Abstract
Inactivation of the tumor suppressor PTEN gene is found in a variety of human cancers and in cancer predisposition syndromes. Recently, PTEN protein has been shown to possess phosphatase activity on phosphatidylinositol 3,4,5-trisphosphate, a product of phosphatidylinositol 3-kinase. We have identified a homolog of PTEN in Caenorhabditis elegans and have found that it corresponds to the daf-18 gene, which had been defined by a single, phenotypically weak allele, daf-18(e1375). By analyzing an allele, daf-18(nr2037), which bears a deletion of the catalytic portion of CePTEN/DAF-18, we have shown that mutation in daf-18 can completely suppress the dauer-constitutive phenotype caused by inactivation of daf-2 or age-1, which encode an insulin receptor-like molecule and the catalytic subunit of phosphatidylinositol 3-kinase, respectively. In addition, daf-18(nr2037) dramatically shortens lifespan, both in a wild-type background and in a daf-2 mutant background that normally prolongs lifespan. The lifespan in a daf-18(nr2037) mutant can be restored to essentially that of wild type when combined with a daf-2 mutation. Our studies provide genetic evidence that, in C. elegans, the PTEN homolog DAF-18 functions as a negative regulator of the DAF-2 and AGE-1 signaling pathway, consistent with the notion that DAF-18 acts a phosphatidylinositol 3,4,5-trisphosphate phosphatase in vivo. Furthermore, our studies have uncovered a longevity-promoting activity of the PTEN homolog in C. elegans.
Human PTEN, also called MMAC1 or TEP1, is encoded by a tumor suppressor gene located on chromosome 10q23 (1–3). Mutation or deletion of the PTEN gene has been found in a variety of human cancers, such as glioblastoma, endometrial tumors, and prostate cancer and in familial cancer predisposition syndromes (4). Recently, it has been shown that mice carrying a homozygous PTEN gene deletion are embryonic lethal, whereas mice heterozygous for the deletion show hyperproliferation in the prostate, skin, and colon, reminiscent of the features of human Cowden’s disease and Bannayan–Ruvalcaba–Riley syndrome (5, 6). These studies indicate that PTEN is a bona fide tumor suppressor.
The PTEN protein contains a sequence motif, HCXXGXGRXG, that is highly conserved among members of the protein tyrosine phosphatase family. PTEN protein can dephosphorylate phosphotyrosyl and phosphoseryl/threonyl residues on protein substrates in vitro (3, 7). Recently, it has been shown that PTEN can dephosphorylate phosphatidylinositol 3,4,5-trisphosphate (PIP3) (8). PIP3 is generated by phosphatidylinositol 3-kinase (PI 3-kinase), which is activated by binding to ligand-engaged receptor tyrosine kinases (9). PIP3 then acts as a second messenger to activate a variety of signaling molecules, one of which is Akt. Akt, also called PKB, is a serine/threonine kinase whose activation requires binding to PIP3 through its pleckstrin homology domain and subsequent recruitment to the plasma membrane (10). In human tumor cells, overexpression of PTEN leads to decreased levels of PIP3 and inactivation of Akt (11–17). Conversely, in mouse Pten−/− cells in which the Pten gene has been genetically deleted, increased levels of PIP3 and enhanced activation of Akt are observed (18, 19). Mouse Pten−/− cells show decreased apoptosis (18, 19) and accelerated cell cycle progression (19). These studies suggest that mammalian PTEN functions as a phosphatase for the inositol phospholipid PIP3. The role of PTEN as a tumor suppressor has been attributed to its ability to modulate two important cellular processes: cell cycle progression and apoptosis. However, it remains unclear whether PIP3 is the major in vivo substrate for PTEN, because dephosphorylation of protein substrates (e.g., FAK, a protein tyrosine kinase associated with focal adhesions) or inhibition of the mitogen-activated protein kinase pathway have also been reported (20, 21). In addition, the lethality of the homozygous PTEN gene deletion in mice makes it difficult to carry out a detailed analysis of the biological processes affected by the complete PTEN deficiency in mammals.
Caenorhabditis elegans provides a model system in which powerful genetic analysis can be used to investigate the major signaling pathways and the physiological processes regulated by a PTEN homolog in a living organism. Here we report that the C. elegans homolog of the mammalian PTEN tumor suppressor is involved in dauer formation and lifespan regulation and that CePTEN is encoded by the daf-18 gene. The dauer state is essentially a state of hibernation that is normally only entered under conditions of starvation or overcrowding. The insulin receptor-like protein DAF-2 and the PI 3-kinase homolog AGE-1 are also known to function in both dauer formation and lifespan (22, 23). Inactivation of either daf-2 or age-1 causes animals to form dauer larvae rather than progressing continuously through the four larval stages and into adulthood (24). In addition, daf-2 or age-1 animals that reach adulthood have a dramatically extended lifespan, indicating that signaling triggered by the insulin receptor-like tyrosine kinase has a profound influence on the aging process in C. elegans (22, 25–28). Both the constitutive dauer formation and lifespan extension phenotypes observed in daf-2 or age-1 mutants requires the activity of daf-16, which encodes two fork–head group transcription factors (25, 27, 29–32). Another gene that has been suggested to function downstream of daf-2 and age-1 is daf-18 (27, 28, 30). The sole original mutation, daf-18(e1375), was isolated as a mutant that failed to form dauer larvae even under starvation conditions (33). Studies of daf-18 have been hampered by the fact that this sole allele only partially suppresses certain daf-2 or age-1 mutant phenotypes, and because this allele by itself has a weak phenotype (27–30, 33).
