Abstract
In the nematode Caenorhabditis elegans, an insulin receptor signaling pathway regulates adult life span and developmental arrest at the dauer larval stage. Here we show that the unc-64 and unc-31 genes also function in this pathway. These two genes are involved in mediating Ca2+-regulated secretion. Mutations in unc-64 and unc-31 increase adult life span and cause constitutive dauer formation. Both phenotypes are suppressed by mutations in daf-16, which also suppresses other mutations in this pathway. We present evidence that the site of action of unc-64 is neuronal, suggesting that a neurosecretory signal regulates life span and dauer formation.
In Caenorhabditis elegans, genes that regulate life span have been identified by their effects on developmental arrest at the dauer larval stage (1). The dauer larva is an alternative third-stage larva that forms in response to unfavorable environmental conditions and is specialized for long-term survival (2). Mutations in many genes that cause constitutive dauer formation have been isolated. These genes define at least three parallel pathways that regulate dauer formation (3, 4). One of these, an insulin receptor signaling pathway, also regulates adult life span. Mutations in daf-2, which encodes a homolog of the insulin receptor (5), and age-1, which encodes a homolog of the phosphatidylinositol-3-OH kinase catalytic subunit (6), cause adults to live twice as long as wild types and cause constitutive dauer formation (1, 4). Both phenotypes are suppressed by mutations in daf-16, which encodes a Fork head transcription factor (7–10). The current model for this pathway is that activation of the DAF-2 insulin receptor by its ligand (not yet identified) leads to activation of AGE-1, which generates the 3-phosphoinositide second messengers PIP2 and PIP3. These second messengers activate downstream signaling molecules such as the AKT-1 and AKT-2 kinases (11), which ultimately antagonize the activity of DAF-16. DAF-16 presumably acts in the nucleus to control transcription.
The unc-64 and unc-31 genes were originally identified in a screen for mutants affecting locomotion (12). unc-64 and unc-31 encode homologs of syntaxin and CAPS, proteins that mediate Ca2+-regulated secretion (13–16). Here we show that unc-64 and unc-31 also regulate life span and dauer formation through the insulin receptor pathway.
MATERIALS AND METHODS
Strains.
All strains were derivatives of the wild-type strain N2 and were cultured as described (12).
Dauer Formation Assays.
Parents were allowed to lay eggs for 4–6 hours at room temperature, and progeny were incubated at the assay temperature. Animals were counted ≈43 hours later at 27° and ≈103 hours later at 15°. Small differences in temperature around 27° can make significant differences in the percentage of dauers formed (data not shown), so each set of assays included all of the relevant strains. The data compiled in Tables 1 and 3 combine the results from multiple assays in which the mean temperature probably varied slightly. Actual values varied from assay to assay, but the relative values of different strains were consistent in all experiments. For the dauer counts of unc-64(md1259)/unc-64(e246) and controls, unc-64(e246 or md1259)/+ or wild-type males were mated to dpy-18(e364) unc-64(e246) hermaphrodites at 20° for 1 day. The mated hermaphrodites were allowed to lay eggs for 1 day at 20° and then were shifted to 26.8°. Non-Dpy dauers and non-dauers were counted the following day. Dauers and non-dauers were scored as Unc or non-Unc to determine the presence of the paternal unc-64 mutation.
Table 1.
