Abstract
Although genetic analysis has demonstrated that members of the winged helix, or forkhead, family of transcription factors play pivotal roles in the regulation of cellular differentiation and proliferation, both during development and in the adult, little is known of the mechanisms underlying their regulation. Here we show that the activation of phosphatidylinositol 3 (PI3) kinase by extracellular growth factors induces phosphorylation, nuclear export, and transcriptional inactivation of FKHR1, a member of the FKHR subclass of the forkhead family of transcription factors. Protein kinase B (PKB)/Akt, a key mediator of PI3 kinase signal transduction, phosphorylated recombinant FKHR1 in vitro at threonine-24 and serine-253. Mutants FKHR1(T24A), FKHR1(S253A), and FKHR1(T24A/S253A) were resistant to both PKB/Akt-mediated phosphorylation and PI3 kinase-stimulated nuclear export. These results indicate that phosphorylation by PKB/Akt negatively regulates FKHR1 by promoting export from the nucleus.
Members of the winged helix, or forkhead, family of transcription factors are characterized by the presence of a conserved 100-aa DNA binding, or forkhead, domain (1). In addition to their roles during normal development, recent studies have demonstrated that members of the winged helix family may participate in neoplasia (2, 3). The consistent involvement of members of the FKHR subclass of the winged helix family in chromosomal translocations found in human cancer suggests that members of this subclass may play a critical role in the regulation of cellular proliferation and/or differentiation (4–7). Thus, an understanding of the mechanisms underlying their regulation may provide insight into the role of not only the FKHR subclass, but also the larger winged helix family in normal and neoplastic development. Here we sought, by biochemical and genetic analyses, to determine whether the activity of FKHR1, a member of the FKHR subclass of forkhead-related proteins, is subject to regulation (W.H.B., W.K.C., and K.C.A., unpublished observation).
MATERIALS AND METHODS
Materials.
The pSG5-P110wt, pSG5-P110CAAX, pSG5-P85wt, and pSG5-P85ΔN-SH2 expression vectors were obtained from Julian Downward (Imperial Cancer Research Fund, London). pCMV5-HA-Akt, pCMV6-myrAkt-HA, pCMV5-HA-Akt(K179M), and pCMV5-HA-Akt(T308A/S473A) were obtained from Philip Tsichlis (Thomas Jefferson Medical College, Philadelphia). Leptomycin B was a kind gift from Barbara Wolff-Winiski (Novartis Forschungsinstitut, Vienna).
FKHR1 Constructs.
Hemagglutinin (HA) epitope-tagged mouse FKHR1 (FKHR1-HA) was generated by using PCR as described (9). Briefly, the HA epitope CYPYDVPDYASC was inserted at the carboxyl terminus of FKHR1 immediately before the translational termination codon. The HA oligonucleotide included a BamHI site after the translation stop to facilitate insertion of FKHR1-HA into the mammalian expression vector pcDNA3.1 (CLONTECH). FKHR1(T24A)-HA, FKHR1(S253A)-HA, and FKHR1(T24A/S253A)-HA also were generated by using PCR. All altered forms of FKHR1 were sequenced to confirm that no other changes had occurred during the modification process.
A series of carboxyl-terminal deletions of FKHR1 were generated by PCR. The PCR products were ligated in-frame to the gene encoding green fluorescent protein (GFP) in pEGFP-N1 (CLONTECH). Mutation of leucine-375 to alanine also was carried out by PCR. All constructs were sequenced to ensure that no spurious mutations had occurred and that the fusion between FKHR1 and GFP was correct. CV1 cells were transiently transfected with each construct, and the fusion protein was visualized by using epifluorescence.
Cell Culture.
The CV1 cell line was maintained by passage in high-glucose DMEM supplemented with 10% FCS, 100 mg/ml penicillin, and 100 mg/ml streptomycin. For immunofluorescence, 7.5 × 104 cells per well were seeded onto sterile coverslips in 6-well dishes. Transfections were carried out by using a modified calcium phosphate protocol as described (9). Cotransfections of pCNDA3.1-P110CAAX (or P85ΔN-SH2) and FKHR1 constructs were carried out at a ratio of 3:1 (P110 or P85/FKHR1). Pretreatments where indicated were as follows: 20 nM leptomycin B for 6 hr, 50 μM LY290042 (Calbiochem) for 60 min, and 10 nM wortmannin (Calbiochem) for 60 min before fixation. The media, alone or with drugs as indicated, were changed 1–2 hr before fixation. For serum starvation, CV1 cells transfected with pcDNA3.1-FKHR1-HA were allowed 24 hr of recovery in DMEM + 10% FCS, then washed extensively with DMEM without added serum and subsequently maintained in serum-free conditions for an additional 24 hr before fixation and staining. For kinetic analysis of relocalization, CV1 cells transiently transfected with FKHR1-HA were treated as described for serum starvation. After 24 hr of serum starvation, the cells’ serum-free media were exchanged for media containing 50 nM insulin-like growth factor I (GIBCO/BRL). Cells were washed and fixed at the times indicated.
