Abstract
The products of the human leukocyte antigen subtypes HLA-B*2705 and HLA-B*2709 differ only in residue 116 (Asp vs. His) within the peptide binding groove but are differentially associated with the autoimmune disease ankylosing spondylitis (AS); HLA-B*2705 occurs in AS-patients, whereas HLA-B*2709 does not. The subtypes also generate differential T cell repertoires as exemplified by distinct T cell responses against the self-peptide pVIPR (RRKWRRWHL). The crystal structures described here show that pVIPR binds in an unprecedented dual conformation only to HLA-B*2705 molecules. In one binding mode, peptide pArg5 forms a salt bridge to Asp116, connected with drastically different interactions between peptide and heavy chain, contrasting with the second, conventional conformation, which is exclusively found in the case of B*2709. These subtype-dependent differences in pVIPR binding link the emergence of dissimilar T cell repertoires in individuals with HLA-B*2705 or HLA-B*2709 to the buried Asp116/His116 polymorphism and provide novel insights into peptide presentation by major histocompatibility antigens.
Keywords: X-ray structure, major histocompatibility antigen, peptide binding modes, ankylosing spondylitis, residue 116
Introduction
MHC class I antigens are cell surface glycoproteins consisting of a polymorphic transmembrane heavy chain (HC) noncovalently bound to β2-microglobulin (β2m; reference 1). The HC forms a groove with six pockets (A–F) for binding of proteolytic fragments derived from self- and nonself-proteins (2, 3). These peptides are presented to TCRs located on CTLs and initiate a cellular immune response leading to the destruction of the antigen-presenting cell if the peptide is derived from a nonself-protein. Several MHC class I alleles are associated with autoimmune diseases, and allele-specific binding of foreign or self-peptides by MHC class I molecules may be crucial to disease pathogenesis (4–8).
The association of the HLA class I allele HLA-B27 with ankylosing spondylitis (AS) and related spondyloarthropathies is among the strongest observed for any HLA gene (9, 10). Population analyses have shown that most of the common HLA-B27 subtypes, including the frequent B*2705 allele, are disease associated, whereas two subtypes, B*2706 and B*2709, are not (11). The B*2705 protein differs in only a single HC amino acid (Asp116) from B*2709 (His116) (12). This residue is located at the floor of the F pocket, which accommodates the COOH-terminal side chain of the bound peptide, and both subtypes can be distinguished by the peptides presented in vivo (13, 14). Despite some differences, in particular with regard to the COOH-terminal peptide residue, the majority of the peptides constituting the B*2705 repertoire were found to overlap with those from B*2709 and vice versa (14).
A self-peptide (pVIPR, previously termed VIP1R400–408, RRKWRRWHL) with potential arthritogenic properties (15) is derived from vasoactive intestinal peptide type 1 receptor and shows high sequence homology to a peptide from the latent membrane protein 2 (pLMP2, residues 236–244, RRRWRRLTV) of EBV (16). pVIPR can be presented by both subtypes, but the resulting T cell repertoires are distinct; B*2709 individuals very rarely possess pVIPR-specific T cells, whereas such T cells are more abundant in peripheral blood of healthy B*2705 persons and are very frequently present in patients suffering from AS (15). This indicates efficient thymic elimination of T cells specific for HLA-B27:pVIPR complexes in B*2709- but not in B*2705-positive individuals. Although these findings do not prove a causative role of pLMP2 and pVIPR in AS, they open the possibility to correlate structural properties of these subtypes with the retention (in B*2705) or elimination (in B*2709) of pVIPR-specific T cells.
The two crystal structures described here show that B*2705 accommodates pVIPR in two different conformations, of which one is identical to that seen when complexed with B*2709, whereas the other diverges substantially. Combined with functional studies in which pVIPR-specific CTLs were used, these findings suggest a molecular explanation for inappropriate T cell selection by individuals with a disease-associated HLA-B27 subtype.
Materials and Methods
HLA-B27–positive Donors.
Six patients with AS and one healthy donor were enrolled for this paper. HLA-B27 typing was performed using the HLA-B27 high resolution kit (Dynal). Five patients and the healthy donor were HLA-B*2705 positive, and one patient (LV) was HLA-B*2702 positive. Lymphoblastoid cell lines from these individuals were generated by in vitro transformation of B cells using the standard type 1 EBV isolate B95.8 (17).
Generation of pVIPR-specific Cytotoxic T Lymphocyte Lines.
PBMCs were isolated on a gradient of lymphoprep and depleted of the CD4+ fraction by Dynabeads M-540 CD4 (Dynal). Cells were incubated at 2 × 104 cells/well in 96-well flat-bottom microplates, stimulated at an 0.5:1 stimulator/responder ratio with autologous EBV-transformed B cells prepulsed overnight with 8.5 μM pVIPR, and γ irradiated (200 Gy). They were grown in RPMI 1640 medium containing 10% heat-inactivated pooled human serum, 2 mM l-glutamine, 10 U/ml penicillin, and 100 μg/ml of streptomycin and in the presence of 20–100 U/ml of human rIL-2 (Boehringer). 10 d later, cytotoxic T lymphocyte lines were restimulated as aforementioned. After 1 wk, they were tested for pVIPR specificity in a standard 51Cr-release assay using as targets T2 cells transfected with either B*2705 cDNA (T2-B*2705) or B*2709 cDNA (T2-B*2709), pulsed with 70 μM of the peptide, or used untreated.
Expression, Purification, and Crystallization of the HLA-B27:pVIPR Complexes.
HLA-B27:pVIPR protein complexes were produced as described previously (18, 19). The pVIPR peptide (RRKWRRWHL) was obtained by standard solid phase synthesis and purified by HPLC (Alta Bioscience). The purified protein complexes were used for crystallization at concentrations of 20 mg/ml (B*2705:pVIPR) and 16 mg/ml (B*2709: pVIPR), respectively, in TBS buffer (10 mM Tris HCl, pH 7.5, and 150 mM NaCl).
Using hanging drop vapor diffusion, crystals suitable for X-ray diffraction experiments were obtained from a PEG 8000 pH screen. As observed earlier for similar HLA-B27 complexes, streak seeding was indispensible to produce large single crystals. For B*2705:pVIPR, drops made from 1 μl of precipitant solution (0.1 M Tris HCl, pH 8.0, and 16% PEG 8000) and 1 μl of protein solution produced prismatic crystals with a maximum size of 400 × 250 × 250 μm. Using glycerol as cryoprotectant, the best dataset in terms of resolution (1.47 Å) was obtained at the BL2 beamline at BESSY-II, Berlin, Germany. In the case of B*2709:pVIPR, drops made of 1 μl of precipitant solution (0.1 M Tris HCl, pH 8.5, and 19% PEG 8000) and 1 μl of protein solution yielded platelike crystals with a maximum size of 300 × 200 × 100 μm. Cryo-cooled crystals of this complex (glycerol as cryoprotectant) allowed collection of a 2.2-Å resolution dataset at the X13 beamline at Deutsches Elektronen Synchrotron. Both datasets were collected at 100 K and processed with the HKL package (see Table I; reference 20).
Table I.