Mutations in unc-64 and unc-31 cause constitutive dauer formation and increase life span
| Genotype | Percent of dauers at 26.8 ± 0.2°C* | Days lived at 20°C† |
|---|---|---|
| N2 (wild-type) | 12 ± 10 | 22 ± 5 (42) |
| unc-64(e246) | 99 ± 2 | 39 ± 12 (46) |
| unc-31(e928) | 93 ± 11 | 29 ± 12 (72) |
| daf-16(m27) | 6 ± 5§ | 18 ± 3 (47) |
| daf-16(m27); unc-64(e246) | 5 ± 4§ | 20 ± 5 (47) |
| daf-16(m27); unc-31(e928) | 8 ± 6§ | 19 ± 5 (104) |
| daf-5(e1385); unc-64(e246) | 99 | |
| daf-5(e1385); unc-31(e928) | 100 | |
| +/unc-64(e246)‡ | 9 | 23 ± 5 (14) |
| unc-64(e246)/unc-64(e246)‡ | 100 | 44 ± 14 (9) |
| unc-64(md1259)/unc-64(e246)‡ | 94 | 38 ± 8 (17) |
| unc-31(u280) | 100 | 29 ± 10 (40) |
The top part of the table gives the mean ± SD of five independent experiments at temperatures ranging between 26.6 and 27.0°C. Each experiment had at least 100 animals of each strain. The middle two lines give the mean of two experiments at 26.6 and 27.0°C. The bottom part of the table gives the results from a single experiment at 26.8°C.
The top part of the table combines the data from two independent experiments and gives the mean ± SD for the entire data set. No strain differed significantly from one experiment to the other (all P values > 0.15). The bottom part of the table gives the results from a single experiment. The number in parentheses is the number assayed.
This unc-64(e246) chromosome also carries dpy-18(e364).
Partial dauers (21).
Table 3.
unc-64 acts in the nervous system
| Relevant genotype | Expression* | Percent of dauers at 26.8 ± 0.2°C† |
|---|---|---|
| N2 (wild-type) | – | 12 ± 12 |
| unc-64(e246) | – | 100 ± 0 |
| unc-64(e246); saEx357‡ | Neurons | 70 ± 20 |
| unc-64(e246); saEx358‡ | Intestine | 99 ± 1 |
Predominant tissue in which the transgene is expressed as determined by fusion of gfp to the identical promoter (data not shown). saEx357 fuses unc-64 to the aex-3 promoter, which is expressed strongly in all neurons and weakly in intestine. saEx358 fuses unc-64 to the elt-2 promoter, which is expressed in the intestine and parts of the pharynx.
Results are the mean ± SD of five independent experiments at temperatures ranging between 26.6 and 27°C. Each experiment had at least 50 animals of each strain.
These strains carry a chromosomal lin-15(n765) mutation and lin-15(+) in the extrachromosomal array.
Life Span Assays.
Animals were assayed as described (8). For the data presented in Table 2 and Fig. 2, animals were grown at 15° until the L4 or young adult stage to bypass the dauer stage and then were shifted to 20° for the remainder of the experiment. All other experiments were performed exclusively at 20°. Both unc-64 and unc-31 mutants grew to the adult stage at approximately the same rate as N2. unc-31 mutant animals are defective in egg-laying (17) and frequently were killed by the internal hatching of larvae. Such animals were excluded from the assay results. All P values were determined by the Mann–Whitney U test by using instat 2.01 software (http://www.graphpad.com/instat3/instat.htm).
Table 2.
Mutations in unc-64 and unc-31 do not enhance the dauer and life span phenotypes of daf-2 mutants
| Genotype | Percent of dauers at 15°C* | Days lived at 20°C† |
|---|---|---|
| N2 (wild-type) | 0 | 24 ± 7 (20) |
| unc-63(e246) | 1 | 33 ± 14 (27) |
| unc-31(e928) | 0 | 35 ± 8 (29) |
| daf-2(e1370) | 0 | 57 ± 9 (31) |
| daf-2(e1370) unc-64(e246) | 1 | 59 ± 13 (37) |
| daf-2(e1370); unc-31(e928) | 1 | 47 ± 9‡ (62) |
| unc-64(e246); unc-31(e928) | 67 | 43 ± 11 (36) |
Results are the mean of two independent experiments at 15°C. More than 200 animals of each strain were assayed. We attempted dauer counts at 20°C, but daf-2, daf-2 unc-64, and daf-2; unc-31 all formed a high percentage of dauers, making interpretation of synergy difficult.
Worms were grown at 15°C until they were at the L4 or young adult stage and then were shifted to 20°C. Results are the mean ± SD from a single experiment. The number in parentheses is the number assayed.