Immunohistochemistry.
Immunofluorescent detection of FKHR1-HA was carried out as described (10). Anti-HA mAb (16B12, Berkeley Antibody, Richmond, CA) was used at 1:1,000. Lissamine rhodamine-conjugated donkey anti-mouse secondary antibody was used at 1:200. The average (± SE) number of cells in which FKHR1-HA, FKHR1(T24A)-HA, FKHR1(S253A)-HA, or FKHR1(T24A/S253A)-HA) was localized to the nucleus or cytoplasm or was uniformly distributed was determined by counting ≥500 FKHR1-HA-expressing cells in each of three independent experiments.
FKHR1 Luciferase Reporter Assays.
CV1 cells were transfected with the reporter 8xFK1tkLuc (8XFK1tkLuc contains a direct repeat of eight FKHR1 binding sites upstream of the herpes simplex virus thymidine kinase minimal promoter and the gene encoding firefly luciferase), pCMV-β-galactosidase (β-gal), and a number of additional constructs as indicated in the individual figures. In all experiments the ratio of 8xFK1tkLuc to wild-type or mutant forms of FKHR1-HA was held constant at 1:2. A constant amount of DNA (8 μg) was used in each transfection by the addition of pcDNA3.1 when necessary. Luciferase and β-gal activities were determined as described (9). β-Gal values were used to normalize luciferase values for transfection efficiency. Luciferase values are reported relative to the activity of the 8xFK1tkLuc reporter alone. Each reported value represents the average of duplicate plates from at least two independent experiments.
Cell Labeling.
CV1 cells transiently transfected with either FKHR1-HA, FKHR1(S253A)-HA, or FKHR1(T24A/S253A)-HA were extensively washed 48 hr posttransfection with phosphate-free DMEM supplemented with 10% dialyzed FCS (1,000 MW cutoff, Sigma). Cells were labeled for 3 hr at 37°C in phosphate-free DMEM supplemented with 10% dialyzed FCS containing 1.5 mCi/ml 32PO4. Labeled cell lysates were prepared, and FKHR1-HA, FKHR1(S253A)-HA, and FKHR1(T24A/S253A) were immunoprecipitated as described (11) with the exception that lysis was carried out in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) supplemented with protease and phosphatase inhibitors and with 2 mM EDTA added to inhibit phosphorylation during the lysis. Phosphoamino acid analysis and tryptic-phosphopeptide mapping was carried out by using chloromethyl ketone-trypsin (Worthington) as described (12). The first (electrophoretic) dimension was performed at 1 kV for 25–35 min at pH 1.9. The second dimension was generated by ascending chromatography in phosphochromo buffer until the solvent front was ≤2.5 cm from the top of the chromatography plate.
Akt in Vitro Kinase Assays.
Plates (6 cm or 10 cm) containing CV1 cells transiently transfected with either pcDNA3.1 (mock), pCMV-myrAkt-HA, pCMV-HA-Akt(K67A), or pCMV-HA-Akt(T308A/S473A) plasmids were rinsed with ice-cold PBS. Cells were lysed and immunoprecipitates were prepared as described (11). In vitro kinase reactions were performed for 40 min at 30°C with constant shaking in 30-μl reaction volumes containing 500 ng histone 2B (H2B), glutathione S-transferase (GST), or GST-FKHR1 as substrates, and 10 μCi [α-32P]ATP as described (13). The kinase reactions were stopped by the addition of SDS/PAGE loading buffer and boiling for 5 min. The kinase reactions were resolved by 15% SDS/PAGE (H2B and GST) or 7.5% SDS/PAGE (GST-FKHR1). The SDS/PAGE gels were washed 5 min in dH2O, dried, and subjected to autoradiographic exposure.