We have genetically characterized a daf-18 mutation that contains a deletion in the phosphatase catalytic domain of CePTEN/DAF-18. Our results establish the role of the C. elegans PTEN homolog in the daf-2 and age-1 signaling pathway that regulates both longevity and dauer development.
MATERIALS AND METHODS
Strain Constructions.
The daf-18(nr2037) mutation was backcrossed five times against the wild-type Bristol N2 strain. The presence of the nr2037 allele was identified by PCR analysis of genomic DNA using the primers F6 (5′-CTATTGAAGGAGGACTAACACAGGC-3′) and R3 (5′-GCCAACGAAGT-GCTAAATCGAC-3′). A diagnostic 0.6-kb or 1.6-kb fragment is indicative of the presence of the nr2037 or wild-type allele, respectively. The presence of the wild-type allele was further confirmed by PCR using the primers F3 (5′-GATTGGTGTCTACGTGGAACGG-3′) and R3, which produce a 0.8-kb fragment only in the wild-type strain. The daf-2(e1370); daf-18(nr2037) double mutant strain was constructed by crossing daf-18(nr2037) males with daf-2(e1370) hermaphrodites. Homozygous daf-2(e1370) F2 progeny were identified by their dauer-constitutive phenotype at 25°C. These animals were allowed to exit from the dauer state by shifting to 20°C, and progeny were identified as daf-18(nr2037) homozygotes by using PCR. Sequencing of genomic DNA confirmed that the daf-2(e1370) allele was, in fact, homozygous. To construct the age-1(m333); daf-18(nr2037) double mutant, daf-18(nr2037) males were mated with age-1(m333)/mnC1[dpy-10(e128) unc-52(e444)] hermaphrodites. F2 progeny that were age-1/age-1; daf-18/+ were identified based on their ability to produce progeny consisting of dauers and very slowly growing non-dauers at a 3:1 ratio. The slow-growing non-dauers were age-1(m333); daf-18(nr2037) candidates containing maternally provided daf-18(+) activity. In the next generation, the maternal daf-18(+) contribution was absent, and the progeny regained a normal growth rate. These animals were confirmed to be daf-18(nr2037) homozygotes by using PCR analysis. The presence of the age-1(m333) allele in the double mutant strain was confirmed by backcrossing with wild-type males and observing the reappearance of the age-1(m333) dauer-constitutive phenotype in some of the descendents of all cross-progeny. Two independent strain isolates were used to assay lifespan and dauer phenotype for all strains constructed in this study, and similar results were obtained for both isolates in all cases. The following strains were obtained from the C. elegans Genetic Center: CB1370, daf-2(e1370); DR722, age-1(m333)/mnC1[dpy-10(e128) unc-52(e444)]; and CB1375, daf-18(e1375).
Allelic Sequencing.
Genomic DNA was amplified by using PCR, and the PCR fragments were directly sequenced. For all of the alleles, sequencing was performed on both DNA strands. The daf-18(nr2037) allele contains a deletion that removes nucleotides 650–1,639 (in reference to the initiation codon as +1). daf-18(e1375) contains a 30-bp insertion at nucleotide 2,852. This mutation was found in strain CB1375.
cDNA Characterization.
cDNA clones for daf-18 (yk400b8, yk43e5, and yk181h9) were obtained from Yuji Kohara (National Institute of Genetics, Mishima, Japan). The cDNA clone with the largest insert, yk400b8, was fully sequenced.
Genomic Rescue.
A 13.6-kb BamHI–SalI genomic fragment, containing 1.3 kb of upstream sequence, 5 kb of daf-18 coding region, and 7.3 kb of downstream sequence, was derived from cosmid T07A9. The ΔBspHI derivative was constructed by deleting the internal 620-bp BspHI–BspHI fragment, resulting in the removal of exons I and II of the daf-18 gene. Each construct was injected into the daf-2(e1370); daf-18(nr2037) double mutant strain at 25 ng/μl together with the transformation marker plasmid pRF4 (100 ng/μl) (34). Stable transgenic lines were obtained by following germ-line transmission of the Roller phenotype. Multiple independent transgenic lines have been established for both daf-18(+) or daf-18(−) transgenes. Progeny from these stable lines were used to examine daf-18 rescue activity by scoring the reappearance of the dauer phenotype at 25°C. Data were pooled from three independent stable lines for each transgene.
Lifespan Assay.
Lifespan experiments were performed essentially as described (25). For assays performed at 25°C, eggs were hatched and raised at 15°C. At the L4 or young adult stage, animals were transferred to new plates and shifted to 25°C. The day of the temperature shift was counted as day 0 in the lifespan assay. For assays performed at 20°C, eggs were hatched and raised at 20°C. At the L4 or young adult stage, animals were transferred to new plates (day 0) and maintained at 20°C. Animals were transferred to new plates once every day during their reproductive period and then once every 3 days after the end of their egg-laying period. The animals were scored as dead the day they failed to respond to a light touch with a platinum wire. The daf-18(nr2037) mutant has a low penetrance protruding vulva phenotype (≈8%) which can lead to vulval ruptures and the death of the animal. For other strains, animals with progeny that hatched internally were observed at low frequencies. All of these animals were excluded in the final data analysis for survival curve and mean lifespan, because they obviously did not die of old age.
