Abstract
Cdc20/fizzy family proteins are involved in activation of the anaphase-promoting complex/cyclosome, which catalyzes the ubiquitin-dependent proteolysis of cell cycle regulatory proteins such as anaphase inhibitors and mitotic cyclins, leading to chromosome segregation and exit from mitosis. Previous work has shown that human Cdc20 (hCdc20/p55CDC) associates with one or more kinases. We report here that Cdc20-associated myelin basic protein kinase activity peaks sharply in early M phase (embryonic cells) or in G2 phase (somatic cells). In HeLa cells, Cdc20 is associated with the kinase aurora2/Aik. Aurora2/Aik is a member of the aurora/Ipl1 family of kinases that, like Cdc20, previously has been shown to be localized at mitotic spindle poles and is involved in regulating chromosome segregation and maintaining genomic stability. The demonstration that Cdc20 is associated with aurora2/Aik suggests that some function of Cdc20 is carried out or regulated through its association with aurora2/Aik.
Accurate segregation of sister chromatids during mitosis is critical for production of two viable, nontumorigenic daughter cells. Mitosis is initiated by the mitotic cyclin/cdk complexes and ends with the ubiquitin-dependent destruction of the cyclin subunits and other inhibitors of the M/G1 transition. This destruction pathway consists of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), a ubiquitin ligase (E3), and the 26S proteasome that degrades ubiquitinated proteins (1, 2). For cyclins and certain other mitotic targets, the E3 activity is provided by the anaphase promoting complex/cyclosome (APC/C) and is regulated during the cell cycle (refs. 3–5 and reviewed in refs. 6–8). Activation of the APC/C is initiated, after a lag phase, by cyclin B/cdk1 and appears to involve additional kinases (9, 10). APC/C activation has been correlated with phosphorylation of APC/C subunits Cdc16, Cdc27, and Tsg24 by Plk1 (10–15). In addition, the APC/C receives inhibitory phosphorylations through protein kinase A (11, 16, 17).
Cdc20/Fizzy (Fzy) family proteins also are required for activation of the APC/C (10, 18–25). Mutants of Cdc20 were described first in Saccharomyces cerevisiae, where cdc20− cells are defective in several microtubule-mediated processes (26–28). Cdc20 homologs in other organisms include slp1+ in fission yeast, Fzy in Drosophila, X-Fzy in Xenopus, and p55CDC/hCdc20 in humans (18, 23, 29, 30). In Drosophila, Fzy mutants fail to separate sister chromatids or degrade mitotic cyclins, a finding that led to the hypothesis that Cdc20 is required for activation of the APC/C (18, 31). Cdc20 has been shown to play a critical, but not fully understood, role in mitotic activation of the APC/C (10, 20, 22, 24, 25, 32). Cdc20 also has been implicated in the DNA damage and spindle assembly checkpoints. Cdc20 binds Mad2 along with Mad1 and Mad3, components of the spindle assembly checkpoint pathway (21, 29, 33–36). When Cdc20 is complexed with Mad2, the APC/C cannot be activated. It appears that once Mad2 is released, Cdc20 permits APC/C activation (22, 23).
A second Cdc20-related set of proteins includes Cdh1/Hct1 in S. cerevisiae, Fzr in Drosophila, X-Fzr in Xenopus, and Hs-Fzr/hCdh1 in human (refs. 10, 19, 32, and 37; I. Dawson, personal communication). In animal cells, this second set of proteins, collectively termed fizzy-related (Fzr), is proposed to maintain the active state of the APC/C in G1, after Cdc20 destruction (19, 20). In yeast, both Cdc20 and Cdh1 act during mitosis, but at different times, and they have been proposed to function as specificity factors for different APC/C substrates (25, 32, 38).
In mammalian cells, Cdc20 proteins have one or more associated kinase activities (30). Here, we report that human Cdc20 associates with aurora2/Aik, a member of the aurora/Ipl1 family of kinases that, like Cdc20, localizes to the mitotic spindle poles and has been implicated previously in pathways regulating chromosome segregation (39–47). We find that Cdc20-associated aurora2/Aik activity peaks during G2 phase in somatic cells and is significantly lower in both unperturbed mitotic cells collected by mitotic shake-off and in nocodazole-arrested M-phase cells, in which the APC/C activity is suppressed by checkpoint control mechanisms. It is possible that Cdc20 could act as a targeting subunit for aurora2/Aik. Alternatively, aurora2/Aik could function by phosphorylating Cdc20, thereby influencing the activity of Cdc20. These possibilities now can be tested in a variety of experimental systems.