RESULTS AND DISCUSSION
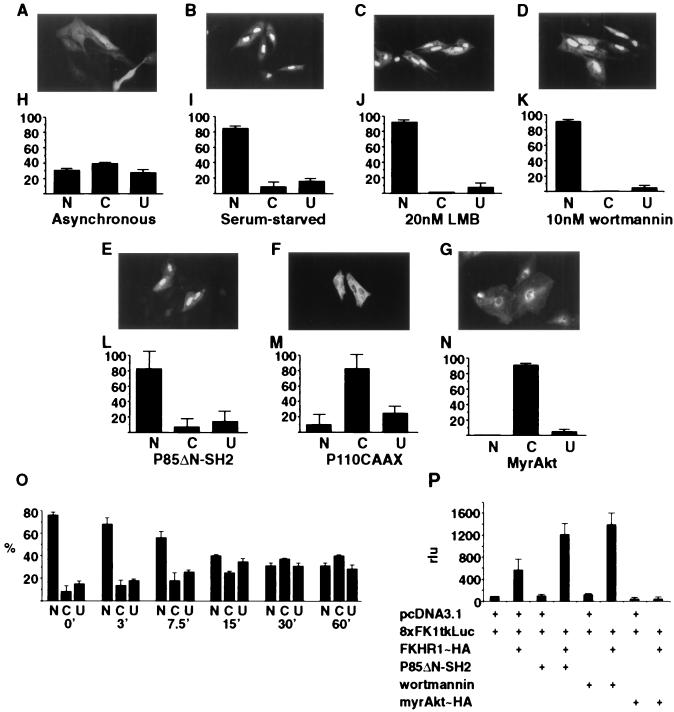
Asynchronously growing CV1 cells transfected with a HA epitope-tagged version of mouse FKHR1 (FKHR1-HA) displayed three distinct populations with respect to localization. In 20% of the expressing cells FKHR1-HA was localized to the nucleus, 30% of the cells exhibited a uniform staining throughout, and ≈50% of the cells exhibited a pattern of localization in which FKHR1-HA was excluded from the nucleus (Fig. 1 A and H).
Figure 1.
Nuclear/cytoplasmic shuttling and transcriptional activity of FKHR1-HA in transiently transfected cells. (A–G) Detection of FKHR1-HA by anti-HA immunofluorescent or epifluorescent staining. (H–N) Quantification of the nuclear (N), cytoplasmic (C), and uniform (U) localization of FKHR1-HA or FKHR1-GFP. Immunolocalization of FKHR1-HA in CV1 cells. (A and H) Asynchronous culture. (B and I) Culture in the absence of serum for 24 hr. (C and J) Treatment with 20 nM leptomycin B (LMB) for 6 hr before fixation and staining. (D and K) Treatment with 10 nM wortmannin for 3 hr before fixation. (E and L) Cotransfection of FKHR1-HA with pSG5-P85ΔN-SH2. (F and M) Cotransfection of FKHR1-HA with pcDNA3.1-P110CAAX. (G and N) Cotransfection of FKHR1-GFP with pCMV6-myrAkt-HA. (O) Kinetic analysis of FKHR1-HA relocalization induced by 50 nM insulin-like growth factor I. (P) Localization of FKHR1-HA directly affects the expression of a FKHR1-responsive reporter. CV1 cells transfected with the FKHR1-responsive reporter 8XFK1tkLuc and cytomegalovirus (CMV)-β-gal, together with FKHR1-HA, P85ΔN-SH2, and myrAkt-HA as indicated. Cells transfected with 8XFK1tkLuc, CMV-β-gal, and FKHR1-HA also were treated with 10 nM wortmannin. Luciferase acivities of the individual samples were normalized to the activity of the 8XFK1tkLuc reporter in the absence of any other construct.
The observation of three distinct patterns (nuclear, cytoplasmic, and both) of localization of FKHR1 suggested the possibility that its intracellular localization might be regulated by extracellular growth signals or cell cycle progression. This hypothesis was tested by determining the localization of FKHR1-HA in serum-starved cells. CV1 cells transiently transfected with FKHR1-HA subsequently were maintained in serum-free medium for 24 hr before fixation. Under conditions of serum starvation FKHR1-HA was restricted to the nucleus in greater than 90% of the cells (Fig. 1 B and I). Moreover, treatment of serum-starved cells expressing FKHR1-HA with either 50 nM insulin-like growth factor I (Fig. 1O) or 10% serum (data not shown) for periods of time as short as 15–30 min was sufficient to cause export of FKHR1-HA from the nucleus in 70% of cells.
The growth factor stimulation of FKHR1-HA nuclear export could reflect either a passive process, such as inhibition of DNA binding leading to a size-based exclusion from the nucleus, or an active process. To distinguish between these possibilities, CV1 cells transiently transfected with constructs expressing FKHR1-HA were treated with leptomycin B (LMB), which quantitatively blocks Crm1-mediated nuclear export through high affinity binding to Crm1 (14–16). This binding prevents the interaction of Crm1 with leucine-rich nuclear-export sequence (NES)-containing target proteins, thereby preventing their export (11). Asynchronously growing CV1 cells treated with 20 nM LMB showed a retention of FKHR1-HA in the nucleus (Fig. 1 C and J), indicating that the serum-stimulated export of FKHR1-HA from the nucleus is an active process likely involving Crm1.
