Abstract
Allelic exclusion is established in development through a feedback mechanism in which the assembled immunoglobulin (Ig) suppresses further V(D)J rearrangement. But Ig expression sometimes fails to prevent further rearrangement. In autoantibody transgenic mice, reactivity of immature B cells with autoantigen can induce receptor editing, in which allelic exclusion is transiently prevented or reversed through nested light chain gene rearrangement, often resulting in altered B cell receptor specificity. To determine the extent of receptor editing in a normal, non-Ig transgenic immune system, we took advantage of the fact that λ light chain genes usually rearrange after κ genes. This allowed us to analyze κ loci in IgMλ+ cells to determine how frequently in-frame κ genes fail to suppress λ gene rearrangements. To do this, we analyzed recombined VκJκ genes inactivated by subsequent recombining sequence (RS) rearrangement. RS rearrangements delete portions of the κ locus by a V(D)J recombinase-dependent mechanism, suggesting that they play a role in receptor editing. We show that RS recombination is frequently induced by, and inactivates, functionally rearranged κ loci, as nearly half (47%) of the RS-inactivated VκJκ joins were in-frame. These findings suggest that receptor editing occurs at a surprisingly high frequency in normal B cells.
Keywords: receptor editing, recombining sequence recombination, immune tolerance, B lymphocytes, V(D)J rearrangements
The fact that virtually all B cells express a single H and L chain prompted many studies to elucidate the underlying mechanism. One process that clearly contributes to allelic exclusion is the imprecision of V(D)J rearrangement that generates a maximum of one in-frame rearrangement per three attempts (1), but more active feedback processes are also involved. Classic studies showing the ability of a H chain transgene (2, 3) or an L chain transgene (4, and for review see reference 5) to mediate feedback suppression of H and L chain rearrangements, respectively, established important paradigms that have been widely accepted. But in the case of L chain allelic exclusion, this paradigm was weakened by an increasing number of “exceptions”, in which ongoing L chain rearrangement occurred despite expression of functional κ chain (6–9). Studies with autoantibody transgenic (Tg)1 mice suggested that many of the exceptions to the feedback regulation model of L chain allelic exclusion could be explained by postulating self-tolerance– induced receptor editing (10–15). In addition, recent in vitro studies (16–18) and analyses of autoantibody Ig knock-in mice (19, 20) have shown that L chain gene receptor editing can be an important mechanism of B cell tolerance. Despite these findings, it is unclear how frequently receptor editing is used for tolerance induction in normal, non-Ig Tg autoreactive B cells, in part because the extent of autoreactivity in the preselected B cell repertoire is unknown.
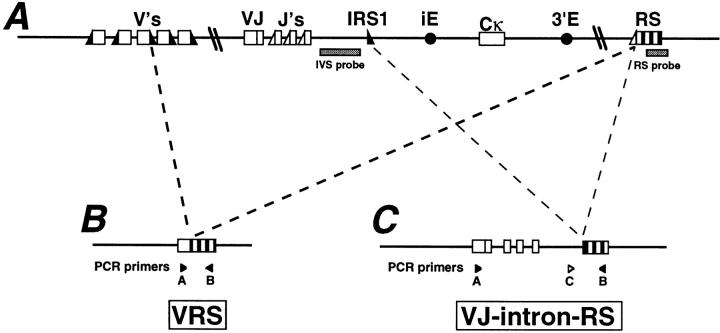
The organization of the κ locus, with arrangement of Vκ genes in both sense and antisense transcriptional orientations, the absence of D region gene segments, and the presence of several Jκ gene segments facilitates sequential, nested Vκ-to-Jκ rearrangement attempts (for review see reference 21). In developing B cells, these secondary rearrangements can both rescue receptor expression in cells that fail to assemble in-frame L chains (1, 22) and rescue autoreactive B cells from tolerance elimination by replacing rearranged κ genes with new ones that alter specificity (for review see reference 23). Another way that the organization of the κ locus promotes receptor editing is suggested by the existence of the conserved element known as recombining sequence (RS) in the mouse (or the homologous “κ deleting element” in humans; reference 24). RS is located ∼25 kb downstream of the Cκ exon (25) and has no coding function (26), but undergoes V(D)J recombinase-dependent rearrangement that inactivates the κ locus by deletional rearrangements in cis (26–28) (see Fig. 1). In an autoantibody knock-in model system, RS rearrangements can inactivate functional κ genes (20), but the extent of RS-mediated receptor editing in normal B cells remains unknown.
Figure 1.
