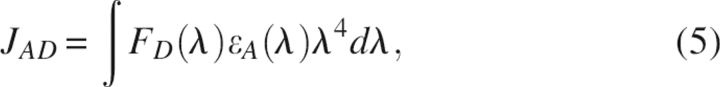 |
where E is the efficiency of energy transfer, R is the distance between donor and acceptor, and R0 is the Förster distance at which the transfer efficiency (and the donor lifetime) is 50%. ϕ is the fluorescence quantum yield of the donor, κ is the dipole–dipole orientation factor, n is the refractive index of the medium between the donor and acceptor, and JAD is the overlap integral between the donor fluorescence spectrum and the acceptor absorption spectrum. JAD can be calculated by numerical summation from the corrected fluorescence spectrum of the donor and the absorption spectrum of the acceptor as described by the equation:
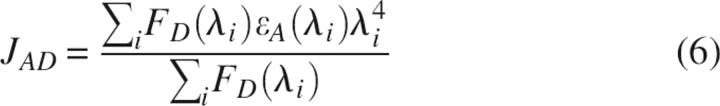 |
with a unit3M−1.
The efficiency of energy transfer (E) can be calculated by the equation:
 |
where FDA and FD are the donor fluorescence intensities in the presence and absence of energy transfer (Cantor and Schimmel 1980; Lakowicz 1983; Liang and Liu 2006). All data were analyzed with the program Prism (GraphPad Software Inc).
Reactivation of guanidine-denatured GAPDH or lysozyme
Reactivation of guanidine-denatured GAPDH or lysozyme assisted by TF with or without various concentrations of bis-ANS, or with bis-ANS-labeled TF, was performed exactly as described (Huang et al. 2000, 2002). The reactivation yield of GAPDH or lysozyme in the presence of TF without bis-ANS were set as 100%. The parameter Ki characterizing the inhibition efficiency was obtained from the equation:
 |
where A is the measured activity, Ar is a calculated constant describing the activity of the TF-bis-ANS complex, Ki is the inhibition constant, m is a proportionality constant, and C is the concentration of bis-ANS (Lookene et al. 2003).
Photoincorporation of bis-ANS into TF
TF (30 μM) was mixed with 200 μM bis-ANS in 1 mL buffer containing 0.1 M sodium phosphate (pH 7.5), and 2 mM EDTA. Samples were illuminated at 254 nm in a Hitachi F-4500 spectrofluorimeter with stirring at room temperature for 60 min. Unbound bis-ANS was removed by applying the samples to a Sephadex G-25 column. Photolabeled proteins were visualized on a UV-light box (Seale et al. 1995). The concentration of bound bis-ANS was determined by the absorbance at 385 nm and the concentration of TF was determined by Bradford assay using nonlabeled TF as standard. The products were analyzed by 15% native-PAGE and MALDI-TOF MS.
Limited proteolysis
TF and bis-ANS-labeled TF were subjected for different time periods to limited proteolysis by α-chymotrypsin in buffer containing 0.1 M sodium phosphate (pH 7.5), and 2 mM EDTA at room temperature. The products were separated on a 18% SDS–polyacrylamide gel as described (Li and Zhou 2000).
Identification of bis-ANS linking sites
A total of 0.2 mg of bis-ANS-labeled TF was digested with trypsin (1:40 w/w) at 25°C overnight to complete proteolysis. The peptides were separated on a C18 reverse-phase column (4.6 mm × 25 cm, 5 μm, Shimadzu) at a flow-rate of 1 mL/min using a Shimadzu HPLC system (Shimadzu CTO-10ASVP). The mobile phase consisted of water with 0.1% TFA (A) and acetonitrile with 0.1% TFA (B). A 40-min linear gradient from 0% to 100% B was typically used.
The eluate was monitored at 220 nm for general peptides and 385 nm for bis-ANS-labeled peptide. The purified bis-ANS-labeled peptide was further analyzed by MALDI-TOF MS (Shimadzu AXJMA-CFR).
Docking of bis-ANS into the TF molecules
TF PDB data 1w26 (Ferbitz et al. 2004) and 1t11 (Ludlam et al. 2004), and the programs Hex 4.5 and Yasara were used to dock bis-ANS into the TF crystal structures.