Epistasis studies have revealed that DAF-16, a Caenorhabditis elegans forkhead family member closely related to FKHR1, is negatively regulated by a PI3 kinase-activated signal transduction pathway (17–19). To assess the role of the PI3 kinase pathway in the regulation of FKHR1 nuclear exclusion and transcriptional activity, we inhibited PI3 kinase activation by using the specific chemical inhibitors wortmannin and LY294002 (20). Treatment of FKHR1-HA-expressing CV1 cells with 10 nM wortmannin (Fig. 1 D and K) or 50 μM LY294002 (data not shown) effectively blocked export of FKHR1-HA from the nucleus. Additionally, cotransfection of FKHR1-HA with a deletion mutant of the P85 subunit of PI3 kinase (P85ΔN-SH2), previously shown to inhibit activation of PI3 kinase in a dominant negative fashion (21), also effectively blocked the nuclear export of FKHR1-HA (Fig. 1 E and L). In comparison, cotransfection of FKHR1-HA with either constitutively active forms of the P110 subunit of PI3 kinase (P110CAAX) (22) or Akt/PKB (myrAkt-HA) (11) resulted in the exclusion of FKHR1-HA from the nucleus of virtually all cells (Fig. 1 F, G, L, and M). These data demonstrate that the nuclear export of FKHR1 in response to serum, or insulin-like growth factor I, requires activation of the PI3 kinase signal transduction pathway. To address the functional significance of PI3 kinase-mediated exclusion of FKHR1 from the nucleus, the transcriptional activity of FKHR1, under conditions promoting nuclear exclusion (e.g., coexpression of myrAkt-HA) or import (e.g., treatment with wortmannin), was determined.
Coexpression of P85ΔN-SH2 and FKHR1-HA in asynchronous CV1 cells resulted in a 2-fold increase in FKHR1-dependent transcriptional reporter activity in comparison to cells expressing FKHR1-HA alone (Fig. 1P). Similar levels of activation were observed in FKHR1-HA-expressing CV1 cells upon treatment with 10 nM wortmannin (Fig. 1P). In contrast with the FKHR1-HA-dependent transcriptional activation observed with P85ΔN-SH2 coexpression and wortmannin treatment, coexpression of myrAkt-HA completely inhibited the transcriptional activity of FKHR1-HA (Fig. 1P). These data suggest that by promoting the exclusion of FKHR1 from the nucleus the PI3 kinase signal transduction pathway effectively inhibits FKHR1-mediated transcription. Moreover, these results provide insight into the mechanism underlying the observation from C. elegans, that PI3 kinase (AGE1) and PKB/Akt (Akt1 and Akt2) signaling abrogates DAF16 activity.
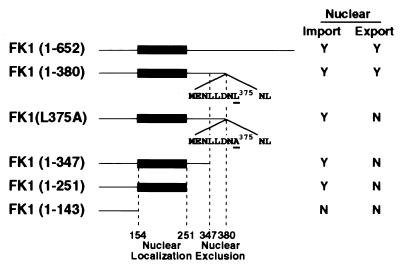
To identify those region(s) that were required for FKHR1 nuclear import/export, a series of carboxyl-terminal deletions were fused in-frame to the gene encoding GFP and transiently expressed in CV1 cells. Localization of these fusion proteins demonstrated that amino acids 147–251 and 347–380 of FKHR1 were required for nuclear import and export, respectively (Fig. 2). The demonstration that the winged helix domain of FKHR1 (amino acids 147–251) is required for nuclear import shows that, in a manner similar to the winged helix family member HNF3β (23), the DNA binding domain of FKHR1 contributes both DNA binding and nuclear localization functions. The presence of a leucine-rich sequence (M368ENLLDNLNL377), which conforms to the consensus leucine-rich NES (LXXXLXXLXL and LXXLXXXLXL) (24) in the region (amino acids 347–380) required for exclusion from the nucleus, suggested that this sequence might be required for nuclear export of FKHR1. Mutation of leucine-375 to alanine (M368ENLLDNANL377) blocked export of the FKHR1-GFP fusion (Fig. 2) protein, suggesting that the sequence M368ENLLDNLNL377 is required for export of FKHR1 from the nucleus. Similar mutations within the leucine-rich NESs present in human T-lymphotrophic virus type I Rex (25) and the c-Abl kinase (24) have been shown to prevent nuclear exclusion of these proteins.
Figure 2.
Immunolocalization of FKHR1-GFP fusion proteins. CV1 cells were transiently transfected with a series of FKHR1-GFP fusion proteins with carboxyl-terminal deletions of FKHR1 and examined by fluorescence microscopy. The regions identified as being required for either nuclear localization (amino acids 147–251) or nuclear exclusion (amino acids 347–380) are indicated. The sequence of the putative leucine-rich NES (M368ENLLNLNL377) is indicated.