Data Collection and Refinement Statistics
| HLA-B*2705:pVIPR | HLA-B*2709:pVIPR | ||
|---|---|---|---|
| Data Collection | |||
| Space group | P21 | P21 | |
| Unit cell (Å) | a = 51.3, b = 81.8, c = 65.6, β = 107.6° |
a = 51.2, b = 81.6, c = 65.3, β = 107.5° |
|
| Resolution (Å)a | 50.0–1.47 (1.53–1.47) | 29.1–2.20 (2.28–2.20) | |
| Unique reflectionsa | 84,476 (8,185) | 25,005 (2,495) | |
| Redundancya | 4.2 (3.3) | 3.1 (3.0) | |
| Completeness (%)a | 97.6 (95.2) | 95.8 (96.8) | |
| I/σIa | 13.3 (2.2) | 9.7 (4.1) | |
| Rsym a , b | 0.078 (0.314) | 0.123 (0.346) | |
| Refinement | |||
| Rcryst a , c | 0.128 (0.184) | 0.188 (0.21) | |
| Rfree a , d | 0.178 (0.286) | 0.244 (0.27) | |
| Heavy chain | No. atoms/average B factor [Å2] | 2,355/18.7 | 2,275/36.9 |
| β2m | No. atoms/average B factor [Å2] | 864/22.8 | 850/40.8 |
| Peptide | No. atoms/average B factor [Å2] | 159/16.5 | 100/35.2 |
| Water | No. molecules/average B factor [Å2] | 721/35.9 | 342/50.2 |
| Estimated overall coordinate error (Å)e |
0.06 | 0.22 | |
| Rmsdf from ideal geometry | |||
| Bond length (Å) | 0.01 | 0.01 | |
| Bond angles (°) | 1.4 | 1.3 | |
| PDB entry code | 1ogt | 1of 2 |
Values in parentheses refer to the highest resolution shell.
Rsym = Σh Σi ∣ Ih,i − <Ih> ∣/ΣhΣiIh,i.
Rcryst = Σh ∣ Fo − Fc∣/Σ Fo Working set; no σ cut-off applied.
Rfree is the same as Rcryst, but calculated on 5% of the data excluded from refinement.
Estimated overall coordinate error based on Rfree as calculated by REFMAC 5.0.
Root mean squared deviation from target geometries.
Structure Determination.
The structure of HLA-B*2709:pVIPR was determined by molecular replacement using peptide-stripped B*2709:m9 (PDB code 1k5n) as a search model and program molrep (CCP4; reference 21). After rigid body refinement using refmac (22), the initial model was improved with ARP/wARP (23), and water molecules were included. Further improvement of the structure was achieved by iterative cycles of manual rebuilding using O (24) and restrained maximum-likelihood refinement with refmac comprising isotropic B factor adjustment. After translation, libration, and screw rotation refinement (25), the R value converged at 0.188 (Rfree, 0.244).
As the two HLA-B27:pVIPR complexes crystallized isomorphously, initial phases for B*2705:pVIPR were calculated from peptide-stripped B*2709:pVIPR with His116 replaced by alanine. This initial model was subjected to rigid body refinement followed by simulated annealing and energy minimization using CNS (26). ARP/wARP was used for improvement of the model and incorporation of water molecules and pVIPR in canonical conformation. After manual rebuilding, inclusion of alternate protein conformations, and atom positional refinement (refmac), there was still residual electron density in the peptide binding groove, to which an additional, noncanonical conformation of pVIPR could be fitted with an estimated occupancy of 0.5 for both peptide chains. Subsequent optimization of atomic displacement parameters decreased the R factor by 2.5% (Rfree, 2%) compared with isotropic refinement, justifying this procedure. Evaluation of anisotropy by parvati (27) showed the expected statistical distribution of 0.52 ± 0.14 for all atoms of the structure. The R value converged at 0.128 (Rfree 0.178). The equimolar distribution of the two pVIPR conformations in the B*2705 molecule was confirmed by an occupancy refinement performed with CNS. With the p6α conformation (see Results) omitted from the atomic coordinates, the occupancy of the central part of the peptide refined to occupancies ∼0.5, whereas that of p1, p2, p8, and p9 stayed with full occupancy. Similar results were obtained when only the p6α conformation was refined.
In both structures, radiation-decarboxylated Asp and Glu that showed negative difference electron density were assigned occupancies <1.0. The final models include all 276 residues of the HC, 100 residues of β2m (all 99 residues plus NH2-terminal Met), all nonhydrogen atoms of the pVIPR peptide, water, and glycerol (from the cryoprotectant). All ϕ/ψ angles lie in allowed regions of the Ramachandran plot (procheck; reference 28). Both structures were validated with whatcheck (29). Statistics are compiled in Table I. Superimpositions were performed using profit (30). Figures were generated using Grasp (31), povray (www.povray.org), molscript (32), povscript1 (33), and raster3D (34) together with a graphical interface (moldraw) developed by N. Sträter (Institut für Kristallographie, Freie Universität Berlin, Berlin, Germany; unpublished data). Molecular surfaces were calculated with MSMS (35).
The atomic coordinates and structure amplitudes have been deposited in the Protein Data Bank (accession codes 1ogt [B*2705:pVIPR] and 1of 2 [B*2709:pVIPR]).
Results
General Features of the HLA-B27:pVIPR Structures.
HLA-B*2705:pVIPR and B*2709:pVIPR crystallized isomorphously (same space group and comparable unit cell constants), showing the typical MHC class I immunoglobulin-like folds (1) and refined to values of Rcryst = 12.8% and Rfree = 17.8% at 1.47 Å and Rcryst = 18.8% and Rfree = 24.4% at 2.20 Å resolution, respectively (Materials and Methods and Table I). In both structures, pVIPR is bound in the common canonical conformation (Fig. 1 A) found in other HLA-B27 molecules, exploiting all six pockets of the peptide binding groove for interaction with the HC (19, 36, 37). However, B*2705:pVIPR features an additional, grossly different noncanonical peptide conformation (Fig. 1 B). Both conformations found in B*2705 are present in a 1:1 ratio as shown by occupancy refinement. Despite this peculiarity in B*2705, the structures of HC and β2m of the two subtypes are practically indistinguishable (Cα root mean square [rms] deviation 0.2 Å), including almost all side chain atoms. The atoms contributing to the binding groove occupy the same positions in B*2705 as in B*2709, and extreme atomic displacement factors (thermal anisotropy), which could mask multiple conformations were also not detected for B*2705. It has to be emphasized that, due to the isomorphous crystal structures, the intermolecular crystal contacts in both B*2705:pVIPR conformations are comparable, demonstrating that the two different binding modes of pVIPR found only in B*2705 are an intrinsic feature due to the presence of Asp116 and not a crystallographic artifact.
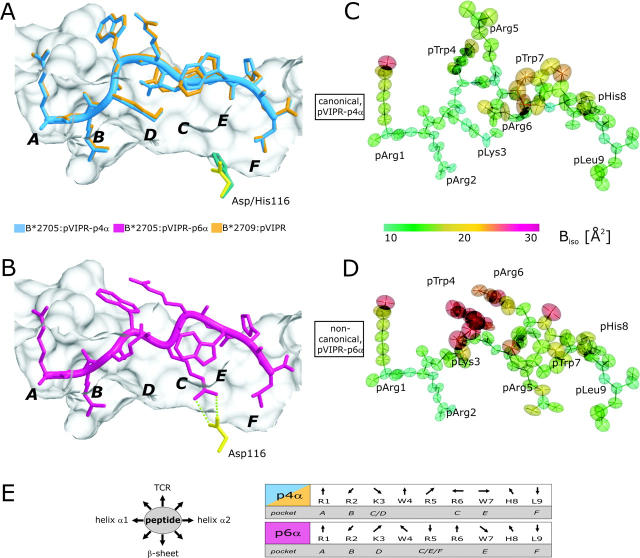
Figure 1.
pVIPR conformations, atomic displacement ellipsoids, and B factors. (A) Superimposition of the canonical pVIPR conformations (p4α) found in B*2705 (blue) and B*2709 (gold). (B) The noncanonical pVIPR conformation (p6α, pink) observed only in B*2705. The peptides are viewed from the side of the α2 helix together with a molecular surface covering the floor and back of the binding groove. The subtype-specific residue 116 is indicated also (Asp116, yellow; His116, turquoise); the bidentate salt bridge to Asp116 is drawn with green dotted lines in B. The binding pockets A–F are shown in bold letters. Atomic displacement ellipsoids for pVIPR-p4α and -p6α in C and D are colored according to the equivalent isotropic temperature factors B (Å2) (see color bar). (E, left) Schematic description of side chain orientation when looking from the NH2 to the COOH terminus of pVIPR. Bottom of peptide binding groove indicated by “β-sheet” and side for T cell recognition by “TCR”. (E, right) The orientation of the peptide side chains in the p4α and p6α conformations as in E (left). The respective binding pockets (A–F) are indicated as well. It is clear from this representation and Fig. 1 (A and B) that the two pVIPR conformations show major differences only from pLys3 to pTrp7.