Old daf-2; unc-31 animals carry hatched but apparently dead L1 larvae in their body. This does not kill the worms immediately, but it could reduce life span and makes it difficult to interpret the life span data for this strain.
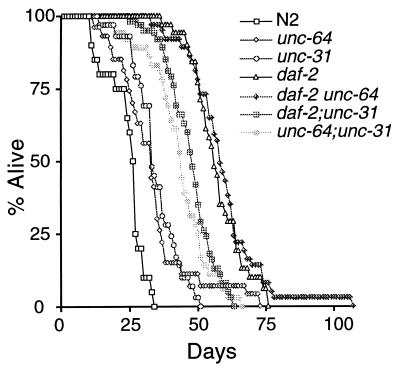
Figure 2.
Mutations in unc-64 and unc-31 do not enhance the longevity of daf-2 mutants. Animals were grown at 15° to allow development past the dauer stage and then were shifted to 20°. unc-64; unc-31 had a significantly longer life span than either unc-64 (P = 0.0002) or unc-31 (P = 0.0005). The daf-2 unc-64 double mutant and daf-2 were not significantly different (P = 0.5139). The maximum life span of daf-2 unc-64 was extended to >100 days by a single animal. We have not investigated whether this has any possible significance. This figure uses the same data set as Table 2.
unc-64 Transgenic Constructs.
The unc-64 B cDNA from pTX4 (14) was first modified to delete an upstream and out-of-frame ATG in the 5′ untranslated region. An ≈400-bp fragment of the 5′ end of the gene was amplified by PCR, was digested with XbaI and HpaI, and was ligated to pTX4 digested with XbaI and HpaI. This resulted in a deletion of ≈100 bp of the unc-64 5′untranslated region, including the out-of-frame ATG. The construct was sequenced to confirm that no undesired mutations had been generated by PCR. The resulting cDNA was cloned into vectors containing the hsp16–2, aex-3, and elt-2 promoters. unc-64(e246); lin-15(n765) animals were injected with lin-15(+) DNA (18) as a transgenic marker at 120 ng/μl and the appropriate unc-64 construct at 20 ng/μl. For each construct, several transgenic lines were generated. Similar dauer phenotypes were seen for the different lines of a given construct, so only one line was analyzed in more detail to give the results seen in Table 3.
RESULTS AND DISCUSSION
Developmental arrest of C. elegans at the dauer larval stage increases as the temperature increases (19, 20). Mutations in many genes cause constitutive dauer formation at 25°C. Of these, only daf-2 and age-1 mutants have an increased life span and are suppressed by mutations in daf-16. Though originally identified by their uncoordinated movement phenotypes (12), we found that the unc-64(e246) and unc-31(e928) mutations caused constitutive dauer formation at 27° but not at 25° (Table 1 and data not shown). An unc-64(e246); unc-31(e928) double mutant formed dauers at temperatures <25° (Table 2). These two results suggest that unc-64 and unc-31 regulate dauer formation but have weaker effects individually than the previously characterized dauer-constitutive genes. There are multiple genetic pathways regulating dauer formation (3, 4). To determine the pathway in which unc-64 and unc-31 function, we built double mutants between unc-64 or unc-31 and mutations that suppress each of the pathways. The dauer-constitutive phenotypes of unc-64 and unc-31 were completely suppressed by mutations in daf-16 (Table 1), and the unc-64; unc-31 double mutant also was suppressed completely by a mutation in daf-16 (data not shown). In contrast, unc-64 and unc-31 were not suppressed by mutations in daf-5 (Table 1), which suppresses mutations (e.g., daf-7) that disrupt the parallel type β transforming growth factor pathway. These data place unc-64 and unc-31 in the genetic pathway of the daf-2 and age-1 genes.