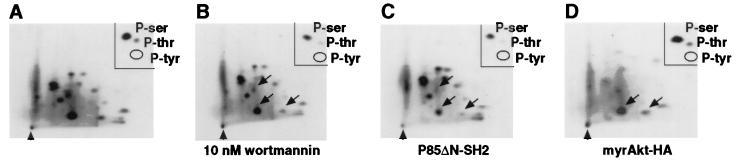
Because protein phosphorylation is a widely used mechanism by which the activity of transcription factors are either positively or negatively regulated (26, 27), we next assessed the phosphorylation state of FKHR1-HA in asynchronous CV1 cells. CV1 cells expressing FKHR1-HA were metabolically labeled with 32PO4, and the FKHR1-HA protein was immunoprecipitated. Incorporation of 32P into FKHR1-HA provided evidence that FKHR1-HA is phosphorylated in vivo (data not shown). Two-dimensional mapping of tryptic fragments revealed phosphorylation at several positions (Fig. 3A), and phosphoamino acid analysis showed that FKHR1-HA is phosphorylated on both serine and threonine (Fig. 3A, Inset). Although the overall level of phosphorylation of FKHR1-HA was largely unaffected by treatment with 10 nM wortmannin (data not shown), phosphorylation of one major and two minor peptides appeared to by slightly reduced (Fig. 3B, arrows). Cotransfection of P85ΔN-SH2 with FKHR1-HA resulted in a decrease in the phosphorylation of FKHR1-HA (Fig. 3A, lane 3), and two-dimensional tryptic phosphopeptide mapping revealed a disproportionate reduction in the same peptides affected by wortmannin treatment (Fig. 3C, arrows). Finally, cotransfection of the constitutively active myrAkt-HA with FKHR1-HA resulted in a relative increase in phosphorylation of the major phosphopeptide affected by either P85ΔN-SH2 expression or wortmannin treatment (Fig. 3D). Taken together, these results strongly suggest that the phosphorylation of FKHR1-HA by the PI3 kinase-activated signal transduction pathway regulates its subcellular localization.
Figure 3.
Phosphorylation of FKHR1-HA in asynchronously growing CV1 cells. (A–D) Two-dimensional tryptic phosphopeptide maps of 32P-labeled wild-type FKHR1-HA immunoprecipitated from metabolically labeled CV1 cells either treated or cotransfected as indicated. Thin layer cellulose plates were run as indicated (horizontal: electrophoresis anode to left; vertical: chromatography thin layer analysis). The position of the origin is indicated (arrowhead). (Insets) Phosphoamino acid content of each of the corresponding 32P-labeled FKHR1-HA immunoprecipitates. The locations of 32P-labeled phospho-serine (P-ser) and phospho-threonine (P-thr) present on FKHR1-HA are indicated. No phospho-tyrosine (P-tyr) was detected. (A) FKHR1-HA. (B) FKHR1-HA immunoprecipitated from metabolically labeled CV1 cells treated with 10 nM wortmannin. Arrows indicate those phosphopeptides that were reduced. (C) FKHR1-HA immunoprecipitated from metabolically labeled CV1 cells cotransfected with FKHR1-HA and P85ΔN-SH2. Arrows indicate those phosphopeptides that were reduced. (D) FKHR1-HA immunoprecipitated from metabolically labeled CV1 cells cotransfected with FKHR1-HA and myrAkt-HA. Arrows indicate those phosphopeptides that were increased.
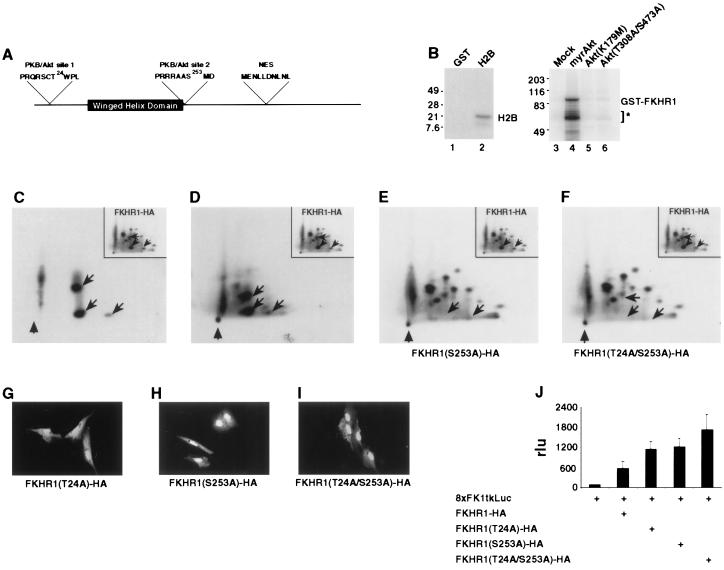
Several recent studies have demonstrated that PKB/Akt plays a central role in the PI3 kinase signal transduction pathway (28–34). Because PI3 kinase signaling regulates FKHR1-HA localization (this work), it was of interest to note that FKHR1 contains the two sequences RPRSCT24W and RRRAAS253M, which bear a close resemblance to the consensus sequence that is phosphorylated by PKB/Akt (13) (R X R Y Z S/T Hyd, where X is any residue, Y and Z are any small side residues except Gly, and Hyd is a bulky hydrophobic side-chain residue) (Fig. 4A). Both of these potential PKB/Akt phosphorylation sites are conserved among the members of the FKHR subclass (8, 17–19, 35).
Figure 4.