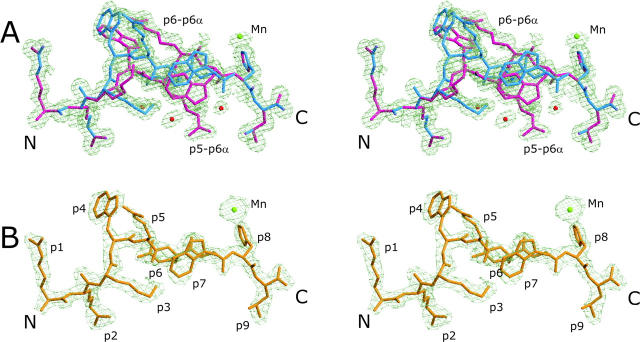
In addition, there is a metal binding site with an octahedral coordination sphere formed by pHis8-Nɛ, His197-Nɛ (of a symmetry-related HC), and four water molecules. The cation is probably Mn2+ as inferred from coordination distances (38). The electron density and B factors of Mn2+ (19.7 Å2 in B*2705 and 42.3 Å2 in B*2709; the B values are different due to the different resolutions) (Fig. 2 , electron densities for Mn2+) suggest that the site is fully occupied. The presence of Mn2+ is a purification artifact that originates from a lysis buffer.
Figure 2.
Final electron density of pVIPR conformations in B*2705 and B*2709. Stereo images of the final 2Fo-Fc electron density contoured at 1 σ level, displayed in light green. The two B*2705:pVIPR conformations are shown in A, the respective B*2709 complex is shown in B. The peptides are color coded as in Fig. 1 (A and B): water molecules as red and Mn2+ as green spheres.
HLA-B27 Subtype-dependent Peptide Conformations.
The different conformations of pVIPR in the binding groove of B*2705 are reflected in the backbone torsion angles of the peptide. In both structures, pVIPR is bound in the canonical conformation found regularly in crystal structures of MHC class I–nonapeptide complexes with main chain torsion angles (φ, ψ) of the extended β-strand type and only residue p4 (pTrp4) in right-handed α-helical conformation (Fig. 1 A, φ − 85.2° and ψ − 32.6°). The α-helical turn at position p4 results in a prominent kink directing the main chain “away” from the floor of the binding groove toward the solvent. Residues of pVIPR in this conformation (pVIPR-p4α) interact with all six pockets (Fig. 1 E, pockets A–F) provided by the binding groove, as found commonly for nonapeptides (1).
The additional peptide conformation found in B*2705 exhibits a pArg6 in α-form (Fig. 1 B, φ − 106.4° and ψ + 34.7° designated pVIPR-p6α) and both pVIPR-p4α and pVIPR-p6α are half occupied (Materials and Methods and Table I). pVIPR-p4α is virtually identical in both subtypes (Fig. 1 A, rms deviation 0.1 Å for nonhydrogen atoms). Between the p4α peptides in B*2705 and B*2709, there is only one minor difference in the orientation of the pArg6 guanidinium group as it is engaged in different hydrogen bonds.
Conformation-dependent HLA-B27:pVIPR Interactions.
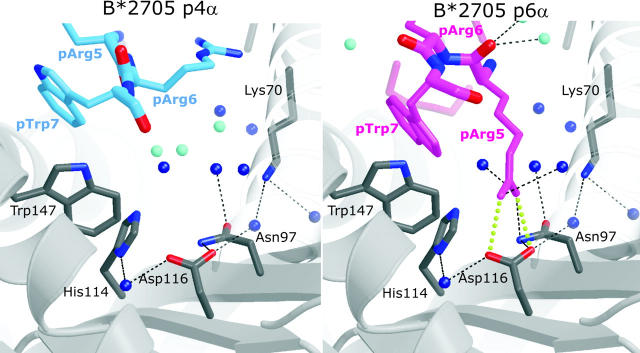
At the peptide NH2- and COOH-termini, pArg1, pArg2, pHis8, and pLeu9 occupy identical positions in pVIPR-p4α and -p6α (Fig. 1, A, B, and E and Fig. 2 A). The interactions of pArg1 and pArg2 with A and B pocket residues correspond to those observed for the B*2709:s10R complex (PDB code 1JGD; reference 37); the side chain of pArg1 is sandwiched between the side chains of HC Arg62 (α1-helix) and Trp167 (α2-helix). In addition, the side chains of Arg62 and Glu163 (α2-helix) are linked by a water-mediated salt bridge (Fig. 3, A and B , bridge) that covers the deeply embedded pArg2. At the COOH terminus, pHis8 is solvent exposed as well (Figs. 2 and 3), and pLeu9 is accommodated in the F pocket. The carboxy group of pLeu9 forms the common polar interactions with Tyr84-Oη, Thr143-Oγ, and Lys146-Nζ (1), and the aliphatic pLeu9 side chain forms hydrophobic interactions with the side chains of Leu81, Leu95, Tyr123, and Trp147 (19). Because the side chain of pLeu9 is too short to engage in any direct contacts with Asp116 (B*2705) or His116 (B*2709), there are no significant differences in the F pockets between both subtypes.
Figure 3.
