Abstract
Cotranscriptional cleavage mediated by a hammerhead ribozyme can affect alternative splicing if interposed between an exon and its intronic regulatory elements. This has been demonstrated using two different alternative splicing systems based on α-tropomyosin and fibronectin genes. We suggest that there is a requirement for intronic regulatory elements to be covalently attached to exons that are in turn tethered to the elongating polymerase. In the case of the alternatively spliced EDA exon of the fibronectin gene, we demonstrate that the newly identified intronic downstream regulatory element is associated with the splicing regulatory protein SRp20. Our results suggest that targeted hammerhead ribozyme cleavage within introns can be used as a tool to define splicing regulatory elements.
Keywords: alternative splicing regulation, RNA polymerase II, α-tropomyosin gene, fibronectin gene, ribozyme
INTRODUCTION
Mammalian protein coding genes are transcribed by RNA polymerase II (Pol II) and usually contain relatively short exon sequences separated by very long introns. The data from alternative splicing microarrays combined with data sets from expressed sequence tags (ESTs) indicate that up to 73% of human genes are alternatively spliced (Johnson et al. 2003). The accuracy of splicing is defined by consensus splice sites and regulatory elements such as exon and intron splicing enhancers and silencers. These regulatory elements are recognized by members of the SR and hnRNP family of proteins. Interaction between these proteins and spliceosomal components establishes a “splicing code” that defines the nature and level of a particular spliced isoform (Matlin et al. 2005; Blencowe 2006). An additional level of splicing regulation occurs through coupling transcription to pre-mRNA processing. Thus, the C-terminal domain of Pol II large subunit provides a platform for recruitment of RNA processing factors to act on the emerging pre-mRNA transcript (Maniatis and Reed 2002; Kornblihtt et al. 2004). The importance of splicing accuracy and efficient regulation is exemplified by the fact that many human genetic diseases arise through the misregulation of alternative splicing (Caceres and Kornblihtt 2002; Faustino and Cooper 2003).
As most human introns are very long (average 3 kb, but frequently greater than 10 kb), transcription of such extensive RNA sequence will accumulate vast amounts of pre-mRNA near the site of transcription. This might interfere with the ability of the splicing machinery to locate splice sites within a mass of unspliced RNA. Therefore, removal of long introns from the pre-mRNA transcript is likely to be a critical process requiring careful regulation.
We have previously demonstrated that intron 2 of the constitutively spliced β-globin transcript can be cotranscriptionally cleaved with no effect on mRNA levels or the accuracy and efficiency of pre-mRNA splicing of exons flanking the cleaved intron (Dye et al. 2006). In this study we initially extend the analysis of exon tethering by showing that cotranscriptional cleavage of the first intron or both introns together has no effect on β-globin mRNA synthesis. This result confirms that transcribed exons are tethered to the elongating Pol II transcription complex so that they are unaffected by disruption of continuity in the growing RNA chain. We then examine the effect of intronic cotranscriptional cleavage on the regulation of alternative splicing in two different alternatively spliced minigenes based on the rat α-tropomyosin (α-TM) and human fibronectin genes. We show that in contrast to constitutively spliced introns, cotranscriptional cleavage between an exon and its regulatory elements can affect alternative splicing. These observations imply a requirement for continuity of the intron transcript between such elements that is essential for alternative splicing of both α-TM and fibronectin EDA minigenes. Also using the EDA exon minigene, we demonstrate that the hammerhead ribozyme can be used as a tool for mapping intronic regulatory elements that affect alternative splicing. By this approach we have identified a novel negative regulatory element (downstream regulatory element [DRE]) located downstream from the EDA exon that mediates its effect through association with the splicing factor, SRp20.