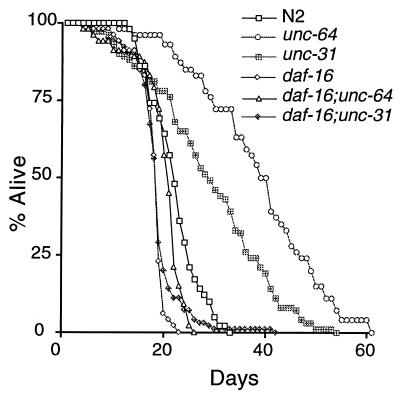
Because unc-64 and unc-31 affect the same dauer pathway as daf-2 and age-1, we tested whether they also regulate adult life span. We found that both the unc-64(e246) and unc-31(e928) mutants had significantly longer life spans than the wild type (Table 1, Fig. 1), although the life span extension was not as great as for daf-2 and age-1. These weaker life span phenotypes are consistent with the weaker dauer phenotypes of the unc-64 and unc-31 mutants. Furthermore, daf-16(m27) suppressed the life span extension of both unc-64(e246) and unc-31(e928) (Table 1 and Fig. 1). Thus, unc-64 and unc-31 appear to be new genes in the daf-2/age-1 pathway regulating dauer formation and life span.
Figure 1.
unc-64 and unc-31 mutants have increased life spans and are suppressed by daf-16. Assays were performed at 20°. unc-64 and unc-31 have significantly longer life spans than N2 (P < 0.0001 for unc-64 and P = 0.0004 for unc-31). This figure uses the same data set as Table 1.
All of the results presented so far were with a single allele of each gene. To determine whether the dauer and life span phenotypes of these mutants were allele-specific, we examined an additional allele of each gene. An unc-31(u280) amber mutant (17) had dauer and life span phenotypes virtually identical to the unc-31(e928) mutant (Table 1). The unc-31(e928) mutation is a deletion that removes most of the unc-31 gene, so it is presumably null (15). The unc-64(e246) mutation is a missense mutation (13, 14). Null alleles of unc-64 result in larval lethality (14) and thus cannot be examined for dauer and life span phenotypes. unc-64(md1259), a mutation in a splice site resulting in viable but unhealthy animals (14), caused constitutive dauer formation on its own at 27°, but broods were asynchronous and difficult to count, so we examined the phenotypes of unc-64(md1259) in trans to unc-64(e246), which is fully recessive on its own. unc-64(md1259)/unc-64(e246) animals had dauer and life span phenotypes that were only slightly weaker than those of unc-64(e246) homozygotes (Table 1), indicating that these phenotypes are not allele-specific. We also built double mutants of unc-64(e246) and unc-31(e928) with daf-16(mgDf50), a deletion of almost all of the daf-16 coding sequence and presumed null allele (9). As with the non-sense mutation daf-16(m27) (10), the dauer-constitutive and long-lived phenotypes were completely suppressed by daf-16(mgDf50) (data not shown).
The unc-64 and unc-31 genes could act in a linear pathway with daf-2 and age-1 upstream of daf-16 or in a parallel branch. To help distinguish these possibilities, we built double mutants of unc-64(e246) and unc-31(e928) with the daf-2(e1370) mutation. If unc-64 and unc-31 act in a linear pathway with daf-2, the double mutants should have phenotypes similar to the daf-2 single mutant, but, if unc-64 and unc-31 act in parallel to daf-2, the double mutants might have stronger phenotypes, as seen for double mutants between daf-2 and mutations in the parallel type β transforming growth factor pathway (9). Because the daf-2 single mutant forms 100% dauers at 25° (4, 21), we measured dauer formation at 15°. As shown in Table 2, the double mutants do not have stronger dauer-constitutive phenotypes. In contrast, double mutants of unc-64 or unc-31 with daf-7(e1372) form 100% dauers at 15°, demonstrating that unc-64 and unc-31 are capable of enhancing the dauer-constitutive phenotype of mutants in a parallel pathway. Similar to the lack of synergy seen for the dauer-constitutive phenotype, the life spans of daf-2 unc-64 and daf-2;unc-31 were similar to or shorter than that of the daf-2 single mutant (Table 2 and Fig. 2). Thus, we could find no evidence that unc-64 and unc-31 act in parallel to daf-2. In contrast, both the dauer and life span phenotypes of an unc-64; unc-31 double mutant were stronger than those of either single mutant (Table 2 and Fig. 2), suggesting that unc-64 and unc-31 are partially redundant.