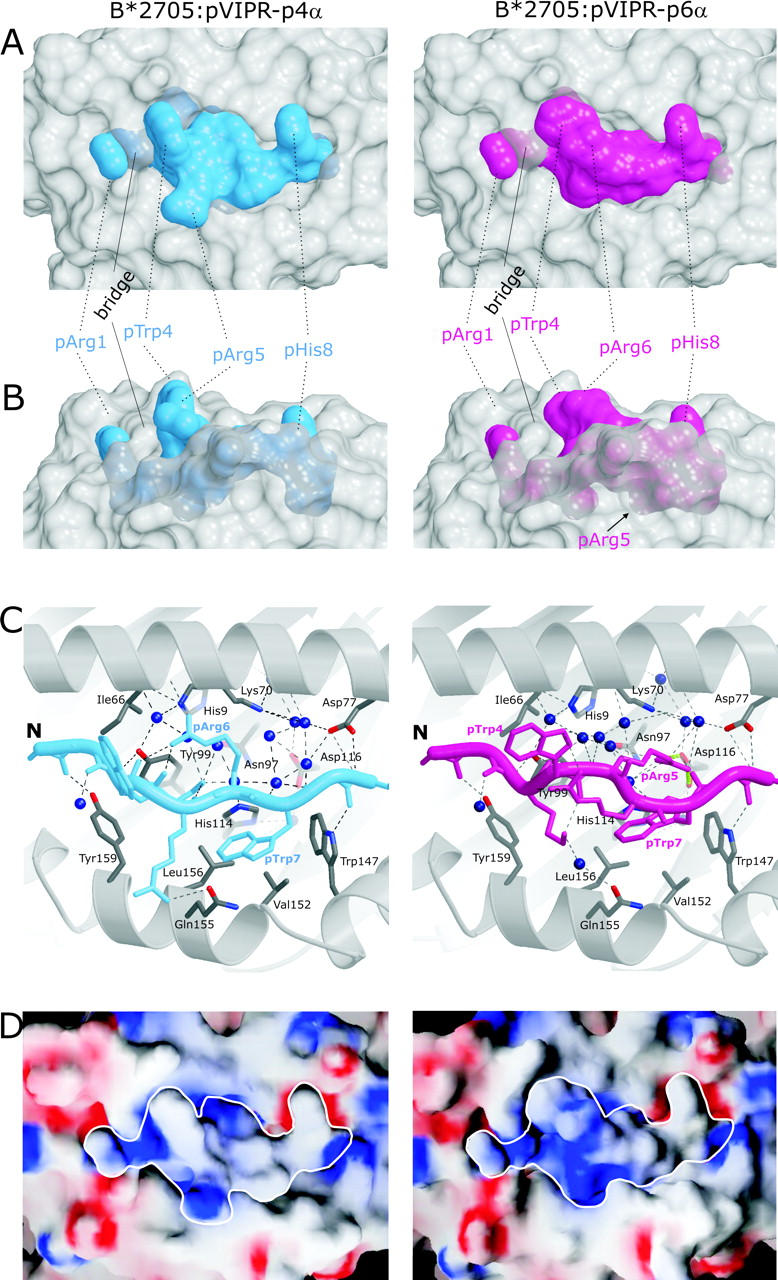
Molecular surfaces and contacts of pVIPR in the p4α and p6α conformations. (A and B) Molecular surfaces show the central part of the B*2705 peptide binding groove in gray and the pVIPR peptide in the p4α and p6α conformations (color coded as in Fig. 1 [A and B]). The binding groove has been rendered semi-transparent, allowing also the inspection of buried side chains exhibiting conformational differences. In A, the view is TCR-like, straight onto the peptide, whereas in B (rotated by 90° about a horizontal axis), the view is through the α2 helix. The center section of the peptide shows clear shape differences between the p4α and p6α conformations. (C) pVIPR hydrogen bonding in p4α and p6α, color coded as in Fig 1 (A and B). Only side chains with different binding modes (residues p3–p7) are shown. The binding groove's secondary structure is represented as gray spirals (α helices) and arrows (β strands) together with selected interacting residues (carbon atoms, gray; oxygens, red; and nitrogens, blue). Hydrogen bonds are depicted as black broken lines, the pArg5–Asp116 bidentate salt bridge is depicted as green dotted lines, and water molecules are depicted as dark blue spheres. (D) Electrostatic surfaces of both pVIPR conformations. Red indicates negative, blue indicates positive surface charge, and gray areas are uncharged. The view is looking straight onto the binding groove as in A. The border of the peptides is highlighted in white.
In contrast to the NH2- and COOH-terminal residues, the central sections of pVIPR-p4α and -p6α, pLys3 to pTrp7, which are principally accessible for recognition by a TCR, differ drastically with maximal disparity at pArg5, 3.7 Å for Cα, and 16.5 Å for Nη2 (Table II and Figs. 1, A, B, and E and Fig. 2 A and Figs. 3 and 4) . In pVIPR-p4α, side chains of pTrp4 and pArg5 are exposed to solvent, but in pVIPR-p6α, pArg5 is locked within the binding groove, its guanidinium group forming a bidentate salt bridge with the B*2705 subtype-specific Asp116 at the interface of the C, E, and F pockets (Figs. 1 B and 2 C and Figs. 3 and 4). In pVIPR-p6α, residues pTrp4 and pArg6 flanking pArg5 are maximally exposed to solvent and stacked (Fig. 1 B and Fig. 2). As a consequence of the structural differences around pArg5, the solvent-accessible surface areas of pVIPR-p4α and pVIPR-p6α have different size (160 Å2 for pVIPR-p4α and 124 Å2 for pVIPR-p6α), shape (Fig. 3, A and B), and charge distribution (Fig. 3 D) so that several prerequisites for differential recognition by a TCR are provided.
Table II.
Comparison of Peptide Coordination in the p4α and p6α Binding Modes of B*2705:pVIPR
| p4α conformation
|
p6α conformation
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Peptide residue |
Atom
|
Contact residue |
Distance
|
Interaction
|
Atom
|
Contact residue |
Distance
|
Interaction
|
| Å | Å | |||||||
| pArg1 | all contacts formed by pArg1-p4α are identical to those observed in -p6α conformation | |||||||
| pArg2 | all contacts formed by pArg2-p4α are identical to those observed in -p6α conformation | |||||||
| pLys3 | ||||||||
| pLys3N * | Tyr99OH * | 3.02 | H bond | pLys3N * | Tyr99OH * | 2.96 | H bond | |
| pLys3O ‡ | pArg6NH1 ‡ | 3.00 | H bond | pLys3O * | Tyr99OH * | 3.43 | H bond | |
| pLys3NZ ‡ | pArg5O ‡ | 2.78 | H bond | pLys3NZ ‡ | pTrp4O ‡ | 2.82 | H bond | |
| pTrp4 | solvent exposed | solvent exposed | ||||||
| pTrp4O ‡ | pArg6NH2 ‡ | 3.00 | H bond | pTrp4O ‡ | pLys3NZ ‡ | 2.82 | H bond | |
| pTrp4 ‡ | pArg6 ‡ | ∼3.3 | v.d. Waals | |||||
| pArg5 | solvent exposed | |||||||
| pArg5NH1 § | Gln155OE1 § | 3.20 | H bond | pArg5NH1 * | Asp116OD1 * | 3.04 | salt bridge | |
| pArg5O ‡ | pLys3NZ ‡ | 2.78 | H bond | pArg5NH2 * | Asp116OD2 * | 3.10 | salt bridge | |
| pArg6 | solvent exposed | |||||||
| pArg6NH1 ¶ | Ile66O ¶ | 2.74 | H bond | pArg6 ‡ | pTrp4 ‡ | ∼3.3 | v.d. Waals | |
| pArg6NH1 ‡ | pLys3O ‡ | 3.00 | H bond | |||||
| pArg6NH2 ‡ | pTrp4O ‡ | 3.00 | H bond | |||||
| pTrp7 | ||||||||
| pTrp7 § | Val152 § | ∼3.5 | v.d. Waals | pTrp7 § | Leu156 § | ∼3.5 | v.d. Waals | |
| pHis8 | all contacts formed by pHis8-p4α are identical to those observed in -p6α conformation | |||||||
| pLeu9 | all contacts formed by pLeu9-p4α are identical to those observed in -p6α conformation | |||||||
Only direct contacts are included, and water-mediated interactions are omitted. Interacting atoms are specified for polar interactions only because v.d. Waals contacts are too numerous and less specific.
β-sheet floor.
Intrapeptide contacts.
Helix α2.
Helix α1.
Figure 4.
Conformation-dependent peptide contacts with F-pocket residues. The F-pocket architecture and intermolecular interactions in B*2705:pVIPR-p4α (left) and -p6α (right), with relevant part of the peptide shown (same color code as in Fig. 1 [A and B]). Fully occupied water molecules are shown in dark blue and partially occupied ones (related to a specific peptide conformation) are in turquoise. The space occupied by pArg5-p6α is filled by water molecules in the p4α binding mode. The view is looking along the binding groove with the peptide COOH terminus in front.
As both p4α and p6α are roughly half occupied in the B*2705 structure (Materials and Methods), approximate energetic equivalence of the two conformations seems likely. This is suggested by the temperature factors for the peptide in the two conformations (Fig. 1, C and D) that are very similar: in B*2705, the average (isotropic) B factor for all peptide atoms is 14.8 Å2 (range 10.0–25.9 Å2) for p4α and 17.4 Å2 (range 10.8–31.9 Å2) for p6α. The highest flexibility found for pTrp4-p6α (∼30 Å2) is still relatively low on an absolute scale, and marked atomic displacement as indicated by large B factors is only found in the side chains of pTrp4, pArg6, and pTrp7 for both p4α and p6α (Fig. 1, C and D).