RESULTS
Ribozyme cleavage within β-globin intronic sequences does not affect splicing
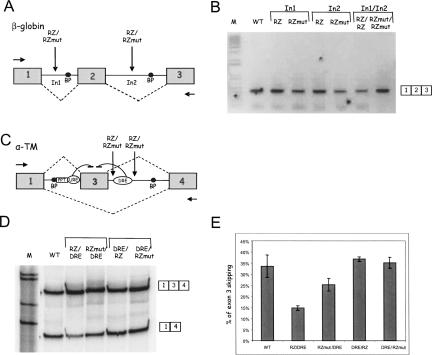
Our previous studies have demonstrated that β-globin intron 2 RNA sequence can be cotranscriptionally cleaved with no affect on pre-mRNA splicing of exons flanking the cleaved intron (Dye et al. 2006). To investigate this further, we inserted a 52-nucleotide (nt) hammerhead ribozyme (RZ) and its inactive version, containing a single point mutation (Samarsky et al. 1999) (RZmut), at different locations within the β-globin minigene construct (Fig. 1A). In particular, we placed it in intron 1 creating βIn1-RZ and βIn1-RZmut constructs. We also generated a double ribozyme construct containing RZ sequences within both intron 1 and intron 2 sequences (βIn1-RZ/In2-RZ and βIn1-RZmut/In2-RZmut). Following transfection of these minigene constructs into HeLa cells, their splicing profiles were analyzed by RT-PCR. As shown (Fig. 1B), the splicing pattern of all analyzed constructs was unaffected. This suggests that cleavage within the β-globin intronic sequences does not affect its constitutive splicing pattern, most probably due to the tethering of transcribed exonic sequences to the elongating Pol II transcription complex.
FIGURE 1.
Cotranscriptional intron cleavage between exon and DRE affects α-TM alternative splicing. (A) Diagram of β-globin minigene (βWT). β-globin exons 1, 2, and 3 are shown as boxes, and introns are shown as lines. Branch point (BP) sequences of exons 3 and 2 are black ovals. Splicing pattern is represented by diagonal dashed lines. Arrows indicate location of primers used in RT-PCR analysis. Ribozyme (RZ) and mutant ribozyme (RZmut) sequences (Samarsky et al. 1999) were positioned in intron 1 (In1) and intron 2 (In2) of βWT. (B) RT-PCR analysis of total RNA from HeLa cells expressing different minigenes. Minigenes were transfected into HeLa cells, and the resulting RT-PCR products were resolved on 1% agarose gels. The splicing product 1–2-3 is indicated. (C) Diagram of α-TM minigene (WT). α-TM exons 1, 3, and 4 are boxes and introns are lines. Branch point (BP) sequences of exons 3 and 4 are black ovals. Splicing patterns are represented as diagonal dashed lines. In most cells the generated splicing product is 1–3-4, while in smooth muscle (SM) cells it is 1–4. PPT, URE, and DRE are negative regulatory elements involved in the repression of exon 3 in SM cells. Minigene contains six tandem URE copies to increase skipping of exon 3 in SM and nonmuscle cells (Gromak and Smith 2002). Arrows indicate location of primers used in RT-PCR analysis. Ribozyme (RZ) and mutant ribozyme (RZmut) sequences (Samarsky et al. 1999) were positioned either side of DRE. (D) RT-PCR analysis of total RNA from HeLa cells expressing different minigenes. Minigenes were transfected into HeLa cells, and the resulting RT-PCR products were resolved on 6% polyacrylamide gels. The splicing products 1–3-4 and 1–4 are indicated. (E) Histogram showing the percentage of α-TM exon 3 skipping. Data are expressed as average percentage of exon 3 skipping (±SD) based on three independent experiments.
Ribozyme cleavage between α-TM exon 3 and intronic DRE
To determine whether or not intronic cotranscriptional cleavage can affect alternative splicing we employed the α-tropomyosin (α-TM) minigene (Gromak and Smith 2002). This construct recapitulates splicing regulation of the α-TM gene and contains exons 1, 3, and 4 surrounded by their intronic regulatory sequences (Fig. 1C). Although the α-TM minigene lacks the mutually exclusive exon 2, this does not interfere with the splicing regulation of exon 3 (Gooding et al. 1994, 1998). Exon 3 of α-TM is predominantly incorporated into the spliced mRNA in most cell types, except for smooth muscle (SM) tissue. Repression of exon 3 in SM cells is mediated by a number of negative regulatory elements surrounding this exon: the polypyrimidine tract (PPT), upstream and downstream regulatory elements (URE and DRE) (Gooding et al. 1994; Perez et al. 1997). In this study, we used constructs containing six copies of the URE element, which enhances exon 3 skipping in nonmuscle cells (i.e., HeLa) (Gromak and Smith 2002).