Acknowledgments
The present study was supported in part by the Chinese Ministry of Science and Technology (G1999075608, 2006CB500703, 2006CB910903), the Chinese National Natural Science Foundation (30470363, 30620130109, 30670428), and a CAS Knowledge Innovation Grant (KSCX2-SW214-3). We thank San-Bo Qin for his help with molecular docking analysis, the proteomic platform of the Institute of Biophysics, Chinese Academy of Sciences, for the purification and mass analysis of the bis-ANS-labeled peptide and protein, and Dr. Fuquan Yang for his suggestions on the revision of this manuscript.
Footnotes
Reprint requests to: J.M. Zhou, National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Chaoyang District, Beijing 100101, China; e-mail: zhoujm@sun5.ibp.ac.cn; fax: 86-10-6484-0672.
Abbreviations: ANS, 1-anilinonaphthalene-8-sulfonate; bis-ANS, 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid dipotassium salt; DTT, dithiothreitol; GAPDH, D-glyceraldehyde-3-phosphate dehydrogenase; GdnHCl, guanidine hydrochloride; GSH, glutathione; GSSG, oxidized glutathione; MALDI-TOF, Matrix assisted laser desorption–ionization time-of-flight; MS, mass spectrometry; PAGE, polyacrylamide gel electrophoresis; PMSF, phenylmethylsulfonyl fluoride; PPIase, peptidyl-prolyl cis-trans isomerase; SDS, sodium dodecyl sulfate; TF, trigger factor; TNS, 2-(p-toluidino)naphthalene-6-sulfonate.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062623707.
References
- Baram D., Pyetan, E., Sittner, A., Auerbach-Nevo, T., Bashan, A., and Yonath, A. 2005. Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action. Proc. Natl. Acad. Sci. 102: 12017–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C.R. and Schimmel, P.R. 1980. Biophysical chemistry. W.H. Freeman and Company, San Francisco, CA.
- Deuerling E. and Bukau, B. 2004. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit. Rev. Biochem. Mol. Biol. 39: 261–277. [DOI] [PubMed] [Google Scholar]
- Ferbitz L., Maier, T., Patzelt, H., Bukau, B., Deuerling, E., and Ban, N. 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431: 590–596. [DOI] [PubMed] [Google Scholar]
- Genevaux P., Keppel, F., Schwager, F., Langendijk-Genevaux, P.S., Hartl, F.U., and Georgopoulos, C. 2004. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 5: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U. and Hayer-Hartl, M. 2002. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858. [DOI] [PubMed] [Google Scholar]
- Hesterkamp T. and Bukau, B. 1996. Identification of the prolyl isomerase domain of Escherichia coli trigger factor. FEBS Lett. 385: 67–71. [DOI] [PubMed] [Google Scholar]
- Hesterkamp T., Hauser, S., Lutcke, H., and Bukau, B. 1996. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc. Natl. Acad. Sci. 93: 4437–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterkamp T., Deuerling, E., and Bukau, B. 1997. The amino-terminal 118 amino acids of Escherichia coli trigger factor constitute a domain that is necessary and sufficient for binding to ribosomes. J. Biol. Chem. 272: 21865–21871. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Merz, F., Rutkowska, A., Zachmann-Brand, B., Deuerling, E., and Bukau, B. 2006. Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J. Biol. Chem. 281: 6539–6545. [DOI] [PubMed] [Google Scholar]
- Houry W.A. 2001. Chaperone-assisted protein folding in the cell cytoplasm. Curr. Protein Pept. Sci. 2: 227–244. [DOI] [PubMed] [Google Scholar]
- Huang C.Y. 1982. Determination of binding stoichiometry by the continuous variation method: The Job plot. Methods Enzymol. 87: 509–525. [DOI] [PubMed] [Google Scholar]
- Huang G.C., Li, Y., Zhou, J.M., and Fischer, G. 2000. Assisted folding of D-glyceraldehyde-3-phosphate dehydrogenase by trigger factor. Protein Sci. 9: 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.C., Chen, J.J., Liu, C.P., and Zhou, J.M. 2002. Chaperone and antichaperone activities of trigger factor. Eur. J. Biochem. 269: 4516–4523. [DOI] [PubMed] [Google Scholar]
- Kramer G., Patzelt, H., Rauch, T., Kurz, T.A., Vorderwulbecke, S., Bukau, B., and Deuerling, E. 2004a. Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli . J. Biol. Chem. 279: 14165–14170. [DOI] [PubMed] [Google Scholar]
- Kramer G., Rutkowska, A., Wegrzyn, R.D., Patzelt, H., Kurz, T.A., Merz, F., Rauch, T., Vorderwulbecke, S., Deuerling, E., and Bukau, B. 2004b. Functional dissection of Escherichia coli trigger factor: Unraveling the function of individual domains. J. Bacteriol. 186: 3777–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J.R. 1983. Principles of fluorescence spectroscopy. Plenum Press, New York and London, UK.