Scoring Dauer Phenotypes.
The dauer-constitutive phenotype of daf-2(e1370) and age-1(m333) mutants were scored as described (27, 30). For the daf-2(e1370) or daf-18(nr2037) single mutants, daf-2(e1370); daf-18(nr2037) double mutant, and the derived transgenic lines, eggs were laid at 20°C during a 10-hour period and shifted to 25°C. Progeny were scored at 48 hours after the temperature shift and then rescored 24 hours later to confirm the phenotype. For the age-1(m333) single mutant and the age-1(m333); daf-18(nr2037) double mutant, eggs were layed at 20°C during a 10-hour period and maintained at 20°C. Progeny were scored at 48 hours and again at 72 hours after egg-laying. The categories for dauers and partial dauers were defined as described (35).
RESULTS
C. elegans Homolog of PTEN.
We have identified a homolog of PTEN in the C. elegans sequence database generated by the C. elegans Genome Sequencing Consortium (T07A9.6). The DNA sequence of a full-length cDNA clone (yk400b8) revealed a predicted ORF of 962 aa, abbreviated as CePTEN. The genomic structure of the CePTEN gene is shown in Fig. 1A. The amino-terminal region of CePTEN contains the phosphatase catalytic domain, and this region shares 38% sequence identity with the corresponding domain of human PTEN (Fig. 1B). The carboxyl-terminal region of CePTEN has lower levels of sequence homology to human PTEN. The noncatalytic domains of CePTEN and PTEN may play a species-dependent regulatory role.
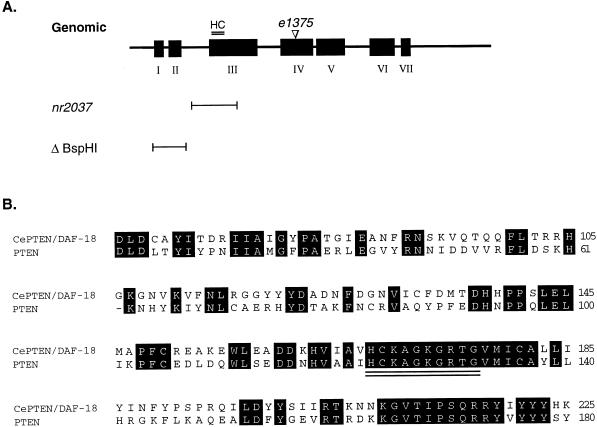
Figure 1.
CePTEN/daf-18 gene structure. (A) Genomic structure. The exons are depicted as black boxes and numbered. The nr2037 allele contains a deletion that covers part of intron II and exon III. The phosphatase catalytic center, HCKAGKGRTG, abbreviated as the HC motif (double underlined), is deleted in the nr2037 mutant. The ΔBspHI deletion construct, in which exon I and II of the CePTEN/daf-18 gene is removed, is used as a control for the genomic rescue experiment (see Table 1). The 30-bp insertion found in the daf-18(e1375) allele is located in exon IV in the noncatalytic domain of CePTEN/DAF-18. This insertion occurs after codon 574, leading to a frameshift and premature stop codon. (B) Amino acid alignment of the amino-terminal phosphatase domain of CePTEN/DAF-18 and human PTEN. The phosphatase catalytic center, HCKAGKGRTG, is double underlined. Identical residues are shown as white letters in black boxes.
To understand the function of CePTEN, we employed a reverse genetics approach. A deletion mutation in the CePTEN gene, nr2037, was identified by using PCR screening from an ordered array of mutagenized worms (kindly provided by Carl Johnson and Leo Liu; NemaPharm, Cambridge, MA). Sequencing of the nr2037 allele revealed a deletion that removes 990 nt of the CePTEN gene spanning parts of intron 2 and exon 3 (Fig. 1A). The nr2037 mutation is predicted to abolish the phosphatase activity of CePTEN, because the deletion removes part of the catalytic domain, including the conserved sequence motif, HCKAGKGRTG, that forms the phosphatase catalytic center. The rest of exon 3 also is unlikely to be translated because of removal of the splice acceptor site at the 5′ end of exon 3.
Strains homozygous for nr2037 are viable and show no obvious abnormalities in development or morphology when grown under well fed conditions. However, at high-saturation growth densities or under starvation conditions, we noticed that nr2037 homozygotes failed to form dauers (a dauer-defective phenotype). The dauer-defective phenotype can be rescued by germ-line transformation with a small genomic fragment containing the wild-type CePTEN gene (see below), indicating that this phenotype results from the nr2037 lesion. These observations indicate that the CePTEN gene is required for dauer larvae formation. Because the PI 3-kinase pathway is known to be involved in dauer formation and because our parallel biochemical studies in mammalian cells have suggested that PTEN acts as a phosphatase for PIP3 (16, 19), we more closely examined nr2037 mutant animals for phenotypes known to be affected by PI 3-kinase signaling, as described below.
CePTEN Is Encoded by the daf-18 Gene.