EXPERIMENTAL PROCEDURES
Isolation of Clam Cdc20 cDNAs.
cDNAs were isolated from a clam oocyte cDNA library in λZAPII, made from unstimulated, full-grown clam oocytes by using the ZAP-cDNA Synthesis Kit (Stratagene). A fragment of clam Cdc20 (FT11) was obtained by thermocycling using primer C20F3 [5′-TIGCI(T/A)(C/G)IGGIGGIAA(C/T)GA(C/T)AA-3′] corresponding to the LASGGNDN sequence conserved in human, Drosophila, and S. cerevisiae Cdc20, along with the vector primer, T7, found at the 3′ end of all the cDNAs. A subclone of FT11 (FT11B3) containing the 3′ end of the ORF was used to screen phage plaques for full-length cDNAs. Three cDNAs yielded identical coding sequences except for a C to A change at nucleotide 466, which does not change the amino acid sequence.
Antibodies.
His6-tagged clam or human (30) Cdc20 proteins were purified from bacteria by using Ni2+-NTA agarose (Qiagen, Chatsworth, CA). The recombinant His6-tagged Cdc20 was solubilized in solutions containing 8 M urea, further purified by SDS/PAGE (48), and used to generate rabbit polyclonal antibodies.
Polyclonal antipeptide antibodies recognizing human aurora2/Aik were the gift of G. Gopalan and P. Donovan (Thomas Jefferson University, Philadelphia) (ref. 41 and unpublished work). Antibodies to human p55CDC and human cyclins were purchased from Santa Cruz Biotechnology. Rabbit polyclonal antibodies to human Cdc16 and Cdc27 were a gift of P. Hieter (Johns Hopkins School of Medicine, Baltimore) (49). Immune blotting was done as in ref. 50.
Oocyte Extracts.
Clam oocytes were collected and washed according to refs. 51 and 52. Oocytes (48,000 oocytes/ml) were fertilized, and 1-ml aliquots were collected at 5-min intervals for protein gel samples; 100-μl aliquots were orcein stained and examined microscopically to determine cell cycle position (52). For extracts, ≈3 ml of packed oocytes (3 × 107 cells) in 200 ml of sea water was activated by fertilization. Extracts were prepared as described (51) from G2-arrested oocytes (unfertilized), GVBD meiosis I (germinal vesicle breakdown visible, early meiosis I), interphase 1 (both polar bodies and pronuclei with decondensed chromosomes visible), early mitosis 1 (condensed chromosomes, occasional metaphase plate), late mitosis 1 (anaphase underway, 5-min postmetaphase), mitosis 1 arrest (colchicine-arrested), interphase 2 arrest (emetine-arrested at the two-cell stage with decondensed chromosomes).
HeLa Cell Extracts.
HeLa cells were cultured, and whole-cell lysates were prepared as described (53). For synchronization, cells were treated with 2 mM thymidine for 18–24 hr, released for 8–10 hr, then treated with thymidine for 16–18 hr. Cells harvested directly from the double-thymidine arrest point were considered G1/S. S phase (≈4 hr postrelease) and mitosis were monitored by bromodeoxyuridine incorporation and Hoechst staining (54). G2 cells (8 hr postrelease) were not incorporating bromodeoxyuridine, and the DNA remained decondensed. Mitotic cells were collected by shake-off once cells showed an increase in the mitotic index (≈8–13 hr postrelease). For nocodazole arrest, cells were treated once with thymidine as above, released for 4 hr in fresh medium, treated with 0.3 μM nocodazole for 16–20 hr, and recovered by shake-off. For G1 cells, the nocodazole-arrested cells isolated were replated in complete medium and incubated 4 hr before harvesting. Cells isolated by shake-off or trypsinization were processed as described (55, 56), with sonication in 20 mM Hepes KOH, pH 7.5, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.5% NP-40, 25 mM NaF, and a mixture of protease inhibitors (Boehringer Mannheim).
Immunoprecipitations.
Immunoprecipitations were done in Bolen-modified RIPA buffer (0.15 M NaCl/0.04 M Mops, pH 7.2/0.001 M EDTA/1% NP-40/1% sodium deoxycholate/0.1% SDS) (53) and typically used >200 μg of clam oocyte or HeLa cell extract per reaction. Extracts, antibodies, and Protein A-Sepharose beads (Amersham Pharmacia CL-4B) were incubated with gentle mixing at 4°C 2–16 hr. For SDS/PAGE, the beads were washed three times in RIPA and resuspended in sample buffer. For human Cdc20, the heavy chain Ig band often interfered with detection by Western blot. To avoid this problem, bead-coupled antibodies were used for immunoprecipitations (48). After washing, bound proteins were eluted in 1 M KCl/RIPA, precipitated (57), dried briefly, and dissolved in sample buffer. For kinase assays of immunoprecipitates, beads underwent additional washes in 50 mM Tris⋅HCl, pH 7.4/10 mM MgCl2. Kinase assay conditions were modified from those described (30), to include 0.1 mM spermidine (clam only), 220 μM ATP, 20 μCi γ-[32P]ATP, and 0.4 mg/ml substrate protein. Reactions were 30 min at 25°C (clam) or 30°C (HeLa). After SDS/PAGE, the gel was dried and analyzed by autoradiography or PhosphorImaging.