Regulation of FKHR1-HA localization by PKB/AKT-mediated phosphorylation. (A) Relative location of the two consensus AKT phosphorylation sites (threonine-24 and serine-253), as well as the putative leucine-rich NES, within FKHR1. The winged helix DNA binding domain also is indicated. (B) PKB/AKT phosphorylates GST-FKHR1 in vitro. Lane 1, GST incubated with myrAkt-HA; lane 2, H2B incubated with myrAkt-HA; lane 3, GST-FKHR1 incubated with immunoprecipitate from mock-transfected cells; lane 4, GST-FKHR1 incubated with myrAkt-HA; lane 5, GST-FKHR1 incubated with Akt(K67A)-HA; lane 6, GST-FKHR1 incubated with Akt(T308A/S473A)-HA. Degradation products of GST-FKHR1 that copurified with the full fusion protein are indicated (∗). These fragments also were phosphorylated by myrAkt-HA. Molecular mass (kDa) markers (×1,000) are indicated. (C) Two-dimensional tryptic phosphopeptide map of GST-FKHR1 phosphorylated in vitro by myrAkt-HA. (Inset) Two-dimensional tryptic phosphopeptide map of FKHR1-HA immunoprecipitated from 32P-labeled CV1 cells. Phosphopeptides from in vitro and in vivo maps, which comigrate, are indicated by arrows. Position of the origin is indicated (arrowhead). (D) Two-dimensional tryptic phosphopeptide map of a mixture of FKHR1 immunoprecipitated from asynchronous CV1 cells metabolically labeled with 32P-inorganic phosphate and FKHR1-HA phosphorylated in vitro by myrAkt-HA. (Inset) Two-dimensional tryptic phosphopeptide map of FKHR1-HA immunoprecipitated from 32P-labeled CV1 cells. Phosphopeptides shared between the in vivo and in vitro maps are indicated by arrows. Position of the origin is indicated (arrowhead). (E) Two-dimensional tryptic phosphopeptide map of FKHR1(S253)-HA immunoprecipitated from asynchronous CV1 cells metabolically labeled with 32P-inorganic phosphate. (Inset) Two-dimensional tryptic phosphopeptide map of FKHR1-HA immunoprecipitated from 32P-labeled CV1 cells. Phosphopeptides absent in the FKHR1(S253A)-HA map are indicated by arrows. Position of the origin is indicated (arrowhead). (F) Two-dimensional tryptic phosphopeptide map of FKHR1(T24A/S253)-HA immunoprecipitated from asynchronous CV1 cells metabolically labeled with 32P-inorganic phosphate. (Inset) Two-dimensional tryptic phosphopeptide map of FKHR1-HA immunoprecipitated from 32P-labeled CV1 cells. Phosphopeptides absent in the FKHR1(T24A/S253A)-HA map are indicated by arrows. Position of the origin is indicated (arrowhead). (G–I) Mutation of PKB/AKT phosphorylation sites alters the localization of FKHR1-HA. (G) Nuclear localization of FKHR1(T24A)-HA in CV1 cells. (H) Nuclear localization of FKHR1(S253A)-HA in CV1 cells. (I) Nuclear localization of FKHR1(T24A/S253A)-HA in CV1 cells. (J) Mutation of the PKB/Akt phosphorylation sites, threonine-24, and/or serine-253 increased the transcriptional activity of FKHR1-HA. Luciferase activities of the individual samples were normalized to the activity of the 8XFK1tkLuc reporter in the absence of any other construct.
To explore whether PKB/Akt can directly phosphorylate FKHR1, a constitutively active form of Akt (myrAkt-HA) was immunoprecipitated from myrAkt-HA-transfected CV1 cells. In vitro kinase reactions then were performed by using purified recombinant GST-FKHR1 fusion protein as a substrate. PKB/Akt immune complexes also were incubated with the known substrate, H2B (positive control), or with GST (negative control). PKB/Akt immune complexes phosphorylated both H2B and GST-FKHR1, whereas no GST phosphorylation was observed (Fig. 4B). In control reactions, GST-FKHR1 incubated with either of two inactive forms of Akt, Akt(K179M) or Akt (T308A/S473A), was not phosphorylated (Fig. 4B). Phosphoamino acid analysis of the PKB/Akt phosphorylated GST-FKHR1 revealed the presence of both phosphoserine and phosphothreonine in GST-FKHR1 (data not shown). Subsequent two-dimensional mapping of tryptic fragments demonstrated that the majority of the in vitro PKB/Akt-mediated phosphorylation was contained in two major phosphopeptides (Fig. 4C), each of which comigrated with phosphopeptides derived from tryptic digests of FKHR1-HA immunoprecipitated from CV1 cells that were metabolically labeled with 32PO4 (Fig. 4D).