Peptide Conformation-dependent Recognition of HLA-B27 Subtypes by T Cells.
Previous analyses of autoreactive CTL lines from individuals typing as B*2705 or B*2709 have indicated that pVIPR-specific CTLs are frequently observed in patients with AS, although they occur also in healthy B*2705 individuals. In contrast, such CTLs are only rarely found in B*2709 individuals (15). We extended these studies, in particular in an attempt to identify CTLs exclusively reactive against B*2705 that might indicate immunogenicity also of B*2705-p6α. The results shown in Table III are based on 39 CTL lines, 17 of which are already published (15) and 22 were newly obtained from six AS patients (five B*2705 and one B*2702) and one B*2705 healthy control. The CTLs are grouped as preferentially recognizing peptide-transporter–deficient T2-B*2705 or T2-B*2709 cells, both subtypes with approximately equal efficiency, or exclusively T2-B*2705. No CTL recognizing only T2-B*2709 was observed, although pVIPR was used at a high concentration (70 μM). This experimental setup was chosen to maximize the chances to detect reactivity genuinely specific for B*2705:pVIPR complexes. Dose–response curves for the reactivity of pVIPR-specific CTLs have already been determined (15); at lower peptide concentrations, T2-B*2709 target cells tended to be lysed at higher efficiency than T2-B*2705 cells.
Table III.
Reactivity of CTLs from HLA-B27–positive Subjects towards the pVIPR Peptide in the Context of Either B*2705 or B*2709 Molecules
| Donora | Number of CTLs |
Specific lysis of T2-B*2709 > 05b |
Specific lysis of T2-B*2709 = 05c |
Specific lysis of T2-B*2709 < 05b |
Specific lysis of T2-B*2705 only |
|---|---|---|---|---|---|
| MP | 19 | 9 | 7 | 3 | 0 |
| EP | 6 | 0 | 4 | 1 | 1 (EP76) |
| AS | 4 | 1 | 3 | 0 | 0 |
| AB | 2 | 0 | 2 | 0 | 0 |
| MD | 4 | 1 | 2 | 1 | 0 |
| LV | 2 | 1 | 0 | 1 | 0 |
| CV | 2 | 0 | 2 | 0 | 0 |
| Total | 39 | 12 (30.7%) | 20 (51.3%) | 6 (15.4%) | 1 (2.6%) |
Five donors (MP, EP, AS, AB, and MD) are HLA-B*2705–positive patients with AS. LV is an HLA-B*2702–positive patient with AS, and CV is a healthy HLA-B*2705–positive individual. The results summarize the data obtained in two cytotoxicity experiments. Background (specific lysis of unpulsed T2-B*2705 and T2-B*2709) was <12%.
The difference in the percentage of specific lysis between T2-B*2705 and T2-B*2709 cells after background subtraction was >20%.
The difference in the percentage of specific lysis between T2-B*2705 and T2-B*2709 cells after background subtraction was <20%.
The majority of the CTLs lysed both T2-B*2705 and T2-B*2709 equally well (Table III). However, there were exceptions showing a clear preference for T2-B*2709 cells. This was not unexpected because it has previously been found that B*2709 antigens present pVIPR more efficiently than B*2705 (15). Only one CTL line was found (EP76) that selectively reacted with pVIPR presented by T2-B*2705 cells. The complete lack of reactivity against the B*2709:pVIPR complex, even at the high peptide concentration used, indicates that EP76 is directed against a structure different from B*2705:pVIPR-p4α because of the extreme structural similarity of B*2709:pVIPR and the p4α binding mode in B*2705 (Fig. 1 A). In contrast, five further CTLs from the AS-patient EP reacted with both T2-B*2705 and T2-B*2709 cells.
However, it must be pointed out that oligoclonal CTL lines were used in these studies. Therefore, it appears possible that noncross-reactive CTL clones may be concealed in the population of cells exhibiting cross-reactivity, resulting in a lower degree of lysis of B*2709 target cells, and leading to an underestimation of the immunogenicity of the p6α conformation in B*2705. 15.4% of the CTL lines lysed B*2705:pVIPR targets more efficiently than T2-B*2709:pVIPR cells (Table III).
Discussion
A fundamental deviation from the single binding mode found in all previously determined MHC class I:peptide complexes is the dual conformation of pVIPR in B*2705. This feature is of considerable interest both from structural and immunological points of view. Only one exception to the single binding mode of peptides has been described so far in the crystal structure of the rat RT1-Aa antigen in complex with the exceptionally long 13-mer peptide MTF-E. Two entirely different MTF-E conformations were found that are possibly associated with crystal packing effects (39). Because they could consequently represent nonphysiological artifacts, the two conformations and the reasons why MTF-E does not adopt a single binding mode are different from those reported here.
The simultaneous occurrence of pVIPR-p4α and pVIPR-p6α in B*2705 may be static or dynamic. If static, the analyzed crystals would be composed of approximately equal amounts of two cocrystallizing asymmetric units: one containing pVIPR-p4α and the other pVIPR-p6α. If dynamic, the peptide would change continually from one conformation to the other within the binding groove, using fixed NH2- and COOH-terminal amino acids with pivot points located at or close to the main chain connections of p2-p3 and p7-p8, respectively (Figs. 2 A and 3 C). Currently, we cannot distinguish between static and dynamic modes of peptide binding, but spectroscopic methods (40) or X-ray analyses of crystals produced at different temperatures may eventually provide an answer.
Thus far, the presentation of a peptide by an MHC molecule in a single, defined conformation has been considered a prerequisite for positive and negative selection of T cells within the thymus (41). The two drastically different binding modes of pVIPR to B*2705 show that this assumption might be an oversimplification. A peptide changing its conformation dynamically within the groove could possibly preclude high-affinity interaction with the two CDR3 regions of a TCR (42, 43), whereas a static conformation might allow a more efficient recognition. The rare occurrence of pVIPR-specific CTL in B*2709 individuals suggests that the B*2709:pVIPR complex can indeed serve in establishing tolerance (15). Conversely, the number of B*2705:pVIPR-p4α and -p6α complexes might fall below the epitope density threshold required for negative selection (44, 45), leading to the frequent presence of autoreactive, pVIPR-specific CTL (15).
The CTL lines analyzed here seem to possess relatively low affinity (15), questioning the presentation of the VIPR-derived peptide under physiological conditions, and suggesting the use of VIPR transfectants in cytotoxicity studies. However, even when testing the reactivity of CTLs against EBV-derived peptides, lysis of target cells is rarely observed (15, 46). At least the viral peptides are surely processed and presented; comparative studies with transfectants are usually unsuccessful. In addition, a COOH-terminal pLeu9 as seen in pVIPR does not present an obstacle for efficient proteasomal processing (47) and transport to the endoplasmic reticulum, as shown by peptide elution studies from B*2705 and B*2709 molecules (14). The high number of pVIPR-specific CTLs in some B*2705 individuals (15) might be explained by selective increase (clonal expansion) due to cross-reaction. This is not unlikely because TCR cross-reactivity is mandatory for the control of infections (48–52). In the case discussed here, the pVIPR cross-reactive nonself antigen could be the EBV-derived pLMP2 (15, 16, 53). Convincing examples of molecular mimicry between viral and self-proteins have already been identified (51, 52).