Sucrose Density Gradients.
Two to three milligrams of clam oocyte extract or HeLa cell extract was used per gradient. Sucrose solutions were made in 50 mM Hepes NaOH, pH 7.3, 1 mM DTT for clam oocyte samples or 50 mM Hepes KOH, pH 7.3, 100 mM KCl, 1 mM MgCl2, 1 mM DTT for HeLa extracts. Five-milliliter gradients were centrifuged 18 hr at 27,000 rpm (68,000 × g) and 4°C in a Sorvall AH650 rotor (4). Twenty-four fractions of 200–220 μl were collected.
RESULTS
Cdc20 Protein Levels Do Not Vary During the Meiotic or Early Mitotic Cycles of Clam Oocytes.
Because the naturally synchronous cell cycles of early embryos provide good opportunities for examining cell cycle stage-specific activities of proteins uncomplicated by the effects of synchronizing reagents, we wanted to examine Cdc20 and the Cdc20-associated kinase activity across the meiotic and mitotic cell cycles of the clam Spisula solidissima. Three independently isolated clam Cdc20 cDNA clones were found to encode the same 522-aa protein (GenBank accession no. AF052575). Clam Cdc20 is very similar to human (48%), Xenopus (49%), and Drosophila (42%) Cdc20 proteins and less similar to Schizosaccharomyces pombe slp1+ (36%) and S. cerevisiae Cdc20 (25%) proteins (see Fig. 6, which is published as supplemental data on the PNAS web site, www.pnas.org). The conservation is particularly strong in the C-terminal region of the protein, which consists of seven WD-40 domains. Clam Cdc20 is less similar to Xenopus and Drosophila fizzy-related (35%) and human Cdh1 (34%), indicating that it is a member of the Cdc20/fizzy group.
Cdc20 protein levels oscillate in somatic yeast and mammalian cells, being highest in G2- and M-phase cells (20, 22, 58). By contrast, immunoblots show that Cdc20 protein levels are roughly constant throughout the meiotic and first two mitotic cell cycles of clam oocytes (Fig. 1). This result extends earlier work with Xenopus eggs, where Cdc20 protein levels were reported to be similar in interphase and M-phase extracts (23). Differences in protein levels across embryonic and somatic cell cycle systems have been seen for other cell cycle regulators such as cyclin E, which is expressed periodically in human cells but is constant in Xenopus early embryonic cell cycles (59, 60).
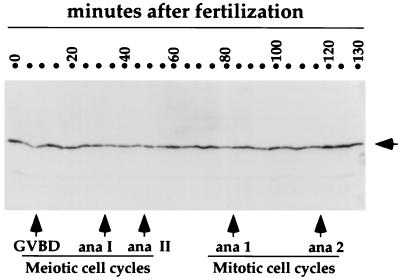
Figure 1.
Clam Cdc20 protein levels are relatively constant across the meiotic and mitotic cell cycles of clam embryos. Oocytes were fertilized at zero min. Aliquots taken at 5-min intervals were pelleted and resuspended in sample buffer. Five microliters (corresponding to ≈1,200 oocytes) of each sample was analyzed by SDS/PAGE, followed by immunoblotting with α-clam Cdc20 antibodies. GVBD is a visual indication of the progression past the G2/M border of meiosis I. Small variations in Cdc20 levels at different time points were not reproducible between sample sets. The arrow indicates the position of clam Cdc20.
Clam Oocyte Cdc20-Associated Kinase Activities.
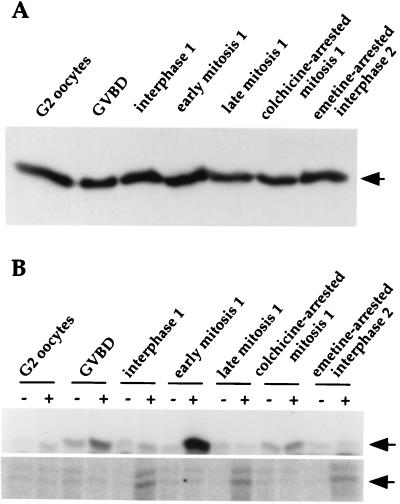
To investigate Cdc20-associated kinase activities at different cell cycle stages, oocytes were fertilized and 10,000 × g supernatants were prepared at the following stages: quiescent oocytes (G2 arrest), GVBD [early meiosis I M phase, just after germinal vesicle (nuclear) breakdown], interphase (S phase, premitosis 1), early mitosis (mitosis 1, preanaphase), late mitosis (mitosis 1, anaphase underway), colchicine-arrested mitosis 1, and interphase arrest (emetine-treated during mitosis 1, collected at interphase 2). All extracts contained Cdc20 protein at approximately the same levels (Fig. 2A). Cdc20 immunoprecipitates were tested for kinase activities toward myelin basic protein (MBP) and α-casein (Fig. 2B). Strong Cdc20-associated MBP kinase activity was seen in early mitotic M-phase extracts. By contrast, embryos arrested in the first mitosis with colchicine, which activates the spindle checkpoint arrest system, showed much lower activity. Oocytes in early meiotic M phase (GVBD) showed intermediate Cdc20-associated kinase activity. Little activity was seen in the late mitotic or interphase extracts. Thus, whereas Cdc20 protein levels are constant across the meiotic and early mitotic cell cycles, Cdc20-associated MBP kinase activity varies: it is low in interphase, peaks sharply in early mitosis, and declines around the time of anaphase onset. These results differed considerably from those obtained by using mammalian cells, where Cdc20-associated MBP kinase activity was reported to be relatively constant across the cell cycle. This point was investigated further by using HeLa cells (see below).