The dauer-defective phenotype of nr2037 animals resembles the phenotype described for daf-18(e1375) (33). daf-18 maps to chromosome IV, in close proximity to the CePTEN gene locus. To determine whether the daf-18(e1375) mutation also affects the CePTEN gene, we sequenced the genomic region of the CePTEN locus in daf-18(e1375) animals and found a 30-bp insertion in exon IV (see Fig. 1A). This insertion occurs downstream of the phosphatase catalytic domain and leads to premature termination of the CePTEN protein. We have adopted the nomenclature DAF-18 for CePTEN and daf-18(nr2037) for the nr2037 allele. The nature of the daf-18(e1375) mutation was recently independently reported by Ogg and Ruvkun (36).
The molecular nature of the nr2037 and e1375 mutations suggests that the nr2037 mutation should completely eliminate the phosphatase activity of CePTEN/DAF-18, whereas the e1375 mutation may only partially reduce CePTEN/DAF-18 function. As described below, our detailed genetic characterization of daf-18(nr2037) strongly supports such a hypothesis.
daf-18(nr2037) Suppresses the Dauer-Constitutive Phenotype of daf-2(e1370).
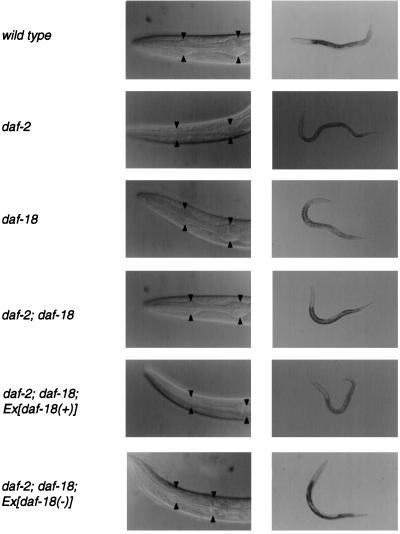
The daf-2 and age-1 signaling pathway is known to be required for normal development through the third larval stage (L3 stage). In molecular terms, appropriate levels of PIP3, produced through the PI 3-kinase AGE-1 and its upstream regulator DAF-2 (a receptor tyrosine kinase of the insulin receptor subfamily), are required for prevention of dauer formation. daf-2(e1370) is a temperature-sensitive mutant that forms dauer larvae when grown at the nonpermissive temperature of 25°C (a temperature-sensitive, dauer-constitutive phenotype) (27, 29). We reasoned that if CePTEN/DAF-18 acts as a phosphatase for PIP3 in vivo, loss of CePTEN/DAF-18 might promote accumulation of PIP3 and balance the effect of insufficient production of PIP3 caused by inactivation of DAF-2. Consequently, in daf-2; daf-18 double mutants, wild-type levels of PIP3 may be restored, thus allowing normal larval development instead of dauer arrest. We therefore constructed the daf-2(e1370); daf-18(nr2037) double mutant strain and tested whether daf-18(nr2037) could suppress the dauer-constitutive phenotype of daf-2(e1370) at the nonpermissive temperature of 25°C. As shown in Table 1, daf-2(e1370) homozygotes form dauer larvae at 100% penetrance in this assay. These dauers are characterized by nonfeeding, reduced locomotion, a dark intestine, and a thin body appearance. In addition, daf-2(e1370) dauer larvae have a characteristic remodeled pharynx (Fig. 2). By contrast, daf-2(e1370); daf-18(nr2037) double mutants develop normally through the L3 stage and into adulthood when assayed in parallel (Table 1). These double mutants do not show the remodeled pharynx that is characteristic of dauer larvae (Fig. 2). The daf-18(nr2037) single mutant also develops normally and has a normal pharynx (Table 1 and Fig. 2). The complete suppression of the daf-2(e1370) dauer-constitutive phenotype at 25°C by the daf-18(nr2037) mutation suggests that CePTEN/daf-18 acts in an antagonistic manner to daf-2.
Table 1.
The daf-18(nr2037) mutation suppresses the dauer-constitutive phenotype of daf-2(e1370)
| Genotype of parent | Transgene | Phenotype of progeny at 25°C, %
|
||||
|---|---|---|---|---|---|---|
| L4 and adult | Dauer | Partial dauer | Other | n | ||
| Wild type | none | 100 | 0 | 0 | 0 | 250 |
| daf-2(e1370) | none | 0 | 100 | 0 | 0 | 376 |
| daf-18(nr2037) | none | 98.5 | 0 | 0 | 1.5 | 200 |
| daf-2(e1370); daf-18(nr2037) | none | 96.5 | 0 | 0 | 3.5 | 310 |
| daf-2(e1370); daf-18(nr2037) | daf-18(+) | 0 | 93.5 | 4.3 | 2.2 | 203 |
| daf-2(e1370); daf-18(nr2037) | daf-18(−) | 99 | 0 | 0 | 1.0 | 205 |
Eggs were collected at 20°C during a 10-hour period and then shifted to 25°C. Phenotypes were scored at 48 hours after the temperature shift and confirmed by rescoring 24 hours later. Partial dauers are defined as those with less constriction or with less darkness of the intestine than typical dauer progeny observed in the daf-2(e1370) strain. “Other” includes animals that died as young larvae or those with grossly abnormal morphology. Wild type, Bristol N2 strain. n, total number of animals scored.