To determine whether or not cotranscriptional cleavage can affect alternative splicing, a 52-nt hammerhead ribozyme (RZ) and its inactive variant (RZmut) (Samarsky et al. 1999) was positioned on either side of the clearly defined regulatory elements of α-TM exon 3. Initially we introduced RZ and RZmut sequences either side of the DRE element and then analyzed RNA extracted from transfected HeLa cells using RT-PCR (Fig. 1D). As expected, neither RZ nor RZmut affected exon 3 splicing when placed downstream of the DRE (DRE/RZ, DRE/RZmut; Fig. 1D,E). This can be explained by the tethering of the exons 3 and 4 to Pol II so that cleavage in the middle of intron 3 does not affect alternative splicing, as demonstrated above and previously (Dye et al. 2006) for the constitutively spliced β-globin gene. However, when RZ rather than RZmut was positioned upstream of the DRE (RZ/DRE), a significant decrease in the amount of exon 3 skipping was observed. Disruption of transcript continuity between the exon and its regulatory element appears to affect alternative splicing. We suggest that tethering may be important not only between adjacent exons and elongating Pol II but also between the intronic regulatory sequences and their adjacent exons. The RZmut sequence upstream of the DRE element (RZmut/DRE) does slightly decrease exon 3 skipping, suggesting that this 52-nt sequence has a nonspecific spacing effect caused by moving the DRE away from the exon 3 (Roberts et al. 1998). Nevertheless, RNA cleavage as in the RZ/DRE affects alternative splicing significantly more than with the RZmut/DRE construct (Fig. 1E).
Ribozyme cleavage between α-TM exon 3 and URE disrupts exon definition
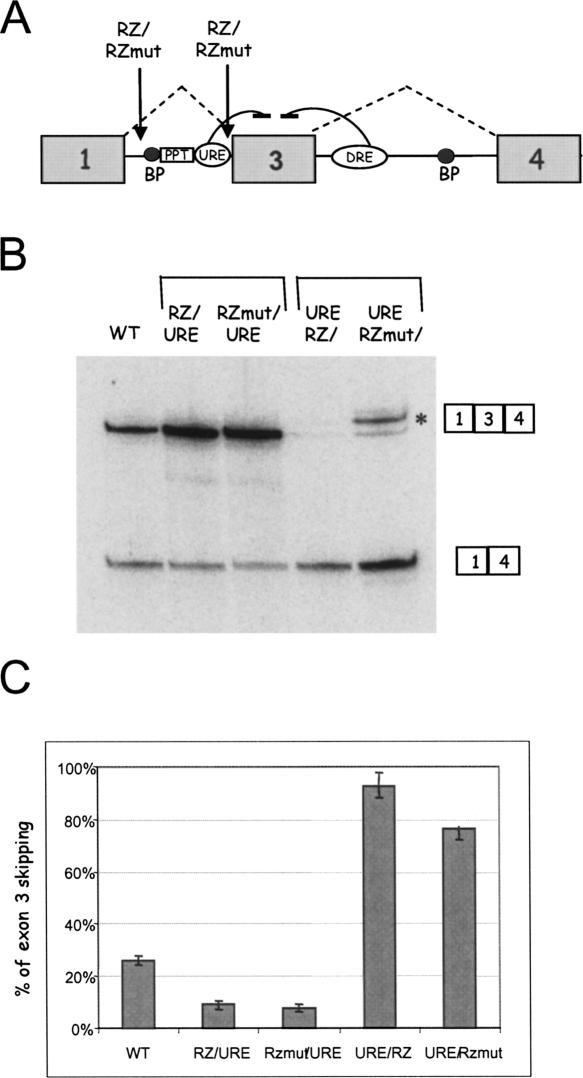
To further test the hypothesis that alternative splicing can be affected by cotranscriptional intronic cleavage, we placed RZ and RZmut sequences upstream and downstream of the α-TM minigene multimerized URE element (Fig. 2A). When upstream, both RZ and RZmut inserts caused a similar decrease in exon 3 skipping (Fig. 2B,C). This result may relate to nonspecific spatial effects. However, with RZ and RZmut placed downstream of the URE, we observed the opposite effect of increased exon 3 skipping. This can be interpreted as a consequence of disruption of the “exon definition” threshold. The branch point (BP) of the endogenous α-TM gene exon 3 is located 175 nt upstream of its 3′ splice site and 301 nt upstream of the exon 3 5′ splice site. This corresponds to the exon definition threshold of 300 nt, above which exon skipping normally occurs (Robberson et al. 1990; Berget 1995). In the URE/RZ and URE/RZmut constructs, this distance increases to 428 nt (due to insertion of both 6URE and RZ sequences), which is likely to be above the exon definition threshold, and so causes exon 3 skipping. Consequently, URE/RZmut construct generates only a small amount of 1–3-4 product, which is comprised of 2 bands, wild-type 1–3-4 product (Fig. 2B, bottom band) and cryptic 1–3-4 product (Fig. 2B, top band, *), due to inclusion of an extra 22 nt of the intronic sequence upstream of exon 3 by activation of a cryptic splice site within the RZ insert. Sequence analysis revealed that this cryptic 3′ splice site is a canonical AG dinucleotide. Placing the RZ sequence downstream of the URE causes almost 100% exon 3 skipping presumably due to both disruption of the exon definition threshold as well as cleavage of RNA between the BP and the 3′ splice site of exon 3. Very little cryptic 3′ splice site usage occurs with the URE/RZ construct. Possibly cotranscriptional cleavage upstream of this splice site blocks the required interaction between splicing factors that recognize exon 3 splicing signals. Although alternative splicing regulation of exon 3 is disrupted due to loss of exon definition, cotranscriptional cleavage in the URE/RZ construct appears to further enhance exon 3 skipping. These data also provide evidence that, with this experimental system, the hammerhead ribozyme cleaves cotranscriptionally as previously described (Dye et al. 2006).