Figure 2.
Clam Cdc20-associated MBP and α-casein kinase activities vary across the cell cycle. (A) Extracts were prepared from: unfertilized oocytes (arrested at G2 of meiosis I), GVBD (meiosis I), interphase 1, early mitosis 1 (preanaphase), late mitosis 1 (anaphase underway), colchicine-arrested mitosis 1, or emetine-arrested interphase 2. Thirty micrograms of each clam extract was analyzed by immunoblotting with anti-clam Cdc20 polyclonal antibodies. The arrow indicates the position of clam Cdc20. (B) Extracts were incubated with preimmune (−) or anti-clam Cdc20 antibodies (+). Immunoprecipitates were assayed for associated kinase activities toward MBP (Upper) or α-casein (Lower). Samples were analyzed by SDS/PAGE followed by autoradiography or PhosphorImaging. Arrows indicate the positions of MBP (Upper) and α-casein (Lower).
A second and different Cdc20-associated kinase activity was seen when immunoprecipitates were assayed by using α-casein as a substrate. This activity was high in interphase, decreased in early M, and reappeared in late mitotic cells (Fig. 2B). These results resemble those reported for mammalian somatic cells (30).
Clam Oocyte Cdc20 Protein Sediments as Part of a Large Complex.
In addition to kinase activities, Cdc20 has been reported to associate with Mad2 and the APC/C (20–22, 33, 35, 36). Sucrose gradient sedimentation analysis of oocyte extracts indicates that Cdc20 is part of a large complex (>19 S), both in interphase and mitotic M-phase cells (Fig. 3). The bulk of Cdc20 sediments more slowly than does the APC/C subunit Cdc16, however, suggesting that the Cdc20-containing complex is mostly distinct from the APC/C. This finding is in agreement with Lorca et al. (23) who reported that the bulk of the Xenopus homolog X-FZY does not coimmunoprecipitate with Cdc27. In somatic cells, some Cdc20 binding to the APC/C is detected (20–22, 33). This difference may indicate that Cdc20 association with the APC/C is less stable or more transient in embryonic cells.
Figure 3.
Clam Cdc20 sediments as part of a large complex in both mitotic and interphase cells. Extracts of interphase 1 or early mitosis 1 clam oocytes were separated on 15%–40% sucrose gradients. Gradient fractions were analyzed by Western blotting with antibodies raised against clam Cdc20 or human Cdc16 (49). Cdc16 is a component of the APC/C, and thus indicates the position of the 1,500-kDa clam oocyte APC/C (4). T is a sample of the total extract loaded on the gradient, and numbers indicate gradient fractions (top to bottom). Size markers are BSA (B; 65 kDa, 4.5 S), catalase (C; 248 kDa, 11.3 S), and thyroglobulin (Th; 670 kDa, 19 S) (4, 12, 17).
During mitosis 1 the clam Cdc16 peak shifts to a slightly larger sedimentation value than in interphase 1 as was reported for the S. pombe APC/C complex (17); by contrast, the position of the Cdc20 peak does not change. The significance of these Cdc16 shifts is not known. The Cdc16 and Cdc20 sedimentation profiles for G2 quiescent oocytes or for meiotic embryo extracts were similar to those from interphase cells (data not shown).
In Human Somatic Cells, Cdc20 Associates with Kinase Aurora2/Aik.
To identify the Cdc20-associated MBP kinase activity that appears late in the cell cycle, we turned to human cells, where a wider range of antibodies was available. Previous work had suggested that Cdc20-associated MBP kinase activity was constant across the human somatic cell cycle (30), but the G2/M cells used in that study were obtained by nocodazole arrest, a treatment that causes cells to accumulate in M phase via the spindle assembly checkpoint pathway (61, 62). In light of our observation in clam oocytes that Cdc20-associated MBP kinase activity was high in naturally synchronous early M-phase cells and low in spindle checkpoint-arrested M-phase cells (Fig. 2B), we re-examined the cell cycle profile of Cdc20-associated MBP kinase activity in human somatic cells.