Unconventional peptide binding modes distantly resembling that observed for the p6α conformation in B*2705:pVIPR have occasionally been found in other MHC:peptide complexes as well (18, 54, 55), but the involvement of a strategic subtype-specific residue at the floor of the binding groove is unprecedented. The detection of p6α in B*2705:pVIPR but not in the closely related B*2709:pVIPR complex is obviously a consequence of the combined presence of the allele-specific residue Asp116 (B*2705) and a specific peptide sequence with Arg at position 5. The larger size and neutral or positive charge of His116 compared with Asp116 would interfere sterically and electrostatically if the pArg5 side chain were bound to the F pocket (Fig. 1 A). The binding of the peptide COOH-terminal pLeu9 to the F pocket brings a further constraint into play: to avoid clashes, it is likely that the side chain of this residue has to be small and hydrophobic, but not positively charged to prevent competition with another basic amino acid (in this instance, pArg5) that contacts Asp116.
The paucity of peptides with pArg5, such as pLMP2, which have been found to be presented by HLA-B27 molecules under natural conditions (14, 16), makes it difficult to provide an estimate of the generality of the p6α conformation, let alone a dual peptide binding mode, in HLA-B27 molecules. We have found a few further peptides derived from self- as well as microbial proteins that have in common not only an Arg residue but even the WRR motif (unpublished data). As it seems conceivable that some of these peptides share the unorthodox structural features of pVIPR when complexed with the B*2705 subtype, analyses with these novel peptides are underway to examine the possibility that the WRR motif represents a sequence with direct involvement in AS pathogenesis. With only very few structures of HLA-B27:peptide complexes solved to date (Results; references 19, 36, 37), it seems premature to engage in a speculation on the prerequisites for a dual peptide conformation.
However, the pVIPR-p6α peptide binding mode and its subtype dependency is not a unique, serendipitous finding. We have observed recently that the sequence-related pLMP2 peptide (16) binds in p4α-like form to B*2709 but in p6α-like conformation to B*2705, with pArg5 again forming a bidentate salt bridge with Asp116 (unpublished data). Therefore, the noncanonical binding mode involving residue 116 as observed for one of the conformations in B*2705:pVIPR and even its subtype dependency can be found in other HLA-B27:peptide complexes as well. Most AS-associated HLA-B27 alleles encode Asp116, whereas this residue is Tyr (B*2706) or His (B*2709) in the two subtypes not associated with AS (7).
A constellation similar to that of the B*2705/B*2709 pair is found in A*68012 (Asp116) and A*6807 (His116; reference 56), or in B*3501 (Ser116) and B*3503 (Phe116; reference 57), but comparative structural studies have not been performed for these proteins. Among further HLA class I genes, approximately half of the HLA-A alleles encode Asp116, and nearly all others encode Tyr116 that occurs also in a smaller number of HLA-B and HLA-C antigens, in which Asp116 is rarely represented (58). However, the mere availability of Asp116 is not sufficient for a basic residue in the middle of a peptide to engage in a p6α-like binding mode. This is exemplified by the HLA-A*0201:MAGE-A4 complex (59) or HLA-B*4402 and B*4403 complexed with an HLA-DP–derived peptide (60): these peptides exhibit Arg either at position 5 or 6 that are solvent exposed and not buried. Other residues on the floor of the binding groove could principally also be involved in noncanonical anchoring modes such as the HC residues 97, 99, and 114, which are, like residue 116, highly polymorphic in HLA class I molecules (61). For example, HLA-B*3501 presents an EBV-derived nonapeptide with pAsp5 contacting Arg97 and Tyr99 (18). Clearly, a nonconventional peptide anchoring could be more frequent than currently envisaged.
Polymorphisms within the MHC class I peptide binding groove as shown here may influence peptide presentation and T cell repertoire selection (1, 4, 7, 15, 62–64), but a dual peptide presentation mode as found in B*2705 that depends on the HLA-B27 subtype-specific amino acid exchange is novel. Distinct pVIPR presentation modes and differential AS-association of B*2705 and B*2709 may be linked in the course of the development of AS (15), although other factors influencing AS initiation and progression have to be taken into consideration, among them possible polymorphisms of HLA-B–linked genes (65–67). Our results support certain models of AS–pathogenesis that can incorporate differential peptide display by HLA-B27 subtypes, such as the arthritogenic peptide theory (4, 7), in particular in the modified form proposed by us (15), or the β2m-deposition hypothesis (68).
HLA subtype-dependent differential presentation of peptides in noncanonical conformations due to HLA class I HC polymorphisms at residue 116 might also be connected to the pathogenesis of other disease states, such as the rate of the progression to AIDS in HIV-positive patients (57) or the outcome of bone marrow transplants between unrelated individuals (69). In conclusion, our results provide an unexpected twist regarding the mode of peptide display as a single, structurally unaltered MHC molecule can present a given peptide in two entirely different binding modes, depending only on a buried HC amino acid polymorphism.
Acknowledgments
The authors thank all patients and the healthy proband for their participation in this paper. We are grateful to P.G. Coulie, J.A. López de Castro, and D.J. Schendel for discussions and comments on the manuscript; to M. Rühl for excellent technical assistance; and to U. Müller, S. Popov, and R. Hillig for help at the synchrotron facilities.
This work was financially supported by the Deutsche Forschungsgemeinschaft (SFB 449), Berliner Krebsgesellschaft, Monika Kutzner-Stiftung, Berlin, Sonnenfeld-Stiftung, Berlin, Fonds der Chemischen Industrie, COFIN 2001, and Istituto Pasteur Fondazione Cenci-Bolognetti. Data collection at Deutsches Elektronen Synchrotron was supported by a European Community grant to the European Molecular Biology Laboratory Outstation in Hamburg.
Abbreviations used in this paper: AS, ankylosing spondylitis; β2m, β2-microglobulin; HC, heavy chain; rms, root mean square.