A clue to unc-64 and unc-31 function in the daf-2/age-1 pathway comes from their molecular identities. unc-64 encodes a homolog of syntaxin, a protein involved in synaptic transmission and other types of Ca2+-regulated secretion (13, 14). unc-31 encodes a homolog of CAPS (Ca2+-dependent activator protein for secretion) (15, 16), a protein required for Ca2+-stimulated peptide secretion from a mammalian neuroendocrine cell line (22). Also, both mutants in C. elegans are resistant to the acetylcholinesterase inhibitor aldicarb, a phenotype characteristic of mutants defective in synaptic transmission (23). Thus, it is likely that the dauer and life span phenotypes of these mutants result from a defect in regulated secretion. An unc-31∷lacZ reporter is expressed exclusively in neurons (15) whereas the UNC-64 protein is expressed in many types of secretory tissues, most prominently in the entire nervous system and intestine (14). Because the expression of these two genes coincides only in neurons, we predicted that their site of action in regulating dauer formation and life span would be neuronal. To further test this idea, we made transgenic animals carrying an unc-64 cDNA fused to tissue-specific promoters and tested for rescue of unc-64(e246) mutant phenotypes. Because there are three distinct unc-64 cDNAs (14), we made a fusion to the hsp16–2 heat shock promoter as a measure of the maximal rescue expected from this particular cDNA. unc-64(e246) animals carrying the hsp16–2∷unc-64 fusion without heat shock showed weak rescue of the dauer-constitutive phenotype at 27° and no detectable rescue of the uncoordinated phenotype. With heat shock (1 hour at 33° during the L1 larval stage), there was stronger rescue of the dauer-constitutive (a mean of 50% dauers in four lines ranging from 25 to 73% vs. 100% for unc-64 control) and uncoordinated phenotypes. However, this rescue was still not complete. We fused the same unc-64 cDNA to the aex-3 promoter, which expresses predominantly in the nervous system (ref. 24 and data not shown), and the elt-2 promoter, which expresses predominantly in the intestine (ref. 25 and data not shown). Animals carrying the aex-3∷unc-64 fusion were partially rescued for the dauer phenotype whereas those carrying the elt-2∷unc-64 fusion were not (Table 3). Thus, unc-64 acts at least partly in the nervous system to regulate dauer formation. Failure to achieve complete rescue by the aex-3∷unc-64 transgene could signify function of unc-64 in additional tissues or could simply reflect somatic loss of the extrachromosomal array, insufficient transgene expression or a need for an alternative isoform. Consistent with these last two possibilities, these animals exhibited virtually no rescue of the uncoordinated phenotype, which is expected to have a neuronal focus.
Given the broad neuronal expression patterns of unc-64 and unc-31, one could argue that the life span and dauer phenotypes of these mutants are caused by indirect effects on nervous system function. In addition to the suppression by daf-16, the following observations support a specific role for unc-64 and unc-31 in the daf-2/age-1 pathway. One possible mechanism for increasing life span is caloric restriction, as has been shown for C. elegans mutants defective in eating (26). However, the unc-64(e246) and unc-31 mutants do not significantly decrease pumping rate (14, 17), and their larval growth rates are approximately the same as that of the wild type. The increased life span is also unlikely to be caused by indirect effects on movement because many unc mutants do not affect life span (26). Finally, the phenotypes are probably not caused by general defects in synaptic transmission because other mutants disrupting general synaptic function do not have dauer-constitutive phenotypes at 27°, including unc-11(e47), unc-13(e51), snt-1(md125), and snb-1(md247) (data not shown). snt-1 and snb-1 encode homologs of the synaptic vesicle proteins synaptotagmin and synaptobrevin (27, 28). Thus, the life span and dauer phenotypes of unc-64 and unc-31 mutants appear to be the result of a defect in some type of Ca2+-regulated secretion that is most sensitive to the elimination of unc-64 and unc-31 function.