- Lee G.J., Roseman, A.M., Saibil, H.R., and Vierling, E. 1997. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16: 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.Y. and Zhou, J.M. 2000. Conformational change of dihydrofolate re ductase near the active site after thiol modification: Detected by limited proteolysis. Biochim. Biophys. Acta 1481: 37–44. [DOI] [PubMed] [Google Scholar]
- Li Z.Y., Liu, C.P., Zhu, L.Q., Jing, G.Z., and Zhou, J.M. 2001. The chaperone activity of trigger factor is distinct from its isomerase activity during co-expression with adenylate kinase in Escherichia coli . FEBS Lett. 506: 108–112. [DOI] [PubMed] [Google Scholar]
- Liang J.J. and Liu, B.F. 2006. Fluorescence resonance energy transfer study of subunit exchange in human lens crystallins and congenital cataract crystallin mutants. Protein Sci. 15: 1619–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S.J., Lin, Y.Z., Zhou, J.M., Tsou, C.L., Wu, P.Q., and Zhou, Z.K. 1990. Dissociation and aggregation of D-glyceraldehyde-3-phosphate dehydrogenase during denaturation by guanidine hydrochloride. Biochim. Biophys. Acta 1038: 240–246. [DOI] [PubMed] [Google Scholar]
- Liu C.P. and Zhou, J.M. 2004. Trigger factor-assisted folding of bovine carbonic anhydrase II. Biochem. Biophys. Res. Commun. 313: 509–515. [DOI] [PubMed] [Google Scholar]
- Liu C.P., Perrett, S., and Zhou, J.M. 2005a. Dimeric trigger factor stably binds folding-competent intermediates and cooperates with the DnaK-DnaJ-GrpE chaperone system to allow refolding. J. Biol. Chem. 280: 13315–13320. [DOI] [PubMed] [Google Scholar]
- Liu C.P., Li, Z.Y., Huang, G.C., Perrett, S., and Zhou, J.M. 2005b. Two distinct intermediates of trigger factor are populated during guanidine denaturation. Biochimie 87: 1023–1031. [DOI] [PubMed] [Google Scholar]
- Lookene A., Zhang, L., Tougu, V., and Olivecrona, G. 2003. 1,1′-bis(anilino)-4-,4′-bis(naphtalene)-8,8′-disulfonate acts as an inhibitor of lipoprotein lipase and competes for binding with apolipoprotein CII. J. Biol. Chem. 278: 37183–37194. [DOI] [PubMed] [Google Scholar]
- Ludlam A.V., Moore, B.A., and Xu, Z. 2004. The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae . Proc. Natl. Acad. Sci. 101: 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T., Ferbitz, L., Deuerling, E., and Ban, N. 2005. A cradle for new proteins: Trigger factor at the ribosome. Curr. Opin. Struct. Biol. 15: 204–212. [DOI] [PubMed] [Google Scholar]
- Merz F., Hoffmann, A., Rutkowska, A., Zachmann-Brand, B., Bukau, B., and Deuerling, E. 2006. The C-terminal domain of E. coli trigger factor represents the central module of its chaperone activity. J. Biol. Chem. 281: 31963–31971. [DOI] [PubMed] [Google Scholar]
- Panda M., Smoot, A.L., and Horowitz, P.M. 2001. The 4,4′-dipyridyl disulfide-induced formation of GroEL monomers is cooperative and leads to increased hydrophobic exposure. Biochemistry 40: 10402–10410. [DOI] [PubMed] [Google Scholar]
- Patzelt H., Kramer, G., Rauch, T., Schonfeld, H.J., Bukau, B., and Deuerling, E. 2002. Three-state equilibrium of Escherichia coli trigger factor. Biol. Chem. 383: 1611–1619. [DOI] [PubMed] [Google Scholar]
- Prasad A.R., Luduena, R.F., and Horowitz, P.M. 1986a. Bis(8-anilinonaphthalene-1-sulfonate) as a probe for tubulin decay. Biochemistry 25: 739–742. [DOI] [PubMed] [Google Scholar]
- Prasad A.R., Luduena, R.F., and Horowitz, P.M. 1986b. Detection of energy transfer between tryptophan residues in the tubulin molecule and bound bis(8-anilinonaphthalene-1-sulfonate), an inhibitor of microtubule assembly, that binds to a flexible region on tubulin. Biochemistry 25: 3536–3540. [DOI] [PubMed] [Google Scholar]