Site-directed mutagenesis was used to critically address the role of PKB/Akt-mediated phosphorylation of FKHR1 in vivo. Mutants of FKHR1-HA in which threonine-24 [FKHR1(T24A)-HA] or serine-253 [FKHR1(S253A)-HA] were changed to alanine were immunoprecipitated from 32PO4-labeled CV1 cells. Tryptic mapping of FKHR1(S253A)-HA revealed that two of the phosphopeptides initially observed in maps of FKHR1-HA were now absent (Fig. 4E). Similarly, tryptic mapping of FKHR1(T24A/S253A) revealed the absence of three phosphopeptides initially observed in maps of FKHR1-HA (Fig. 4F). Tryptic mapping of FKHR1(T24A)-HA revealed that a single phosphopeptide initially observed in the map of FKHR1-HA was now absent (data not shown). Immunolocalization of FKHR1(T24A)-HA and FKHR1(S253A)-HA revealed that mutation of either residue to alanine resulted in a retention of FKHR1-HA in the nucleus in >90% of expressing cells (Fig. 4 G and H). Mutation of threonine-24 together with serine-253 FKHR1(T24A/S253A)-HA further enhanced this nuclear localization (Fig. 4I). Nuclear localization of these mutants could not be overcome by the coexpression of P110CAAX (data not shown). Finally, the transcriptional activity of all three mutant forms was 2- to 3-fold greater that that observed for wild-type FKHR1-HA (Fig. 4J).
PI3 kinases play critical roles in the regulation of cellular proliferation, differentiation, and survival (36). Cellular responses to the activation of PI3 kinases are largely mediated by the serine/threonine protein kinases, PKBα, PKBβ, and PKBγ (PKBα is also known as c-Akt) (37–41). The importance of the PKB/Akt family of kinases in the regulation of cellular function is underscored by the finding that members of the PKB family have been found to inhibit BAD-mediated apoptosis (42), as well as being overexpressed in breast (PKBα), ovarian (PKBβ), and pancreatic (PKBβ) cancers (43, 44).
CONCLUSIONS
Our results demonstrate that PKB/Akt directly phosphorylates FKHR1, a member of the closely related FKHR subclass of the forkhead family of transcription factors, on at least two residues (threonine-24 and serine-253). Additionally, we show by mutational analysis that phosphorylation of both residues is required to trigger the nuclear export of FKHR1. It is of note that the kinetics of FKHR1 export from the nucleus (this report) mirrors that recently described for serum-stimulated nuclear import of PKB/Akt. We propose that by promoting export from the nucleus, PKB/Akt-mediated phosphorylation effectively inactivates FKHR1 by limiting access to its target genes. The suppression of FKHR1-regulated transcription of a reporter gene by coexpression of a constitutively activated form of Akt (myrAkt-HA) supports this hypothesis. The recent demonstration in C. elegans that AGE-1 (PI3 kinase), as well as Akt-1 and Akt-2, suppress the activity of DAF-16 provides genetic support for this hypothesis. These results suggest that FKHR1, in vertebrates, and DAF-16, in nematodes, plays a critical role in the down-regulation of cellular responses (e.g., proliferation or differentiation) normally elicited by activation of PI3 kinase and PKB/Akt.
The negative regulation of nuclear transcription factors by protein phosphorylation generally is manifested as an inhibition of DNA binding (26, 27). The export of the nuclear transcription factors FKHR1 (this work), NF-AT (45), and Pho4 (8) from the nucleus suggests that these factors may continue to exert an effect on nuclear function in the absence of DNA binding and that sequestration in the cytoplasm is required to inactivate them fully. For instance, the continued interaction of FKHR1 with elements, either gene-specific or general, of the transcriptional machinery in the absence of DNA binding might abrogate cellular responses to one or more extracellular signals. Thus, the growth factor-dependent phosphorylation and nuclear export of FKHR1 may serve as a general paradigm for the regulation of other transcription factors by extracellular signals.
Acknowledgments
We thank Drs. Julian Downward, Philip Tsichlis, and Barbara Wolff-Winiski for providing reagents used in this study. In addition, we thank Drs. Nigel Carter, James Feramisco, Frank Furnari, Tom Hope, Erik Knudsen, Karen Knudsen, Richard Kolodner, Scott Ogg, Gary Ruvkun, Degui Wang, Jean Wang, and Karen Yamamoto for discussions and/or comments on the manuscript. Finally, we thank Drs. Domenico Accili and Michael Greenberg for communicating results before publication. This work was supported in part by grants from the National Institutes of Health and the National Cancer Institute. T.H. is a Frank and Else Shilling American Cancer Society Research Professor.
ABBREVIATIONS
- PI3
phosphatidylinositol 3
- PKB
protein kinase B
- HA
hemagglutinin
- GFP
green fluorescent protein
- β-gal
β-galactosidase
- GST
glutathione S-transferase
- NES
nuclear-export sequence
- CMV
cytomegalovirus
- H2B
histone 2B
Note Added in Proof
Brunet et al. (46) and Kops et al. (47) recently have reported similar results in analyses of human FKHRL1 and AFX, respectively.