Stable transgenic lines were established using the wild-type genomic daf-18 fragment, daf-18(+), or the same fragment with a deletion of exons 1 and 2, daf-18(−). Transgene data are the combined total from three independent lines.
Figure 2.
Morphological comparison of daf-18, daf-2, and daf-2; daf-18 strains. (Left) Nomarski micrographs. The head of the animal is shown. The two bulbs of the pharynx are indicated by the arrowheads. (Right) Brightfield micrographs. Transgenic animals, shown in the Bottom two rows, carry an extrachromosomal array of either the functional daf-18 gene (Ex[daf-18(+)]), or a mutant derivative that removes exons I and II (Ex[daf-18(−)]). The animals with genotypes daf-2(e1370) and daf-2(e1370); daf-18(nr2037); Ex[daf-18(+)] are dauer larvae and appear thinner in the brightfield micrographs and have a constricted pharynx as shown in the Nomarski micrographs. Wild-type animals and animals with genotypes daf-18(nr2037), daf-2(e1370); daf-18(nr2037) and daf-2(e1370); daf-18(nr2037); Ex[daf-18(-)] are non-dauer L3 larvae.
We have used the suppression of daf-2(e1370) to confirm that the phenotypic effects that we observed for daf-18(nr2037) are due to the deletion in the CePTEN gene. We have established stable transgenic lines by using a small genomic fragment that carries essentially only the wild-type CePTEN/daf-18 gene. This genomic fragment rescued the daf-18(nr2037) defect in the daf-2(e1370); daf-18(nr2037) double mutant background, as the presence of the wild-type CePTEN/daf-18 transgene allowed the reappearance of the daf-2(e1370) dauer-constitutive phenotype at 25°C [Table 1, transgene daf-18(+)]. As a control, stable lines established by using a genomic fragment in which exons 1 and 2 of the CePTEN/daf-18 gene were deleted showed no rescue activity when assayed in parallel [Table 1, transgene daf-18(−)]. Nomarski microscope examination of pharyngeal morphology corroborated these results (Fig. 2). These studies showed that the wild-type, but not mutant, CePTEN/daf-18 transgene can completely rescue the daf-18(nr2037) defect, indicating that the phenotype we observed for daf-18(nr2037) is indeed due to the loss of CePTEN/daf-18 gene activity.
daf-18(nr2037) Suppresses the Dauer-Constitutive Phenotype of age-1(m333).
We next asked whether reduced production of PIP3, caused by inactivation of the AGE-1 PI 3-kinase, could be compensated, at least partially, by loss of the putative PIP3 phosphatase CePTEN/DAF-18. age-1 activity is known to be required for normal development through the L3 stage (22, 27). Maternally provided age-1(+) activity is sufficient to allow age-1/age-1 homozygotes to progress through larval stages to adulthood. However, in the absence of the maternal age-1(+) contribution, age-1/age-1 homozygotes arrest as dauer larvae unconditionally (22, 27). We wondered whether the dauer-constitutive phenotype caused by complete loss of age-1 could be suppressed by the daf-18(nr2037) mutation, and we therefore constructed the age-1(m333); daf-18(nr2037) double mutant. As shown in Table 2, in the absence of any maternal age-1 contribution, age-1/age-1 homozygotes form dauers at 100% penetrance. By contrast, age-1(m333); daf-18(nr2037) double mutants develop normally past the L3 stage to adulthood. This is also true for age-1(m333); daf-18(nr2037)/daf-18(e1375) trans-heterozygotes, further confirming the allelism of these two mutations (data not shown). The fact that the age-1(m333) dauer arrest phenotype can be completely suppressed by the daf-18(nr2037) mutation provides genetic evidence that CePTEN/DAF-18 opposes the action of AGE-1.
Table 2.
The daf-18(nr2037) mutation suppresses the dauer-constitutive phenotype of age-1(m333)
| Genotype of parent | Phenotype of Progeny at 20°C, %
|
||||
|---|---|---|---|---|---|
| L4 and adult | Dauer | Partial dauer | Others | n | |
| age-1(m333) | 0 | 100 | 0 | 0 | 410 |
| age-1(m333); daf-18(nr2037) | 100 | 0 | 0 | 0 | 320 |
Eggs were collected at 20°C during a 10-hour period and maintained at 20°C. Phenotypes were scored twice, first at 48 hours, and then again at 72 hours after egg laying. Homozygous age-1(m333)/age-1(m333) hermaphrodites were obtained from age-1(m333)/mnC1[dpy-10(e128) unc-52(e444)] parents. Definitions for the partial dauers or others are as in Table 1. n, total number of animals scored.
daf-18(nr2037) Shortens the Lifespan of C. elegans.