FIGURE 2.
Cotranscriptional intron cleavage between exon and URE affects α-TM alternative splicing due to disruption of exon 3 definition. (A) Diagram of the α-TM minigene. RZ and RZmut were positioned either side of URE. (B) RT-PCR analysis of total RNA from HeLa cells expressing indicated minigenes. (*) Denotes 1–3-4 cryptic splicing product. (C) Histogram showing the percentage of α-TM exon 3 skipping. Data are expressed as average percentage of exon 3 skipping (±SD) based on three independent experiments.
Intronic ribozyme cleavage affects alternative EDA exon splicing
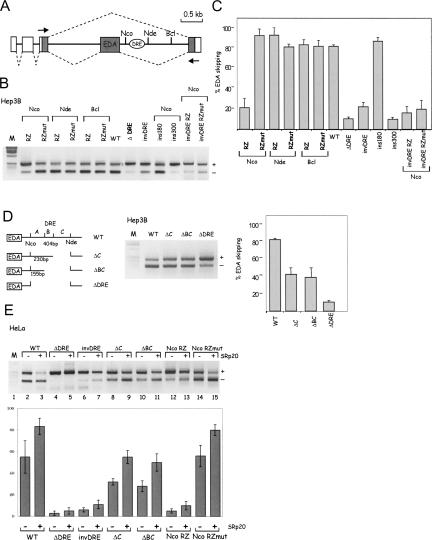
We wished to extend our results on ribozyme cleavage of the α-TM transcript to a different alternative splicing system. We therefore employed the fibronectin EDA minigene in which the alternatively spliced EDA exon is skipped to an 80% level in Hep3B cells. For these experiments we inserted the engineered N117 hammerhead ribozyme (88 bp long) and its catalytically inactive mutant (14A to G) (Yen et al. 2004) in three intronic positions at the indicated NcoI, NdeI, and BclI sites downstream of the alternatively spliced EDA exon (Fig. 3A). The resulting constructs were transiently transfected into Hep3b cells, and the EDA alternative splicing pattern was evaluated by RT-PCR analysis. Positioning of N117 RZ, but not RZmut, at the NcoI site induced a major decrease in the percentage of EDA exon skipping as compared with the parent pEDA (WT) construct (Fig. 3B,C). In contrast, the percentage of EDA exon skipping was unaffected in the Nde RZ or Bcl RZ constructs. We postulate that the position-specific effect of RZ at the NcoI site on EDA skipping is due to the presence of a nearby negative DRE that cannot interact with the Pol II tethered exon when the nascent transcript is cotranscriptionally cleaved.
FIGURE 3.