- Schlunzen F., Wilson, D.N., Tian, P., Harms, J.M., McInnes, S.J., Hansen, H.A., Albrecht, R., Buerger, J., Wilbanks, S.M., and Fucini, P. 2005. The binding mode of the trigger factor on the ribosome: Implications for protein folding and SRP interaction. Structure 13: 1685–1694. [DOI] [PubMed] [Google Scholar]
- Scholz C., Stoller, G., Zarnt, T., Fischer, G., and Schmid, F.X. 1997. Cooperation of enzymatic and chaperone functions of trigger factor in the catalysis of protein folding. EMBO J. 16: 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale J.W., Martinez, J.L., and Horowitz, P.M. 1995. Photoincorporation of 4,4′-bis(1-anilino-8-naphthalenesulfonic acid) into the apical domain of GroEL: Specific information from a nonspecific probe. Biochemistry 34: 7443–7449. [DOI] [PubMed] [Google Scholar]
- Seale J.W., Brazil, B.T., and Horowitz, P.M. 1998. Photoincorporation of fluorescent probe into GroEL: Defining site of interaction. Methods Enzymol. 290: 318–323. [DOI] [PubMed] [Google Scholar]
- Sharma K.K., Kaur, H., Kumar, G.S., and Kester, K. 1998a. Interaction of 1,1′-bi(4-anilino)naphthalene-5,5′-disulfonic acid with α-crystallin. J. Biol. Chem. 273: 8965–8970. [DOI] [PubMed] [Google Scholar]
- Sharma K.K., Kumar, G.S., Murphy, A.S., and Kester, K. 1998b. Identification of 1,1′-bi(4-anilino)naphthalene-5,5′-disulfonic acid binding sequences in α-crystallin. J. Biol. Chem. 273: 15474–15478. [DOI] [PubMed] [Google Scholar]
- Sheluho D. and Ackerman, S.H. 2001. An accessible hydrophobic surface is a key element of the molecular chaperone action of Atp11p. J. Biol. Chem. 276: 39945–39949. [DOI] [PubMed] [Google Scholar]
- Shi L., Palleros, D.R., and Fink, A.L. 1994. Protein conformational changes induced by 1,1′-bis(4-anilino-5-naphthalenesulfonic acid): Preferential binding to the molten globule of DnaK. Biochemistry 33: 7536–7546. [DOI] [PubMed] [Google Scholar]
- Smoot A.L., Panda, M., Brazil, B.T., Buckle, A.M., Fersht, A.R., and Horowitz, P.M. 2001. The binding of bis-ANS to the isolated GroEL apical domain fragment induces the formation of a folding intermediate with increased hydrophobic surface not observed in tetradecameric GroEL. Biochemistry 40: 4484–4492. [DOI] [PubMed] [Google Scholar]
- Stoller G., Rucknagel, K.P., Nierhaus, K.H., Schmid, F.X., Fischer, G., and Rahfeld, J.U. 1995. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 14: 4939–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomic S., Johnson, A.E., Hartl, F.U., and Etchells, S.A. 2006. Exploring the capacity of trigger factor to function as a shield for ribosome bound polypeptide chains. FEBS Lett. 580: 72–76. [DOI] [PubMed] [Google Scholar]
- Valent Q.A., Kendall, D.A., High, S., Kusters, R., Oudega, B., and Luirink, J. 1995. Early events in preprotein recognition in E. coli: Interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 14: 5494–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnt T., Tradler, T., Stoller, G., Scholz, C., Schmid, F.X., and Fischer, G. 1997. Modular structure of the trigger factor required for high activity in protein folding. J. Mol. Biol. 271: 827–837. [DOI] [PubMed] [Google Scholar]
- Zeng L.L., Yu, L., Li, Z.Y., Perrett, S., and Zhou, J.M. 2006. Effect of C-terminal truncation on the molecular chaperone function and dimerization of Escherichia coli trigger factor. Biochimie 88: 613–619. [DOI] [PubMed] [Google Scholar]