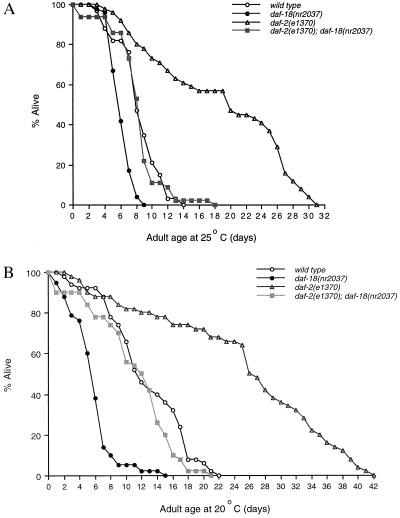
The ability of the daf-18(nr2037) mutation to suppress the daf-2(e1370) dauer-constitutive phenotype is in contrast to the previous report that the daf-18(e1375) mutation could not suppress daf-2(e1370) in such an assay (27, 29). These differences are likely due to daf-18(nr2037) being a null allele, whereas daf-18(e1375) only partially reduces gene function, as one would predict from the molecular nature of these mutations (see Fig. 1). We therefore tested the effect of the stronger daf-18(nr2037) mutation on the lifespan of animals, because the aging process in C. elegans is known to be affected by PI 3-kinase signaling. Our experiments showed that at 25°C, daf-18(nr2037) mutants have a shorter lifespan than wild-type animals (Fig. 3A). The mean lifespan is 6.2 days for daf-18(nr2037) as compared to 8.5 days for wild type, and the reduction of lifespan by the daf-18(nr2037) mutation is ≈30% (Table 3). The maximum lifespan of daf-18(nr2037) animals is also shorter than that of the wild type, 9 days versus 14 days (Table 3).
Figure 3.
Effects of the daf-18(nr2037) mutation on adult lifespans. (A) Lifespans at 25°C. (B) Lifespans at 20°C. The percentage of live animals was plotted as a function of time (days). Wild-type (Bristol N2), daf-18(nr2037), daf-2(e1370), and daf-2(e1370); daf-18(nr2037) strains were compared in parallel. Representatives of two independent sets of experiments are shown. The results were similar in both sets of experiments.
Table 3.
Effect of the daf-18(nr2037) mutation on adult lifespans
| Strain | Lifespan, days
|
|||
|---|---|---|---|---|
| Mean (±SE) | Ratio vs. wildtype | Maximum | n | |
| 25°C* | ||||
| Wild type | 8.5 (±0.3) | 1.0 | 14 | 34 |
| daf-18(nr2037) | 6.2 (±0.2) | 0.7 | 9 | 48 |
| daf-2(e1370) | 18.8 (±0.5) | 2.2 | 31 | 49 |
| daf-2(e1370); daf-18(nr2037) | 8.3 (±0.4) | 1.0 | 18 | 49 |
| 20°C† | ||||
| Wild type | 12.7 (±0.3) | 1.0 | 22 | 50 |
| daf-18(nr2037) | 5.8 (±0.5) | 0.5 | 15 | 42 |
| daf-2(e1370) | 25.3 (±0.5) | 2.0 | 42 | 50 |
| daf-2(e1370); daf-18(nr2037) | 11.1 (±0.5) | 0.9 | 21 | 50 |
Values are calculated using the data from the experiments shown in Fig. 3. The relative ratio of the mean over that of the wild-type (Bristol N2) strain is also shown. All experiments were performed at least two times with similar results obtained. n, total number of animals scored.
The animals were raised at 15°C until the L4 or adult stage and then shifted to 25°C. The day of the shift is counted as day 0 in the adult lifespan assay.
The animals were raised and maintained at 20°C. Young adults or L4s were transferred to new plates to assay for lifespan. The day of the transfer is counted as day 0 in the adult lifespan assay.
When we performed the similar experiment at 20°C, the optimal temperature to culture C. elegans, the lifespan curve of the daf-18(nr2037) mutant was further separated from that of the wild-type strain (Fig. 3B). The mean lifespan for daf-18(nr2037) is 5.8 days, whereas the wild type’s lifespan is 12.7 days (Table 3). The mean lifespan of the daf-18 mutant is 50% shorter than that of the wild type. These studies demonstrate that CePTEN/DAF-18 has a normal function in preventing the onset of aging.
Comparing Fig. 3 A and B, the lifespan curves for daf-18(nr2037) at 25°C and 20°C are almost superimposable. By contrast, the lifespan curve of the wild-type strain is shifted to the left at 25°C. In other words, the lifespan of the wild-type strain is considerably shortened at 25°C, whereas the lifespan of the daf-18(nr2037) mutant is largely unaffected. As discussed below, we suggest that the temperature effect on lifespan may be due to increased production of the inositol phospholipid PIP3 at higher temperature.
Suppression of the daf-18(nr2037) Lifespan Shortening Phenotype by Mutation in daf-2.
We hypothesized that the lifespan-shortening phenotype caused by the daf-18(nr2037) mutation is due to accumulation of higher levels of PIP3 as a result of the loss of the candidate PIP3 phosphatase CePTEN/DAF-18. Accordingly, a decrease in the production of PIP3 caused by inactivation of the DAF-2 receptor tyrosine kinase might be able to restore normal levels of PIP3 in a daf-18(nr2037) background and consequently extend lifespan. We thus compared the lifespan of the daf-18(nr2037); daf-2(e1370) double mutant with that of the daf-18(nr2037) single mutant, the daf-2(e1370) single mutant, and the wild-type strain at 25°C and 20°C. As shown in Fig. 3 and Table 3, the daf-2(e1370) mutation almost doubled the lifespan of the animals, consistent with previous reports (25). Interestingly, we found that the daf-2(e1370) mutation extended the lifespan of daf-18(nr2037) animals at either 25°C or 20°C and restored lifespan to that found for wild type (Fig. 3 A and B, Table 3). These observations suggest that the lifespan-shortening phenotype caused by daf-18(nr2037) depends on the positive signal input from daf-2 gene activity. In addition, daf-18(nr2037) also completely suppressed the long-lived phenotype of daf-2(e1370). The lifespan of the daf-2(e1370); daf-18(nr2037) double mutant is almost identical to that of the wild-type strain at both 25°C and 20°C (Fig. 3, Table 3). The mutual suppression of the daf-18(nr2037) and daf-2(e1370) alleles suggests that their corresponding gene products function in opposite directions in regulating lifespan in C. elegans.