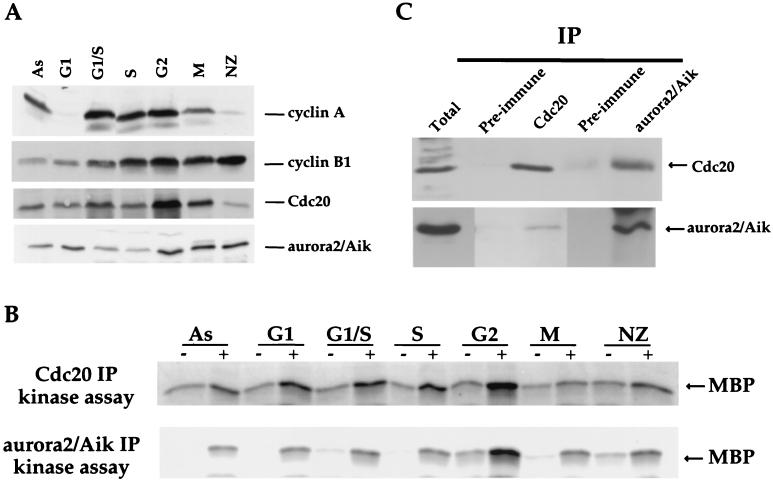
HeLa cells were synchronized and collected at the following cell cycle phases: G1, G1/S, S, G2, M (obtained via mitotic shake-off, which does not depend on the spindle damage checkpoint arrest), and nocodazole-arrested M phase. Samples first were analyzed by immunoblotting to confirm the extent of synchronization (Fig. 4A). As expected from previous work (e.g., refs. 60 and 63), cyclin A levels were very low in G1 cells, rose sharply at G1/S, remained high in G2, dropped during mitosis, and were very low in nocodazole-arrested cells. Cyclin B levels were low in G1 cells, rose gradually across the cell cycle, and were high in both mitotic cells obtained by shake-off and nocodazole-arrested M-phase cells. Cdc20 protein levels increased to a peak in G2, remained high in M phase, and decreased somewhat in nocodazole-arrested M-phase cells.
Figure 4.
Human Cdc20 and aurora2/Aik associate in HeLa cells. (A) HeLa cells were synchronized at the indicated stages and extracts produced as described in the text. Thirty micrograms of each extract was analyzed by Western blotting for cell cycle regulated proteins cyclin A, cyclin B1, hCdc20, or aurora2/Aik. As indicates asynchronously dividing cells. (B) For immunoprecipitate kinase assays, extracts were incubated with either preimmune serum (−) or with antibodies to hCdc20 (+, Upper) or aurora2/Aik (+, Lower); immune complexes were recovered and assayed for kinase activity toward MBP, as described in the text. Phosphorylated MBP was detected by SDS/PAGE followed by autoradiography or PhosphorImaging. (C) Cdc20 or aurora2/Aik immunoprecipitates were prepared as in B, eluted from washed beads, precipitated, and analyzed by SDS/PAGE followed by immunoblotting. Immunoprecipitates were probed for human Cdc20 (Upper) or aurora2/Aik (Lower). Arrows indicate the positions of human Cdc20 and aurora2/Aik.
Cdc20 immune precipitates then were prepared from each stage and assayed for MBP kinase activity. As shown in Fig. 4B, Cdc20-associated MBP kinase activity was low in G1- and S-phase cells, rose during G2, and was low in both cells obtained by mitotic shake-off and nocodazole-arrested M-phase cells. These results argue that in somatic (HeLa) cells Cdc20-associated MBP kinase activity is not constant across the cell cycle but instead peaks during G2. This pattern is consistent with that seen in embryonic (clam) cells: in the first mitotic cell cycle of clam oocytes, Cdc20-associated MBP kinase activity peaks just after cells have entered M phase. Because the early embryonic cell cycles proceed directly from S phase into M phase without an intervening G2 phase, these results are consistent with the idea that Cdc20-associated aurora2/Aik kinase activity peaks at or near the interphase/M border. In agreement with this idea is the observation that Cdc20-associated MBP kinase activity rises as clam oocytes enter M phase of the meiosis I (Fig. 2).
At this point, we recognized several similarities in the behaviors of the Cdc20-associated MBP kinase activity and that of the kinase aurora2 (also termed Aik in humans and IAK1 in mouse; referred to here as aurora2/Aik). First, aurora2/Aik-associated MBP kinase activity peaks in G2/early M-phase cells (41, 42), similar to the profile of the Cdc20-associated MBP kinase activity in both embryonic cells (Fig. 2) and somatic HeLa cells (Fig. 4B). Second, both Cdc20 and aurora2/Aik are localized at centrosomes during G2 and early M phase (21, 41–44, 58, 64). Finally, both Cdc20 and aurora2/Aik function in pathways regulating chromosome segregation (39, 40, 42, 45, 46, 65).