References
- 1.Madden, D.R. 1995. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13:587–622. [DOI] [PubMed] [Google Scholar]
- 2.Saper, M.A., P.J. Bjorkman, and D.C. Wiley. 1991. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J. Mol. Biol. 219:277–319. [DOI] [PubMed] [Google Scholar]
- 3.Matsumura, M., D.H. Fremont, P.A. Peterson, and I.A. Wilson. 1992. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 257:927–934. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin, R., and P. Parham. 1990. Guilt by association: HLA-B27 and ankylosing spondylitis. Immunol. Today. 11:137–142. [DOI] [PubMed] [Google Scholar]
- 5.Szpak, Y., J.C. Vieville, T. Tabary, M.C. Naud, M. Chopin, C. Edelson, J.H. Cohen, J. Dausset, Y. de Kozak, and M. Pla. 2001. Spontaneous retinopathy in HLA-A29 transgenic mice. Proc. Natl. Acad. Sci. USA. 98:2572–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liblau, R.S., F.S. Wong, L.T. Mars, and P. Santamaria. 2002. Autoreactive CD8 T cells in organ-specific autoimmunity: emerging targets for therapeutic intervention. Immunity. 17:1–6. [DOI] [PubMed] [Google Scholar]
- 7.Ramos, M., and J.A. Lopez de Castro. 2002. HLA-B27 and the pathogenesis of spondyloarthritis. Tissue Antigens. 60:191–205. [DOI] [PubMed] [Google Scholar]
- 8.Trudeau, J.D., C. Kelly-Smith, C.B. Verchere, J.F. Elliott, J.P. Dutz, D.T. Finegood, P. Santamaria, and R. Tan. 2003. Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. J. Clin. Invest. 111:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewerton, D.A., F.D. Hart, A. Nicholls, M. Caffrey, D.C. James, and R.D. Sturrock. 1973. Ankylosing spondylitis and HLA-B27. Lancet. 1:904–907. [DOI] [PubMed] [Google Scholar]
- 10.Schlosstein, L., P.I. Terasaki, R. Bluestone, and C.M. Pearson. 1973. High association of an HL-A antigen, W27, with ankylosing spondylitis. N. Engl. J. Med. 288:704–706. [DOI] [PubMed] [Google Scholar]
- 11.Khan, M.A. 2002. Update on spondyloarthropathies. Ann. Intern. Med. 136:896–907. [DOI] [PubMed] [Google Scholar]
- 12.D'Amato, M., M.T. Fiorillo, C. Carcassi, A. Mathieu, A. Zuccarelli, P.P. Bitti, R. Tosi, and R. Sorrentino. 1995. Relevance of residue 116 of HLA-B27 in determining susceptibility to ankylosing spondylitis. Eur. J. Immunol. 25:3199–3201. [DOI] [PubMed] [Google Scholar]
- 13.Fiorillo, M.T., L. Meadows, M. D'Amato, J. Shabanowitz, D.F. Hunt, E. Appella, and R. Sorrentino. 1997. Susceptibility to ankylosing spondylitis correlates with the C-terminal residue of peptides presented by various HLA-B27 subtypes. Eur. J. Immunol. 27:368–373. [DOI] [PubMed] [Google Scholar]
- 14.Ramos, M., A. Paradela, M. Vazquez, A. Marina, J. Vazquez, and J.A. Lopez de Castro. 2002. Differential association of HLA-B*2705 and B*2709 to ankylosing spondylitis correlates with limited peptide subsets but not with altered cell surface stability. J. Biol. Chem. 277:28749–28756. [DOI] [PubMed] [Google Scholar]
- 15.Fiorillo, M.T., M. Maragno, R. Butler, M.L. Dupuis, and R. Sorrentino. 2000. CD8(+) T-cell autoreactivity to an HLA-B27-restricted self-epitope correlates with ankylosing spondylitis. J. Clin. Invest. 106:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks, J.M., R.J. Murray, W.A. Thomas, M.G. Kurilla, and A.B. Rickinson. 1993. Different HLA-B27 subtypes present the same immunodominant Epstein-Barr virus peptide. J. Exp. Med. 178:879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Porto, P., M. D'Amato, M.T. Fiorillo, L. Tuosto, E. Piccolella, and R. Sorrentino. 1994. Identification of a novel HLA-B27 subtype by restriction analysis of a cytotoxic gamma delta T cell clone. J. Immunol. 153:3093–3100. [PubMed] [Google Scholar]
- 18.Menssen, R., P. Orth, A. Ziegler, and W. Saenger. 1999. Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus. J. Mol. Biol. 285:645–653. [DOI] [PubMed] [Google Scholar]
- 19.Hülsmeyer, M., R.C. Hillig, A. Volz, M. Ruhl, W. Schroder, W. Saenger, A. Ziegler, and B. Uchanska-Ziegler. 2002. HLA-B27 subtypes differentially associated with disease exhibit subtle structural alterations. J. Biol. Chem. 277:47844–47853. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. [DOI] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project Number 4. 1994. The CCP4 Suite: Programs for Protein Crystallography. Acta. Crystallogr. D. 50:760–763. [DOI] [PubMed]
- 22.Murshudov, G.N., A.A. Vagin, A. Lebedev, K.S. Wilson, and E.J. Dodson. 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta. Crystallogr. D. 55:247–255. [DOI] [PubMed] [Google Scholar]
- 23.Perrakis, A., R. Morris, and V.S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458–463. [DOI] [PubMed] [Google Scholar]
- 24.Jones, T.A., and M. Kjeldgaard. 1997. Electron-density map interpretation. Methods Enzymol. 277:173–208. [DOI] [PubMed] [Google Scholar]
- 25.Winn, M.D., M.N. Isupov, and G.N. Murshudov. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta. Crystallogr. D. 57:122–133. [DOI] [PubMed] [Google Scholar]
- 26.Brünger, A.T., P.D. Adams, G.M. Clore, W.L. DeLano, P. Gros, R.W. Grosse-Kunstleve, J.S. Jiang, J. Kuszewski, M. Nilges, N.S. Pannu, et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta. Crystallogr. D. 54:905–921. [DOI] [PubMed] [Google Scholar]
- 27.Merritt, E.A. 1999. Expanding the model: anisotropic displacement parameters in protein structure refinement. Acta. Crystallogr. D. 55:1109–1117. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, R.A., M.W. MacArthur, D.S. Moss, and J.M. Thornton. 1995. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. [Google Scholar]
- 29.Hooft, R.W., G. Vriend, C. Sander, and E.E. Abola. 1996. Errors in protein structures. Nature. 381:272. [DOI] [PubMed] [Google Scholar]
- 30.McLachlan, A.D. 1982. Rapid comparison of protein structures. Acta. Crystallogr. A. 38:871–873. [Google Scholar]
- 31.Nicholls, A., K.A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 11:281–296. [DOI] [PubMed] [Google Scholar]
- 32.Kraulis, P.J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946–950. [Google Scholar]
- 33.Fenn, T.D., D. Ringe, and G.A. Petsko. 2003. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J. Appl. Crystallogr. 36:944–947. [Google Scholar]
- 34.Merritt, E.A., and D.J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505–524. [DOI] [PubMed] [Google Scholar]
- 35.Sanner, M.F., A.J. Olson, and J.C. Spehner. 1996. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 38:305–320. [DOI] [PubMed] [Google Scholar]
- 36.Madden, D.R., J.C. Gorga, J.L. Strominger, and D.C. Wiley. 1992. The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC. Cell. 70:1035–1048. [DOI] [PubMed] [Google Scholar]
- 37.Hillig, R.C., M. Huelsmeyer, W. Saenger, K. Welfle, R. Misselwitz, H. Welfle, C. Kozerski, A. Volz, B. Uchanska-Ziegler, and A. Ziegler. 2003. Thermodynamic and structural analysis of peptide- and allele-dependent properties of two HLA-B27 subtypes exhibiting differential disease association. J. Biol. Chem. 10.1074/jbc. M307457200. [DOI] [PubMed]
- 38.Harding, M.M. 2001. Geometry of metal-ligand interactions in proteins. Acta. Crystallogr. D. Biol. Crystallogr. 57:401–411. [DOI] [PubMed] [Google Scholar]
- 39.Speir, J.A., J. Stevens, E. Joly, G.W. Butcher, and I.A. Wilson. 2001. Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa. Immunity. 14:81–92. [DOI] [PubMed] [Google Scholar]
- 40.Uchanska-Ziegler, B., U. Alexiev, R.C. Hillig, M. Hülsmeyer, T. Pöhlmann, W. Saenger, A. Volz, and A. Ziegler. 2003. X-ray Crystallography and Dynamic Studies of HLA-B*2705 and B*2709 Molecules Complexed with the Same Peptide. IHWG Press, Seattle, WA. In press. [Google Scholar]