Cotranscriptional intron cleavage upstream of DRE element affects EDA alternative splicing. (A) Diagram of the fibronectin EDA minigene. Fibronectin and α-globin exons are gray and white boxes, respectively, and introns are lines. The unique restriction sites in the EDA downstream intron, used for RZ insertion, and the two possible alternative splicing products are shown. The arrows indicate primers used in RT-PCR analysis. (B) RT-PCR analysis of total RNA from cells expressing each minigene. Minigenes transfected into Hep3B cells and RT-PCR products resolved on 1.5% agarose gels. RZ and RZmut are the 88-bp active and inactive hammerhead ribozymes (Yen et al. 2004). EDA exon inclusion (+) and exclusion (−) forms are indicated. (M) Marker 1-kb ladder. (C) Histogram showing the percentage of EDA exon skipping (±SD) based on at least three independent duplicate experiments. (D) Diagram indicating deletions introduced within the DRE element of the EDA minigene (left). RT-PCR analysis of the corresponding minigenes in Hep3B cells (middle). The percentage of EDA exon skipping (±SD) is based on at least three independent experiments (right). (E) SRp20 induces EDA exon skipping in DRE-dependent manner and also dependent on RNA continuity between the EDA exon and the DRE. RT-PCR analysis of SRp20 overexpression with indicated minigene constructs in HeLa cells (top). Percentage of EDA exon skipping (±SD) is based on at least three independent experiments (bottom).
To confirm that EDA alternative splicing is regulated by a negative DRE, we made invDRE and ΔDRE minigenes where the potential DRE between the NcoI and NdeI sites was either inverted or deleted. Consistent with the presence of a negative DRE between the NcoI and NdeI sites, both these constructs showed a lower percentage of EDA exon skipping, similar to that seen in the Nco RZ construct (Fig. 3B,C). Interestingly, neither RZ nor RZmut placed at the NcoI site in the invDRE construct had any significant effect on EDA exon skipping, reinforcing the importance of the wt DRE element. Furthermore, a 300-bp spacer sequence, but not 180 bp, between the EDA exon and the DRE also decreases EDA exon skipping (Fig. 3B,C). These experiments suggest that positioning of the ribozyme at the NcoI site interferes with pre-mRNA processing by preventing negative interactions between the EDA exon and the downstream DRE on the nascent transcript.
Human 404-bp DRE demonstrates high evolutionary conservation in mammals (62% identity in Mus, 69% in Rattus, 72% in Canis, 78% in Felis, and 76% in Bos). To further define the role of the DRE element in the splicing regulation of fibronectin EDA minigene, we generated constructs containing progressive deletions of the DRE (Fig. 3D). In comparison with the WT minigene, ΔC construct, containing 3′ end deletion of the DRE, decreased EDA exon skipping by 50%. Further deletion of the region B within the DRE did not significantly change the splicing pattern, whereas additional deletion of the region A decreased EDA exon skipping to only 10% (Fig. 3D). These experiments suggest that the regions A and C within the DRE contain regulatory elements critical for EDA exon skipping.
Intronic splicing regulatory elements can often induce exon skipping through the binding of the SR proteins (Gallego et al. 1997; Dauksaite and Akusjarvi 2002; Simard and Chabot 2002; Buratti et al. 2007). In particular, SRp20 was shown to inhibit EDA exon inclusion through its interaction with the C-terminal domain of PolII (Cramer et al. 1999; de la Mata and Kornblihtt 2006). As SRp20 is the only known member of the SR protein family that induces EDA exon skipping, we decided to test the contribution of the intronic DRE on the SRp20-mediated EDA splicing. The effect of SRp20 can be evaluated in HeLa cells as they show a reduced level of EDA exon skipping (around 55%) compared with Hep3B cells (around 80%) (Fig. 3D,E, WT constructs). As expected, overexpression of SRp20 significantly induced the level of EDA exon skipping in the WT construct (Fig. 3E, lane 3). SRp20 had no significant effect on the ΔDRE minigene and only slightly increased the percentage of the EDA exon skipping in the invDRE construct (lanes 4–7). Both ΔC and ΔBC constructs lacking parts of the DRE were still responsive to SRp20 inhibition of the EDA exon (Fig. 3E, lanes 8–11). Interestingly, in the constructs where RZ/RZmut were inserted in NcoI site of the WT minigene, SRp20 affected EDA exon skipping in the RZmut situation but had little effect with the RZ (Fig. 3E, lanes 12–15). These data suggest that disruption of the continuity between the EDA exon and the DRE at the level of the nascent RNA transcript significantly affects SRp20-mediated splicing regulation of the fibronectin minigene.