DISCUSSION
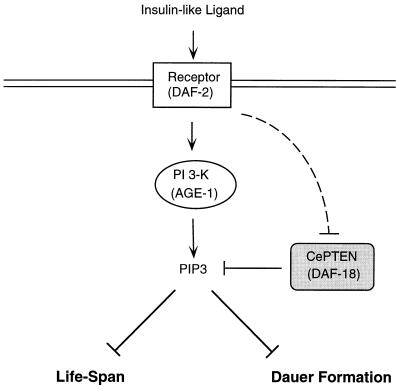
In C. elegans, DAF-2, an insulin receptor-like molecule, and AGE-1, a PI 3-kinase homolog, are involved in a signaling pathway that negatively regulates lifespan and the dauer-formation processes. Our genetic studies have shown that DAF-18, the C. elegans homolog of PTEN, functions in an antagonistic manner to the actions of DAF-2 and AGE-1 (Fig. 4). Our studies strongly support the hypothesis that CePTEN/DAF-18 acts as a physiological phosphatase for PIP3.
Figure 4.
Model for DAF-18 action. AGE-1 is the major signal transducer for the activated DAF-2 insulin receptor-like molecule in regulation of dauer development and lifespan. AGE-1 is predicted to produce PIP3, which negatively regulates lifespan and dauer-formation processes. DAF-18, the PTEN homolog, functions to dephosphorylate PIP3 and thus antagonizes the action of DAF-2 and AGE-1. DAF-18 may also be negatively regulated by DAF-2 (dashed line).
We have analyzed a mutant allele of daf-18, nr2037, that is likely to eliminate all daf-18 function. The nr2037 allele consists of a deletion that removes the phosphatase catalytic domain and likely causes frameshifts of the rest of the protein. This mutation completely suppresses the dauer-constitutive phentotype conferred by a mutation that compromises the function of daf-2. Our studies thus suggest that the production of PIP3 is an essential portion of the DAF-2 signaling pathway for prevention of dauer formation. This is in contrast to the previous reports that the daf-18(e1375) mutation could not suppress the dauer-constitutive phenotype conferred by daf-2(e1370) (27, 29) and that daf-18 RNA interference could only partially suppress the daf-2(e1370) dauer phenotype (36). These differences in the ability to suppress the effect of the daf-2(e1370) mutation could result from a more complete elimination of DAF-18 activity by daf-18(nr2037) than that caused by either the daf-18(e1375) mutation or daf-18 RNA interference. In contrast with its ability to suppress daf-2(e1370), daf-18(e1375) could completely suppress the effects of null alleles of age-1 (e.g., m333 or mg44 allele) on dauer development (27, 30), suggesting that the reduced daf-18 gene activity caused by the daf-18(e1375) mutation is sufficient to restore appropriate PIP3 levels for normal larval development, even in the complete absence of AGE-1 activity. The inability of daf-18(e1375) to suppress daf-2(e1370) may be due to a potential negative regulation of daf-18 by daf-2 (illustrated in Fig. 4). Alternatively, it is possible that the signaling pathway bifurcates downstream of DAF-2, with AGE-1/PI 3-kinase being only one of the pathways involved. If DAF-2 normally inhibits DAF-18 activity, as shown in Fig. 4, in the daf-2(e1370); daf-18(e1375) strain background, the inactivation of daf-2 will cause an increase of daf-18(e1375) gene activity. This increase will lead to further reduction of PIP3 levels, triggering dauer arrest. However, in the age-1(m333); daf-18(e1375) mutant strain, the daf-2 gene activity is still intact, and can inhibit daf-18(e1375). As a consequence, a more potent reduction of daf-18(e1375) activity is achieved, leading to a greater restoration of PIP3 levels and allowing the animal to proceed through normal larval development.
Previous genetic screens have failed to identify mutations in daf-18 as suppressors of the daf-2 dauer-constitutive phenotype. We can account for this based on our observations that maternal daf-18 contributions provide sufficient activity to allow daf-2(e1370); daf-18(nr2037) double mutants to remain dauer-constitutive (data not shown). Such a property of daf-18 may explain the failure to recover daf-18 mutants in the extensive genetic screens performed to date, because many of these screens were not designed to uncover mutants that show maternal rescue activity.