To ask whether aurora2/Aik might represent the Cdc20-associated MBP kinase activity peaking in G2-phase HeLa cells, we first compared the levels of aurora2/Aik and aurora2/Aik-associated MBP kinase activity across the cell cycle with those of Cdc20. Immunoblots showed that aurora2/Aik protein is present at all cell cycle stages examined, showing highest levels in G2, M, and nocodazole-arrested M-phase cells (Fig. 4A). Immunoprecipitation kinase assays showed that aurora2/Aik-associated MBP kinase activity peaks during G2, similar to that seen for Cdc20-associated MBP kinase activity (Fig. 4B). Finally, immunoblots revealed that Cdc20 immune precipitates contained aurora2/Aik, and aurora2/Aik immune precipitates contained Cdc20 (Fig. 4C), directly supporting the idea that Cdc20-associated MBP kinase activity is caused, at least in part, by aurora2/Aik.
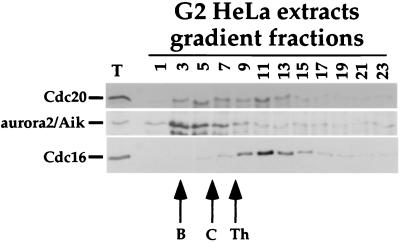
Because clam oocyte Cdc20 appears as a sharply sedimenting, high molecular weight peak on sucrose gradients (Fig. 3), we attempted to ask whether coassociation of Cdc20 and aurora2/Aik also could be demonstrated by gradient fractionation. Unfortunately, the aurora2/Aik antibodies showed no detectable crossreactivity with clam oocyte proteins (not shown). We then investigated this question by using extracts of G2-phase HeLa cells, where Cdc20-associated MBP kinase activity is high. Unlike the discrete peak seen with clam oocytes, HeLa cell Cdc20 sedimented in a very broad peak (Fig. 5). Aurora2/Aik sedimented less broadly, peaking closer to the top of the gradient. Although there is clearly some cosedimentation of Cdc20 and aurora2/Aik, significant portions of each also sediment in different regions of the gradient. Some Cdc20 cosediments with Cdc16, as expected from previous reports of Cdc20 bound to the APC/C; the aurora2/Aik peak is distinct from the Cdc16 peak. These results are consistent with the idea that at least a fraction of Cdc20 and aurora2/Aik associate, and that at least a portion of the Cdc20-associated MBP kinase activity is due to aurora2/Aik. At this point, however, we cannot rule out the possibility that a second kinase also may contribute to Cdc20-associated kinase activity in G2-phase cells.
Figure 5.
Sucrose gradient sedimentation of HeLa cell Cdc20 and aurora2/Aik. Cell lysates from a synchronized population of HeLa G2-phase cells were separated by sedimentation on 15–50% sucrose gradients. Fractions were analyzed by Western blotting as indicated. T is a sample of the total extract loaded on the gradient, and numbers indicate gradient fractions (top to bottom). Molecular weight markers are as in Fig. 3.
DISCUSSION
The main findings of this work are the demonstration that Cdc20’s associated MBP kinase activity peaks late in the cell cycle, and that at least a portion of this activity is the result of its association with the kinase aurora2/Aik. Because Cdc20 and aurora2/Aik both function in pathways that control chromosome segregation during exit from mitosis, the interaction of Cdc20 and aurora2/Aik is likely to be mechanistically important.
The molecular role of Cdc20 in chromosome separation is partly understood. Loss of chromosome cohesion, initiation of anaphase, and sister chromosome segregation are controlled by the APC/C, which requires Cdc20 for the ubiquitin-dependent destruction of Pds1 or functionally related anaphase inhibitors (8, 65). Premature or otherwise inappropriate activation of the APC/C is blocked by the checkpoint pathway protein Mad2, which functionally inactivates Cdc20 until chromosome congression and spindle alignment are completed (22, 23, 65).
By contrast, neither the significance of Cdc20’s association with aurora2/Aik nor the exact role of aurora2/Aik in chromosome segregation is known at this time. Recent work has shown that disregulated expression of aurora2/Aik interferes with accurate segregation of sister chromosomes during exit from mitosis. Overexpression or loss-of-function mutations in aurora2/Aik family members promotes aneuploidy (39, 40, 42, 45, 46), a major cause of tumorigenesis (66, 67). Overexpression of aurora2/Aik (42, 45) or the related Xenopus kinase Eg2 (T. Andresson and J.V.R., unpublished work) in mammalian cells is highly oncogenic. In humans, aurora2/Aik maps to 20q13.2, a chromosomal region that is highly amplified in many breast, bladder, and colon cancers (42, 45, 68). Aurora2/Aik could regulate the function of Cdc20, Cdc20 could act as a targeting subunit for aurora2/Aik, or both. Like Cdc20, members of the aurora2/Aik family are found at the mitotic spindle poles and associated with microtubules (41–44, 64). Cdc20 also is found at kinetochores (21, 22, 58). These localizations place Cdc20 and aurora2/Aik in positions to influence initial spindle structure, to sense spindle integrity, and to control the appropriate initiation of anaphase onset and chromosome segregation.