DISCUSSION
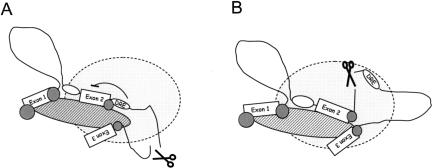
Using the α-TM and EDA minigenes, we have demonstrated that placing a cotranscriptionally cleaved ribozyme in an intron affects alternative splicing. When the ribozyme is positioned in the middle of the intron (outside of the regulatory elements), we did not detect any specific effect on the regulation of alternative splicing (Fig. 4A). However, when it is positioned so that it breaks the continuity between the exon and its regulatory element, we observed significant effects on splicing regulation (Fig. 4B). These results imply a requirement for the continuity of the RNA molecule between intronic regulatory elements and alternatively spliced exons. Consequently, when the RNA tether is broken, misregulation of alternative splicing is observed. These results highlight the importance of coupling between transcription and RNA processing for the regulation of alternative splicing.
FIGURE 4.
Model demonstrating how intronic cotranscriptional cleavage can affect alternative splicing. White ovals are intronic regulatory elements with the DRE being a negative regulatory element essential for repression of exon 2. Gray circles are various RNA processing factors that recognize the nascent RNA transcript. Pol II is depicted as gray area with CTD shown as a hatched oval. (A) When cleavage site is positioned in the middle of the alternatively spliced intron and outside of the regulatory elements, the splicing is unaffected. (B) When RZ cleavage site is engineered to disrupt continuity between exon and regulatory element (but does not affect exon definition threshold), then alternative splicing is perturbed. This suggests that exons and their regulatory elements must be tethered to the elongating RNA Pol II during splicing.
Our results further indicate that alternatively spliced introns are more sensitive to cotranscriptional cleavage than constitutively spliced introns (β-globin). The strong splice signals associated with constitutive introns may also promote more rapid exon tethering to Pol II. In contrast, alternatively spliced introns may require additional intronic regulatory elements for exon tethering that need to be physically linked to their adjacent exons. In the specific case of the fibronectin EDA exon, we show that the activity of the intronic DRE is stimulated by higher SRp20 protein levels. This may suggest that SRp20 associated with the DRE needs to be directly linked to the EDA exon to allow correct splicing regulation. Significantly SRp20 is known to interact with Pol II (de la Mata and Kornblihtt 2006). This is highly consistent with the direct involvement of this factor in tethering the EDA exon and its linked DRE to Pol II.
A number of studies indicate that alterations in transcription elongation rate can affect alternative splicing (de la Mata et al. 2003; Robson-Dixon and Garcia-Blanco 2004). In the EDA system, a reduced transcription elongation rate was shown to favor exon inclusion (de la Mata et al. 2003). This might happen due to a delayed synthesis of the negative DRE identified in this study. Previously, it was demonstrated that insertion of the MAZ4 transcriptional pause element upstream of the α-TM DRE delayed its synthesis by Pol II, and therefore reduced the level of exon 3 skipping. However, the MAZ4 element had no effect when placed downstream of the DRE element in the middle of the intron 3 (Roberts et al. 1998). This is very similar to our results with the α-TM RZ/DRE and DRE/RZ constructs used in this study (Fig. 1D,E). We suggest that delaying the synthesis of the negative regulatory element or disrupting the continuity between this element and the regulated exon affects alternative splicing in a similar manner. This underlines the importance of physical continuity and spatial positioning between the exon and its regulatory sequences at the RNA level for the regulation of alternative splicing.
Our studies using the EDA system demonstrated that hammerhead ribozymes can be used as a tool for defining the splicing regulatory elements in introns. When positioned between an exon and its intronic regulatory elements, a ribozyme can modulate the splicing pattern of the gene. We suggest that, in addition to traditional deletion analysis, ribozymes can be employed to map regulatory elements in alternative splicing systems where introns are long.
Intronic transcript cleavage events can play an important regulatory step in the generation of mature mRNA. In Saccharomyces cerevisiae it has been demonstrated that cotranscriptional cleavage of intronic dsRNA stem–loop structures by RNAse III activity can trigger degradation of unspliced pre-mRNA and lariat introns to regulate the level of mRNA production from such transcripts (Danin-Kreiselman et al. 2003). Recently it has been suggested that miRNAs can be generated cotranscriptionally from intronic sequences (Kim and Kim 2007). The excision of pre-miRNAs from introns involves a specific complex called Microprocessor, containing the endonuclease Drosha and its cofactor DGCR8 (Pasha). We speculate that pre-miRNA processing could occur before splicing of the host intron providing one way of cotranscriptionally cleaving intronic RNA and at the same time coordinating protein and miRNA synthesis from the same transcript. Thus, cotranscriptional excision of regulatory RNAs from introns may have acted as a driving force for the evolution of exon tethering and could also be involved in the regulation of alternative splicing.