Interestingly, we found that daf-18(nr2037) has profound effects on lifespan. Complementary to previous reports that reduced daf-2 or age-1 activity leads to doubling of the lifespan, we observed that daf-18(nr2037) causes up to 50% shortening of lifespan. In comparison, the daf-18(e1375) mutation is reported to have a very modest effect on lifespan, although it can suppress the lifespan extension phenotype caused by daf-2 mutation (27, 28). Such differences could also be attributed to differences in the severity of the daf-18 alleles being assayed. Accordingly, a higher steady-state level of PIP3 in the daf-18(nr2037) mutant may be accumulated than that in the daf-18(e1375) mutant, which in turn leads to the premature aging that we observe. It is interesting to note that in wild-type animals, the onset of aging is accelerated by exposure to higher temperature, as shown by the shift of the survival curve toward shorter lifespans and by the reduction of the mean lifespan of animals at 25°C versus 20°C. Such a shift may be due to increased PIP3 production at higher temperature. The lifespan of the daf-2(e1370) single mutant and the daf-2(e1370); daf-18(nr2037) double mutant also showed temperature dependence. However, in the daf-18(nr2037) single mutant, higher temperature has very little effect on lifespan. It is likely that in the daf-18(nr2037) strain, even at 20°C, the PIP3 level is already above the threshold that is required to activate the aging process. Temperature-dependent production of PIP3 cannot further accelerate this process.
In summary, we have shown that DAF-18, the C. elegans homolog of the PTEN tumor suppressor, functions as a negative regulator of the daf-2 and age-1 signaling pathway. This is consistent with the model that CePTEN/DAF-18 functions as a PIP3 phosphatase in vivo. Our studies have also uncovered CePTEN/DAF-18 as one of the rate-limiting factors that control the onset of aging in C. elegans. Our results thus raise an interesting possibility that in addition to its tumor suppressor function, mammalian PTEN may function to promote longevity in a signal-transduction pathway activated by insulin-like growth factors.
Acknowledgments
We are grateful to Drs. Carl Johnson and Leo Liu at NemaPharm Group at Axys Pharmaceuticles for kindly providing the nr2037 strain. We thank Dr. Yuji Kohara for cDNA clones and Cathy Branda for helpful comments on the manuscript. Some of the strains used in this study were provided by the C. elegans Genetic Center, which is supported by the National Institutes of Health National Center for Research Resources (NCRR). This work was supported by grants from The Patrick and Catherine Weldon Donaghue Medical Research Foundation (H.S.) and the American Cancer Society (M.J.S.). H.S. is a Pew Scholar in the Biomedical Sciences.
ABBREVIATIONS
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- PI 3-kinase
phosphatidylinositol 3-kinase
Note Added in Proof
While this manuscript was being reviewed, Gil et al. also reported the characterization of the daf-18 (nr2037) allele in the dauer pathway (37).
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck P A, Pershouse M A, Jasser S A, Lin H, Yung W K A, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–363. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Li D-M, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 4.Eng C. Int J Oncol. 1998;12:701–710. doi: 10.3892/ijo.12.3.701. [DOI] [PubMed] [Google Scholar]
- 5.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki A, de la Pompa J L, Stambolic V, Elia A J, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 7.Myers M P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 9.Toker A, Cantley L C. Nature (London) 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 10.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 11.Haas-Kogan D, Shalev N, Wong M, Mills G, Yount G, Stokoe D. Curr Biol. 1998;8:1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 12.Furnari F B, Huang H J, Cavenee W K. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- 13.Myers M P, Pass I, Batty I H, Van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies M A, Lu Y, Sano T, Fang X, Tang P, LaPushin R, Koul D, Bookstein R, Stokoe D, Yung W K, et al. Cancer Res. 1998;58:5285–5290. [PubMed] [Google Scholar]
- 15.Li J, Simpson L, Takahashi M, Miliaresis C, Myers M P, Tonks N, Parsons R. Cancer Res. 1998;58:5667–5672. [PubMed] [Google Scholar]
- 16.Li D-M, Sun H. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Senechal K, Neshat M S, Whang Y E, Sawyers C L. Proc Natl Acad Sci USA. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stambolic V, Suzuki A, de la Pompa J L, Brothers G M, Mirtsos C, Sasaki T, Ruland J, Penninger J M, Siderovski D P, Mak T W. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 19.Sun, H., Lesche, R., Li, D.-M., Liliental, J., Zhang, H., GaoJing, Garvarina, N., Mueller, B., Liu, X. & Wu, H. (1999) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 20.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K M. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Tamura M, Yamada K M. J Cell Biol. 1998;143:1375–1383. doi: 10.1083/jcb.143.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris J Z, Tissenbaum H A, Ruvkun G. Nature (London) 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 23.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 24.Riddle D L, Albert P S. In: Genetics and Environmental Regulation of Dauer Larva Development. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 739–790. [PubMed] [Google Scholar]
- 25.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. Nature (London) 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 26.Klass M R. Mech Aging Dev. 1983;22:279–286. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- 27.Larsen P L, Albert P S, Riddle D L. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorman J B, Albinder B, Shroyer T, Kenyon C. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vowels J J, Thomas J H. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottlieb S, Ruvkun G. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 32.Lin K, Dorman J B, Rodan A, Kenyon C. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 33.Riddle D L, Swanson M M, Albert P S. Nature (London) 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- 34.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogg S, Ruvkun G. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 37.Gil E B, Link E M, Liu L X, Johnson C D, Lees J A. Proc Natl Acad Sci USA. 1999;96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]