References
- 1.Kaufmann E, Knöchel W. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 2.Hromas R, Costa R. Crit Rev Oncol Hematol. 1995;20:129–140. doi: 10.1016/1040-8428(94)00151-i. [DOI] [PubMed] [Google Scholar]
- 3.Vogt P K, Li J, Freyaldenhoven B S. Virology. 1997;238:1–7. doi: 10.1006/viro.1997.8846. [DOI] [PubMed] [Google Scholar]
- 4.Barr F G. Curr Top Microbiol Immunol. 1997;220:113–129. doi: 10.1007/978-3-642-60479-9_7. [DOI] [PubMed] [Google Scholar]
- 5.Hillion J, Le Coniat M, Jonveaux P, Berger R, Bernard O A. Blood. 1997;90:3714–3719. [PubMed] [Google Scholar]
- 6.Borkhardt A, Repp R, Haas O A, Leis T, Harbott J, Kreuder J, Hammermann J, Henn T, Lampert F. Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- 7.Arden K C, Anderson M J, Finckenstein F G, Czekay S, Cavenee W K. Genes Chromosomes Cancer. 1996;16:254–260. doi: 10.1002/(SICI)1098-2264(199608)16:4<254::AID-GCC5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 8.Kaffman A, Rank N M, O’Neill E M, Huang L S, O’Shea E K. Nature (London) 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 9.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. In: Current Protocols. Chanda V B, editor. New York: Wiley; 1998. pp. 15.1.1–15.1.5. [Google Scholar]
- 10.Knudsen K E, Arden K C, Cavenee W K. J Biol Chem. 1998;273:20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 11.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 12.van der Geer P, Luo K, Sefton B M, Hunter T. In: Cell Biology: A Laboratory Manual. Celis J E, editor. San Diego: Academic; 1994. pp. 422–446. [Google Scholar]
- 13.Alessi D R, Caudwell F B, Andjelkovic M, Hemmings B A, Cohen P. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 14.Wolff B, Sanglier J J, Wang Y. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 15.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 16.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 17.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 18.Lin K, Dorman J B, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 19.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carpenter C L, Cantley L C. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 21.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson T R, et al. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez C, Jones D R, Rodriguez-Viciana P, Gonzalez-Garcia A, Leonardo E, Wennström S, von Kobbe C, Toran J L, Calvo V, Copin S G, et al. EMBO J. 1998;17:743–753. doi: 10.1093/emboj/17.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian X, Costa R H. Nucleic Acids Res. 1995;23:1184–1191. doi: 10.1093/nar/23.7.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y, Hope T J. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim F J, Beeche A A, Hunter J J, Chin D J, Hope T J. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montminy M. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 27.Karin M, Hunter T. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 28.Alessi D R, Cohen P. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 29.Coffer P J, Jin J, Woodgett J R. Biochem J. 1998;335:1–13. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 31.Blume-Jensen P, Janknecht R, Hunter T. Curr Biol. 1998;8:779–782. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- 32.Cardone M H, Roy N, Stennicke H R, Savlesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 33.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 34.Reif K, Burgering B M, Cantrell D A. J Biol Chem. 1997;272:14426–14433. doi: 10.1074/jbc.272.22.14426. [DOI] [PubMed] [Google Scholar]
- 35.Anderson M J, Viars C S, Czekay S, Cavenee W K, Arden K C. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 36.Toker A, Cantley L C. Nature (London) 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 37.Konishi H, Kuroda S, Tanaka M, Matsuzaki H, Ono Y, Kameyama K, Haga T, Kikkawa U. Biochem Biophys Res Commun. 1995;216:526–534. doi: 10.1006/bbrc.1995.2654. [DOI] [PubMed] [Google Scholar]
- 38.Cheng J Q, Godwin A K, Bellacosa A, Taguchi T, Franke T F, Hamilton T C, Tsichlis P N, Testa J R. Proc Natl Acad Sci USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coffer P J, Woodgett J R. Eur J Biochem. 1991;201:475–481. doi: 10.1111/j.1432-1033.1991.tb16305.x. [DOI] [PubMed] [Google Scholar]
- 40.Jones P F, Jakubowicz T, Pitossi F J, Maurer F, Hemmings B A. Proc Natl Acad Sci USA. 1991;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 42.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 43.Liu A X, Testa J R, Hamilton T C, Jove R, Nicosia S V, Cheng J Q. Cancer Res. 1998;58:2973–2977. [PubMed] [Google Scholar]
- 44.Bellacosa A, de Feo D, Godwin A K, Bell D W, Cheng J Q, Altomar D A, Wan M, Dubeau L, Scambia G, Masciollo V, et al. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 45.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 46.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 47.Kops G J, de Ruiter N D, De Vries-Smits A M, Powell D R, Bos J L, Burgering B M. Nature (London) 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]