MATERIALS AND METHODS
β-globin minigene analysis
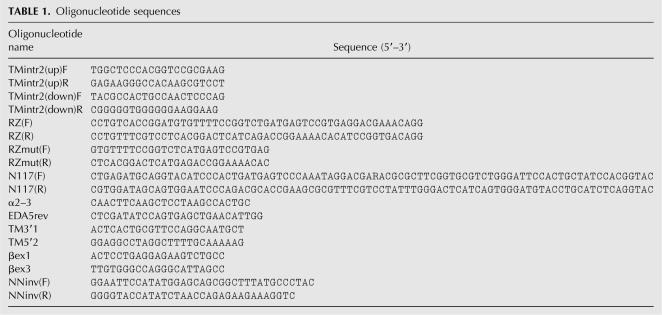
βWT, βIn2-RZ, and βIn2-RZmut minigenes were described previously (Dye et al. 2006). βIn1-RZ was made by insertion of DNA oligonucleotides RZ(F) and RZ(R) containing the 52-nt hammerhead ribozyme sequence (Samarsky et al. 1999) into BbvII site in the intron 1 of βWT. βIn1-RZmut was created by mutation of βIn1-RZ construct using RZmut(F) and RZmut(R) oligonucleotides. βIn1-RZ/In2-RZ (or βIn1-RZmut/In2-RZmut) was created by ligation of the BamHI/BstXI backbone of βIn1-RZ (or βIn1-RZmut) with the BamHI/BstXI fragment from βIn2-RZ (or βIn2-RZmut) construct.
α-TM minigene analysis
6URE α-TM minigene (called WT in these studies) has been described elsewhere (Gromak and Smith 2002). RZ/URE construct was made by amplifying the backbone of WT plasmid with oligonucleotides TMintr2(up)F and TMintr2(up)R and ligating it with the RZ(F) and RZ(R) oligonucleotides. URE/RZ was made by amplifying the backbone of WT plasmid with oligonucleotides TMintr2(down)F and TMintr2(down)R and ligating it with the RZ oligonucleotides (Table 1; Samarsky et al. 1999). RZmut/URE and URE/RZmut constructs were created by mutation of RZ/URE or URE/RZ constructs using RZmut(F) and RZmut(R) oligonucleotides. RZ/DRE was created by inserting RZ into BstEII cut WT. DRE/RZ was made by inserting RZ into EcoRI site downstream of DRE sequence in WT (second EcoRI site upstream of exon 3 BP was mutated). RZmut/DRE and DRE/RZmut were created by mutation of RZ/DRE or DRE/RZ constructs as before. Transient transfections employed 2.5 μL of LipofectAMINE 2000 reagent (Invitrogen) per 35-mm plate with 1 μg of plasmid. Also 0.2 μg of TAT plasmid was cotransfected with β-globin constructs. Cells were harvested after 24 h, and total RNA was isolated using TRIzol reagent (Invitrogen). Reverse transcription reaction was performed with SV3′RT primer (α-TM constructs) or oligo dT primer (β-globin constructs) using SuperScript III reverse transcriptase (Invitrogen) and PCR with radioactively labeled primers TM5′2 and TM3′1 (α-TM) or βex1 and βex3 (β-globin) and quantitated as described previously (Gromak and Smith 2002).
TABLE 1.
Oligonucleotide sequences
pEDA minigene analysis
The pEDA minigene (Muro et al. 1999) contains genomic fibronectin EDA exon, its flanking introns, and part of the −1 and +1 exons embedded in the human α-globin gene under the control of SV40 enhancer. The unique NcoI, NdeI, and BclI sites in fibronectin EDA downstream intron were mutated individually to KpnI sites, and the engineered N117 88-bp long ribozyme from Schistosoma mansoni or its corresponding inactive mutant (N117G, A14 to G substitution) (Yen et al. 2004) was inserted using the following oligonucleotides: N117(F) and N117(R).
The pEDA ΔDRE was created by NcoI/NdeI digestion and ligation of pEDA. InvDRE construct was created by PCR amplification of fibronectin 404-bp NcoI-NdeI fragment with NNinv (F) and NNinv(R) primers and ligated in reverse orientation into NcoI/NdeI-digested pEDA. pBS plasmid sequences of 180 and 300 bp were PCR amplified and cloned in the unique NcoI site to generate pEDAins180 and pEDAins300. RZ and RZmut sequences were inserted into the unique KpnI site of InvDRE to generate InvDRE RZ and InvDRE RZmut minigenes, respectively. The upstream 230 bp and 155 bp of the DRE sequence were amplified with specific primers and inserted in the KpnI/NdeI-digested pEDA to produce ΔC and ΔBC minigenes, respectively.