Our results show that, in human somatic cells, Cdc20-aurora2/Aik kinase activity peaks in G2 phase, whereas genetic and cytological studies indicate that Cdc20 is required for initiation of anaphase (8, 65) and aurora2/Aik is required for accurate separation of sister chromatids (40, 42, 45), both of which are mitotic events. One possible explanation for this difference is suggested by studies with the budding yeast kinase Ipl1, which appears to be the functional homologue of aurora2/Aik (40, 47). Ipl1 is proposed to down-regulate microtubule binding to kinetochores through phosphorylation of the kinetochore protein Ndc10; protein phosphatase 1 counteracts this effect, allowing chromosome attachment to spindle microtubules. Formation of a complete mitotic spindle then is followed by loss of sister chromosome cohesion and anaphase onset (47, 69, 70). In yeast, Cdc20 is required as part of DNA damage checkpoint pathways. In S. cerevisciae, overexpression of Cdc20 prevents arrest at either the DNA damage or spindle assembly checkpoint (34, 35, 71). In S. pombe, mutation of the Cdc20 homolog slp1+ leads to a failure to arrest at the spindle checkpoint (36) or to a defect in restarting the cell cycle and entering mitosis after DNA damage (29). Taken together, these observations suggest that Cdc20-aurora2/Aik complexes could act during G2 to establish conditions required for the timely separation of sister chromatids later in mitosis, after all checkpoint conditions have been satisfied.
Supplementary Material
Acknowledgments
We are especially grateful to Ganesan Gopalan and Peter Donovan for their generous gift of Aik antibodies. We thank Jasminder Weinstein for reagents and advice and Iain Dawson for sharing unpublished results. We are grateful to David Pellman and Holger Bastians for helpful comments on the manuscript. We thank all members of the Ruderman laboratory for support and advice. The clam oocyte cDNA library was made by Ameena Moghe and Ellen Shibuya. This work was supported by the Susan G. Komen Breast Cancer Foundation (D.C.F.), the Wellcome International Prize Research Fellowship (F.M.T.), and National Institutes of Health Grant HD23696 (J.V.R.)
ABBREVIATIONS
- MBP
myelin basic protein
- APC/C
anaphase-promoting complex/cyclosome
- GVBD
germinal vesicle breakdown
Footnotes
Data deposition: The Spisula solidissima (surf clam) Cdc20 nucleotide sequence reported in this paper has been deposited in the GenBank database (accession no. AF052575).
References
- 1.Finley D, Chau V. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 3.King R, Peters J, Tugendreich S M, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 4.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irniger S, Piatti S, Michaelis C, Nasmyth K. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 7.Townsley F, Ruderman J V. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- 8.Peters J. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 9.Lahav-Baratz S, Sudakin V, Ruderman J V, Hershko A. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang G, Yu H, Kirschner M W. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 11.Kotani S, Tugendreich S, Fujii M, Jorgensen P-M, Watanabe N, Hoog C, Hieter P, Todokoro K. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- 12.Peters J M, King R W, Hoog C, Kirschner M W. Science. 1996;274:1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- 13.Descombes P, Nigg E A. EMBO J. 1998;17:1328–1335. doi: 10.1093/emboj/17.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charles J F, Jaspersen S L, Tinker-Kulberg R L, Hwang L, Szidon A, Morgan D O. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita Y M, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. Nature (London) 1996;384:276–279. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]
- 17.Yamada H, Kumada K, Yanagida M. J Cell Sci. 1997;110:1793–1804. doi: 10.1242/jcs.110.15.1793. [DOI] [PubMed] [Google Scholar]
- 18.Dawson I A, Roth S, Artavanis-Tsakonas S. J Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigrist S J, Lehner C F. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 20.Kramer E R, Gieffers C, Holzl G, Hegstschlager M, Peters J M. Curr Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- 21.Kallio M, Weinstein J, Daum J, Durk D, Gorbsky G. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang G, Yu H, Kirschner M W. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorca T, Castro A, Martinez A-M, Vigneron S, Morin N, Sigrist S, Lehner C, Dorée M, Labbé J-C. EMBO J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim H H, Goh P-Y, Surana U. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- 25.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 26.Hartwell L H, Smith D. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sethi N, Monteagudo M C, Koshland D, Hogan E, Burke D J. Mol Cell Biol. 1991;11:5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer R E, Koval M, Koshland D. J Cell Biol. 1989;109:3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto T. Mol Cell Biol. 1997;17:742–750. doi: 10.1128/mcb.17.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstein J, Jacobsen F W, Hsu-Chen J, Wu T, Baum L G. Mol Cell Biol. 1994;14:3350–3363. doi: 10.1128/mcb.14.5.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigrist S, Jacobs H, Stratman R, Lehner C F. EMBO J. 1995;14:4827–4838. doi: 10.1002/j.1460-2075.1995.tb00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwab M, Lutum A S, Seufert W. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 33.Wassman K, Benezra R. Proc Natl Acad Sci USA. 1998;95:11193–11198. doi: 10.1073/pnas.95.19.11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim H H, Surana U. Mol Gen Genet. 1996;253:138–148. doi: 10.1007/s004380050306. [DOI] [PubMed] [Google Scholar]