Insulin release by β cells of the mammalian pancreas is determined by Ca2+-regulated secretion. The simplest model for unc-64 and unc-31 function in regulating life span and dauer formation is that they are involved in the Ca2+-regulated secretion of an insulin-like ligand for the DAF-2 receptor. Although no such ligand has been identified yet, there are a large number of candidate insulin-like genes in the C. elegans genome. We propose that this putative insulin is released from within the nervous system. Mutations in the unc-64 and unc-31 genes would prevent this release and, hence, would prevent activation of the DAF-2 receptor. An alternative model is that unc-64 and unc-31 do not directly mediate secretion of insulin but instead regulate classical neurotransmitter input to the cells that release insulin. Of interest, daf-2 mosaic analysis shows that the DAF-2 receptor functions in neurons and in other tissues to regulate life span and dauer formation (29). Thus, it appears that the nervous system regulates life span at several steps in this pathway.
Acknowledgments
We thank M. Nonet for providing the unc-64 cDNA clone and the unc-64(md1259) mutant; S. Paradis and G. Ruvkun for providing the daf-16(mgDf50) mutant; A. Fire, S. Xu, J. Aynn, and G. Seydoux for providing various vectors; and H. Chamberlin, R. Choy, D. Emu, D. Johnstone, E. Newton, E. Round, and P. Swoboda for comments on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. M.A. is a Howard Hughes Medical Institute Predoctoral Fellow. This work was supported by a National Institutes of Health grant.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. Nature (London) 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 2.Cassada R C, Russell R L. Dev Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- 3.Thomas J H, Birnby D A, Vowels J J. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb S, Ruvkun G. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 6.Morris J Z, Tissenbaum H A, Ruvkun G. Nature (London) 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 7.Larsen P L, Albert P S, Riddle D L. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorman J B, Albinder B, Shroyer T, Kenyon C. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 10.Lin K, Dorman J B, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 11.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa H, Harada S, Sassa T, Yamamoto H, Hosono R. J Biol Chem. 1998;273:2192–2198. doi: 10.1074/jbc.273.4.2192. [DOI] [PubMed] [Google Scholar]
- 14.Saifee O, Wei L, Nonet M L. Mol Biol Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livingstone D. Ph. D. thesis. Cambridge, U.K.: Univ. of Cambridge; 1991. [Google Scholar]
- 16.Ann K, Kowalchyk J A, Loyet K M, Martin T F J. J Biol Chem. 1997;272:19637–19640. doi: 10.1074/jbc.272.32.19637. [DOI] [PubMed] [Google Scholar]
- 17.Avery L, Bargmann C I, Horvitz H R. Genetics. 1993;134:455–464. doi: 10.1093/genetics/134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L S, Tzou P, Sternberg P W. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golden J W, Riddle D L. Proc Natl Acad Sci USA. 1984;81:819–823. doi: 10.1073/pnas.81.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golden J W, Riddle D L. Dev Biol. 1984;102:368–378. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- 21.Vowels J J, Thomas J H. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walent J H, Porter B W, Martin T F J. Cell. 1992;70:765–775. doi: 10.1016/0092-8674(92)90310-9. [DOI] [PubMed] [Google Scholar]
- 23.Miller K G, Alfonso A, Nguyen M, Crowell J A, Johnson C D, Rand J B. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwasaki K, Staunton J, Saifee O, Nonet M, Thomas J H. Neuron. 1997;18:613–622. doi: 10.1016/s0896-6273(00)80302-5. [DOI] [PubMed] [Google Scholar]
- 25.Fukushige T, Hawkins M G, McGhee J D. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- 26.Lakowski B, Hekimi S. Proc Natl Acad Sci USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nonet M L, Grundahl K, Meyer B J, Rand J B. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- 28.Nonet M L, Saifee O, Zhao H, Rand J B, Wei L. J Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apfeld J, Kenyon C. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]