Hep3B and HeLa cells (5 × 106) were grown under standard conditions and transfected with DOTAP reagent with 1.5 μg of plasmid DNA (Pagani et al. 2003). RT-PCR and quantitation of the amplified products (Pagani et al. 2003) used primers α2–3 and EDA5rev. In cotransfection experiments, 0.75 μg of SRp20 encoding plasmid were cotransfected with 1.5 μg of minigene DNA as reported previously (Pagani et al. 2000).
ACKNOWLEDGMENTS
N.G. and N.J.P. are supported by a Wellcome Programme grant to N.J.P.; G.T. and F.P. are supported by an “Associazione Italiana per la Ricerca sul Cancro” grant. We thank Joan Monks and Cristiana Stuani and Erica Bussani for technical assistance and Mick Dye for critical reading of the paper.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.615508.
REFERENCES
- Berget, S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Buratti, E., Stuani, C., De Prato, G., Baralle, F.E. SR protein-mediated inhibition of CFTR exon 9 inclusion: Molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007;35:4359–4368. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, J.F., Kornblihtt, A.R. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Cramer, P., Caceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E., Kornblihtt, A.R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- Danin-Kreiselman, M., Lee, C.Y., Chanfreau, G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell. 2003;11:1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- Dauksaite, V., Akusjarvi, G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 2002;277:12579–12586. doi: 10.1074/jbc.M107867200. [DOI] [PubMed] [Google Scholar]
- de la Mata, M., Kornblihtt, A.R. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat. Struct. Mol. Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- de la Mata, M., Alonso, C.R., Kadener, S., Fededa, J.P., Blaustein, M., Pelisch, F., Cramer, P., Bentley, D., Kornblihtt, A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Dye, M.J., Gromak, N., Proudfoot, N.J. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Faustino, N.A., Cooper, T.A. Pre-mRNA splicing and human disease. Genes & Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Gallego, M.E., Gattoni, R., Stevenin, J., Marie, J., Expert-Bezancon, A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the β-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding, C., Roberts, G.C., Moreau, G., Nadal-Ginard, B., Smith, C.W. Smooth muscle-specific switching of α-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding, C., Roberts, G.C., Smith, C.W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- Gromak, N., Smith, C.W. A splicing silencer that regulates smooth muscle specific alternative splicing is active in multiple cell types. Nucleic Acids Res. 2002;30:3548–3557. doi: 10.1093/nar/gkf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kim, Y.K., Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt, A.R., de la Mata, M., Fededa, J.P., Munoz, M.J., Nogues, G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Reed, R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Matlin, A.J., Clark, F., Smith, C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Muro, A.F., Caputi, M., Pariyarath, R., Pagani, F., Buratti, E., Baralle, F.E. Regulation of fibronectin EDA exon alternative splicing: Possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani, F., Buratti, E., Stuani, C., Romano, M., Zuccato, E., Niksic, M., Giglio, L., Faraguna, D., Baralle, F.E. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- Pagani, F., Stuani, C., Tzetis, M., Kanavakis, E., Efthymiadou, A., Doudounakis, S., Casals, T., Baralle, F.E. New type of disease causing mutations: The example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- Perez, I., Lin, C.H., McAfee, J.G., Patton, J.G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Robberson, B.L., Cote, G.J., Berget, S.M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, G.C., Gooding, C., Mak, H.Y., Proudfoot, N.J., Smith, C.W. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson-Dixon, N.D., Garcia-Blanco, M.A. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 2004;279:29075–29084. doi: 10.1074/jbc.M312747200. [DOI] [PubMed] [Google Scholar]
- Samarsky, D.A., Ferbeyre, G., Bertrand, E., Singer, R.H., Cedergren, R., Fournier, M.J. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. 1999;96:6609–6614. doi: 10.1073/pnas.96.12.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard, M.J., Chabot, B. SRp30c is a repressor of 3′ splice site utilization. Mol. Cell. Biol. 2002;22:4001–4010. doi: 10.1128/MCB.22.12.4001-4010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, L., Svendsen, J., Lee, J.S., Gray, J.T., Magnier, M., Baba, T., D'Amato, R.J., Mulligan, R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]