- 41.Von Boehmer, H., I. Aifantis, F. Gounari, O. Azogui, L. Haughn, I. Apostolou, E. Jaeckel, F. Grassi, and L. Klein. 2003. Thymic selection revisited: how essential is it? Immunol. Rev. 191:62–78. [DOI] [PubMed] [Google Scholar]
- 42.Wu, L.C., D.S. Tuot, D.S. Lyons, K.C. Garcia, and M.M. Davis. 2002. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 418:552–556. [DOI] [PubMed] [Google Scholar]
- 43.Housset, D., and B. Malissen. 2003. What do TCR-pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol. 24:429–437. [DOI] [PubMed] [Google Scholar]
- 44.Cook, J.R., E.M. Wormstall, T. Hornell, J. Russell, J.M. Connolly, and T.H. Hansen. 1997. Quantitation of the cell surface level of Ld resulting in positive versus negative selection of the 2C transgenic T cell receptor in vivo. Immunity. 7:233–241. [DOI] [PubMed] [Google Scholar]
- 45.Delaney, J.R., Y. Sykulev, H.N. Eisen, and S. Tonegawa. 1998. Differences in the level of expression of class I major histocompatibility complex proteins on thymic epithelial and dendritic cells influence the decision of immature thymocytes between positive and negative selection. Proc. Natl. Acad. Sci. USA. 95:5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ong, K.W., A.D. Wilson, T.R. Hirst, and A.J. Morgan. 2003. The B subunit of Escherichia coli heat-labile enterotoxin enhances CD8+ cytotoxic-T-lymphocyte killing of Epstein-Barr virus-infected cell lines. J. Virol. 77:4298–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuon, W., H.G. Holzhutter, H. Appel, M. Grolms, S. Kollnberger, A. Traeder, P. Henklein, E. Weiss, A. Thiel, R. Lauster, et al. 2001. Identification of HLA-B27-restricted peptides from the Chlamydia trachomatis proteome with possible relevance to HLA-B27-associated diseases. J. Immunol. 167:4738–4746. [DOI] [PubMed] [Google Scholar]
- 48.Mason, D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today. 19:395–404. [DOI] [PubMed] [Google Scholar]
- 49.Ueno, T., H. Tomiyama, and M. Takiguchi. 2002. Single T cell receptor-mediated recognition of an identical HIV-derived peptide presented by multiple HLA class I molecules. J. Immunol. 169:4961–4969. [DOI] [PubMed] [Google Scholar]
- 50.Holler, P.D., L.K. Chlewicki, and D.M. Kranz. 2003. TCRs with high affinity for foreign pMHC show self-reactivity. Nat. Immunol. 4:55–62. [DOI] [PubMed] [Google Scholar]
- 51.Panoutsakopoulou, V., M.E. Sanchirico, K.M. Huster, M. Jansson, F. Granucci, D.J. Shim, K.W. Wucherpfennig, and H. Cantor. 2001. Analysis of the relationship between viral infection and autoimmune disease. Immunity. 15:137–147. [DOI] [PubMed] [Google Scholar]
- 52.Lang, H.L., H. Jacobsen, S. Ikemizu, C. Andersson, K. Harlos, L. Madsen, P. Hjorth, L. Sondergaard, A. Svejgaard, K. Wucherpfennig, et al. 2002. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat. Immunol. 3:940–943. [DOI] [PubMed] [Google Scholar]
- 53.Fiorillo, M.T., G. Greco, M. Maragno, I. Potolicchio, A. Monizio, M.L. Dupuis, and R. Sorrentino. 1998. The naturally occurring polymorphism Asp116→His116, differentiating the ankylosing spondylitis-associated HLA-B*2705 from the non-associated HLA-B*2709 subtype, influences peptide-specific CD8 T cell recognition. Eur. J. Immunol. 28:2508–2516. [DOI] [PubMed] [Google Scholar]
- 54.Reid, S.W., S. McAdam, K.J. Smith, P. Klenerman, C.A. O'Callaghan, K. Harlos, B.K. Jakobsen, A.J. McMichael, J.I. Bell, D.I. Stuart, and E.Y. Jones. 1996. Antagonist HIV-1 Gag peptides induce structural changes in HLA B8. J. Exp. Med. 184:2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apostolopoulos, V., M. Yu, A.L. Corper, W. Li, I.F. McKenzie, L. Teyton, and I.A. Wilson. 2002. Crystal structure of a non-canonical high affinity peptide complexed with MHC class I: a novel use of alternative anchors. J. Mol. Biol. 318:1307–1316. [DOI] [PubMed] [Google Scholar]
- 56.Turnquist, H.R., E.L. Schenk, M.M. McIlhaney, H.D. Hickman, W.H. Hildebrand, and J.C. Solheim. 2002. Disparate binding of chaperone proteins by HLA-A subtypes. Immunogenetics. 53:830–834. [DOI] [PubMed] [Google Scholar]
- 57.Gao, X., G.W. Nelson, P. Karacki, M.P. Martin, J. Phair, R. Kaslow, J.J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, et al. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668–1675. [DOI] [PubMed] [Google Scholar]
- 58.Robinson, J., M.J. Waller, P. Parham, N. de Groot, R. Bontrop, L.J. Kennedy, P. Stoehr, and S.G. Marsh. 2003. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic. Acids Res. 31:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hillig, R.C., P.G. Coulie, V. Stroobant, W. Saenger, A. Ziegler, and M. Hulsmeyer. 2001. High-resolution structure of HLA-A*0201 in complex with a tumour-specific antigenic peptide encoded by the MAGE-A4 gene. J. Mol. Biol. 310:1167–1176. [DOI] [PubMed] [Google Scholar]
- 60.MacDonald, W.A., A.W. Purcell, N.A. Mifsud, L.K. Ely, D.S. Williams, L. Chang, J.J. Gorman, C.S. Clements, L. Kjer-Nielsen, D.M. Koelle, et al. 2003. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J. Exp. Med. 198:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reche, P.A., and E.L. Reinherz. 2003. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 331:623–641. [DOI] [PubMed] [Google Scholar]
- 62.Chen, W., J. McCluskey, S. Rodda, and F.R. Carbone. 1993. Changes at peptide residues buried in the major histocompatibility complex (MHC) class I binding cleft influence T cell recognition: a possible role for indirect conformational alterations in the MHC class I or bound peptide in determining T cell recognition. J. Exp. Med. 177:869–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luz, J.G., M. Huang, K.C. Garcia, M.G. Rudolph, V. Apostolopoulos, L. Teyton, and I.A. Wilson. 2002. Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V(β) interactions. J. Exp. Med. 195:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sesma, L., V. Montserrat, J.R. Lamas, A. Marina, J. Vazquez, and J.A. Lopez de Castro. 2002. The peptide repertoires of HLA-B27 subtypes differentially associated to spondyloarthropathy (B*2704 and B*2706) differ by specific changes at three anchor positions. J. Biol. Chem. 277:16744–16749. [DOI] [PubMed] [Google Scholar]
- 65.Okamoto, K., S. Makino, Y. Yoshikawa, A. Takaki, Y. Nagatsuka, M. Ota, G. Tamiya, A. Kimura, S. Bahram, and H. Inoko. 2003. Identification of I kappa BL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am. J. Hum. Genet. 72:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hofmann, M.A., S. Drury, B.I. Hudson, M.R. Gleason, W. Qu, Y. Lu, E. Lalla, S. Chitnis, J. Monteiro, M.H. Stickland, et al. 2002. RAGE and arthritis: the G82S polymorphism amplifies the inflammatory response. Genes Immun. 3:123–135. [DOI] [PubMed] [Google Scholar]
- 67.Fiorillo, M.T., A. Cauli, C. Carcassi, P.P. Bitti, A. Vacca, G. Passiu, F. Bettosini, A. Mathieu, and R. Sorrentino. 2003. Two distinctive HLA haplotypes harbor the B27 alleles negatively or positively associated with ankylosing spondylitis in Sardinia: implications for disease pathogenesis. Arthritis Rheum. 48:1385–1389. [DOI] [PubMed] [Google Scholar]
- 68.Uchanska-Ziegler, B., and A. Ziegler. 2003. Ankylosing spondylitis: a beta2m-deposition disease? Trends Immunol. 24:73–76. [DOI] [PubMed] [Google Scholar]
- 69.Ferrara, G.B., A. Bacigalupo, T. Lamparelli, E. Lanino, L. Delfino, A. Morabito, A.M. Parodi, C. Pera, S. Pozzi, M.P. Sormani, et al. 2001. Bone marrow transplantation from unrelated donors: the impact of mismatches with substitutions at position 116 of the human leukocyte antigen class I heavy chain. Blood. 98:3150–3155. [DOI] [PubMed] [Google Scholar]