- 35.Hwang L H, Lau L F, Smith D L, Mistrot C A, Hardwick K G, Hwang E S, Amon A, Murray A W. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 36.Kim S H, Lin D P, Matsumoto S, Kitazono A, Matsumoto T. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura K, Maekawa H, Shimoda C. Mol Biol Cell. 1998;9:1065–1080. doi: 10.1091/mbc.9.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zachariae W, Schwab M, Nasmyth K, Seufert W. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 39.Glover D, Leibowitz M, McLean D, Parry H. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 40.Francisco L, Wang W, Chan C. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gopalan G, Chan C S M, Donovan P J. J Cell Biol. 1997;138:643–656. doi: 10.1083/jcb.138.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bischoff J R, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roghi C, Giet R, Uzbekov R, Morin N, Chartrain I, LeGuellec R, Couturier A, Doree M, Philippe M, Prigent C. J Cell Sci. 1998;111:557–572. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- 44.Yanai A, Arama E, Kilfin G, Motro B. Oncogene. 1996;14:2943–2950. doi: 10.1038/sj.onc.1201144. [DOI] [PubMed] [Google Scholar]
- 45.Zhou H, Kuang J, Zhong L, Kuo W, Gray J W, Sahin A, Brinkley B R, Sen S. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 46.Schumacher J M, Ashcroft N, Donovan P J, Goldern A. Development (Cambridge, UK) 1998;125:4391–4402. doi: 10.1242/dev.125.22.4391. [DOI] [PubMed] [Google Scholar]
- 47.Biggins S, Severin F F, Bhalla N, Sassoon I, Hyman A A, Murray A W. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 49.Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- 50.Sadler K C, Ruderman J V. Dev Biol. 1998;197:25–38. doi: 10.1006/dbio.1998.8869. [DOI] [PubMed] [Google Scholar]
- 51.Ruderman J V, Sudakin V, Hershko A. Methods Enzymol. 1997;283:614–622. doi: 10.1016/s0076-6879(97)83048-0. [DOI] [PubMed] [Google Scholar]
- 52.Hunt T, Luca F C, Ruderman J V. J Cell Biol. 1992;116:707–724. doi: 10.1083/jcb.116.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Townsley F M, Aristarkhov A, Beck S, Hershko A, Ruderman J V. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pines J. Methods Enzymol. 1997;283:99–112. doi: 10.1016/s0076-6879(97)83010-8. [DOI] [PubMed] [Google Scholar]
- 55.Arvand A, Bastians H, Welford S M, Thompson A D, Ruderman J V, Denny C T. Oncogene. 1998;17:2039–2045. doi: 10.1038/sj.onc.1202129. [DOI] [PubMed] [Google Scholar]
- 56.Bastians H, Townsley F M, Ruderman J V. Proc Natl Acad Sci USA. 1998;95:15374–15381. doi: 10.1073/pnas.95.26.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wessel D, Flugge U I. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 58.Weinstein J. J Biol Chem. 1997;272:28501–28511. doi: 10.1074/jbc.272.45.28501. [DOI] [PubMed] [Google Scholar]
- 59.Howe J A, Newport J W. Proc Natl Acad Sci USA. 1996;93:2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dulic V, Lees E, Reed S I. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 61.Rudner A D, Murray A W. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 62.Sorger P, Dobles M, Tournebize R, Hyman A. Curr Opin Cell Biol. 1997;9:807–814. doi: 10.1016/s0955-0674(97)80081-6. [DOI] [PubMed] [Google Scholar]
- 63.Pines J, Hunter T. Nature (London) 1990;346:706–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 64.Kimura M, Kotani S, Hattori T, Sumi N, Yoshioka T, Todokoro K, Okano Y. J Biol Chem. 1997;272:13766–13771. doi: 10.1074/jbc.272.21.13766. [DOI] [PubMed] [Google Scholar]
- 65.Nasmyth K. Trends Biochem Sci. 1999;24:98–104. doi: 10.1016/s0968-0004(99)01358-4. [DOI] [PubMed] [Google Scholar]
- 66.Cahill D, Lengauer C, Yu J, Riggins G, Willson J, Markowitz S, Kinzler K, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 67.Lengauer C, Kinzler K, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 68.Sen S, Zhou H, White R A. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 69.Bloecher A, Tatchell K. Genes Dev. 1999;13:517–522. doi: 10.1101/gad.13.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sassoon I, Severin F F, Andrews P D, Taba M, Kaplan K B, Ashford A J, Stark M J R, Sorger P K, Hyman A A. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schott E J, Hoyt M A. Genetics. 1998;148:599–610. doi: 10.1093/genetics/148.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.