RS rearrangements inactivate and preserve VκJκ joins. A rearranged, potentially functional κ locus (A) can be silenced by two types of RS recombination: Vκ-RS (B) or VκJκ-intron-RS (C). Type C retains the prior VκJκ join, and the RS recombination event eliminates the known cis acting elements that are critical for efficient rearrangement and expression, thus freezing the locus from further VκJκ recombination. Also shown are the intronic recombination sequence 1 (IRS1) (32), the intronic (iE) and 3′ kappa (3′E) enhancers (35, 36), and the recombining sequence (RS) (27, 28) element. Probes IVS (1) and RS 0.8 (27, 28) are indicated by filled boxes.
One approach to estimate the extent of receptor editing in normal B cells is to analyze V(D)J recombinational remnants that are the predicted residue of editing. In mouse B cells, which contain both κ and λ L chain loci, λ gene rearrangement almost always occurs after κ rearrangement (for review see references 29, 30). Thus, if an appropriate κ gene is not assembled, rearrangement at the λ locus often follows. In λ+ B cells, RS rearrangements usually have deleted the Cκ loci (27, 28, 31) either by recombining to Vκs, through the well characterized heptamer–nonamer recombination signal sequences (Fig. 1 B), or to heptamer sites in the Jκ-Cκ intron (Fig. 1 C) (27, 28, 32). Besides destroying the function of the κ locus, this latter mode of RS recombination has two important effects: first, unlike nested VκJκ recombinations, it eliminates the Cκ-associated cis-acting enhancer elements that are critical for VκJκ expression and rearrangement (33–36), and second, it retains any VκJκ join that was previously adjacent to Cκ. This physiological knockout of regulatory sequences required for κ gene rearrangement thus “freezes” the locus, allowing an analysis of the VκJκ gene that was assembled adjacent to the Cκ exon just before RS and λ gene rearrangement.
In this study, we have isolated such VκJκ joins from a large number of individual IgM+λ+ B cells and determined their nucleotide sequences in order to ascertain the extent to which RS inactivates functional κ genes in a normal, non-Ig Tg immune system. The results indicate that in normal IgM+ B cells RS-mediated receptor editing is induced by and frequently inactivates functionally rearranged κ genes, probably because of immune tolerance.
Materials and Methods
Mice.
Mice homozygous for the targeted deletion of the Jκ– Cκ locus (JCκD/JCκD; a gift from D. Huszar, GenPharm International, San Jose, CA; reference 33) were maintained under specific pathogen–free conditions in the animal care facility at National Jewish Medical and Research Center. JCκD/JCκD mice were bred with B10.D2nSn/J mice to generate B10.D2nSn/ J-JCκD/+ mice (JCκD/+), which were used at 6-8 wk of age.
Cell Sorting and Genomic DNA Isolation.
Splenic cells from JCκD/+ mice were isolated and stained with goat anti–mouse IgM-PE (Caltag Labs., San Francisco, CA) and goat anti–mouse λ-FITC (Fisher Scientific Co., Pittsburgh, PA) and sorted on an ELITE flow cytometer (Coulter Corp., Miami, FL) to collect IgM+λ+ B cells. Genomic DNA was isolated from cells by overnight proteinase K digestion in lysis buffer (100 mM NaCl, 10 mM Tris-Cl, pH 8, 25 mM EDTA, 0.5% SDS) at 55°C, followed by phenol/chloroform extraction and EtOH precipitation.
Analysis of Direct PCR Amplified Ig Rearrangements.
Genomic DNA from sorted cells was used as a template to amplify VκJκ-intron-RS rearrangements. As shown in Fig. 1, primers A (degenerate Vκ framework region [FWR]3; reference 37) and B (RS -101, 5′ ACATGGAAGTTTTCCCGGGAGAATATG 3′) amplified a product of ∼1,450 bps (for Jκ5) using an amplification profile of 1 min at 94°C, 1 min at 58°C and 1 min at 72°C for 30–35 cycles. The resulting VκJκ5-intron-RS products were gel isolated and cloned into the TA II vector (Invitrogen, Carlsbad, CA) and colonies were screened by hybridization using the IVS probe (1). PCR clones were sequenced by the dideoxy termination method (Sequenase; United States Biochemical Corp., Cleveland, OH) using vector-specific forward and reverse primers, as well as antisense Jκ5-specific (5′ CTAACATGAAAACCTGTGTCTTACACA 3′) and RS-specific (5′ AAAGCTACATTAGGGCTCAAATCTGA 3′) primers. Both DNA strands from the PCR clones were sequenced over the VκJκ joins to verify the reading frame.
Production of λ+ Hybridomas.
Splenocytes from B10.D2nSn/J-JCκD/+ mice were cultured in DMEM supplemented with 10% fetal bovine sera plus 50 μg/ml LPS (E. coli LPS; Sigma Chemical Co., St. Louis, MO) for 3 d, then fused with NSO-bcl2 (38) myeloma cells for fusion 1 or SP2/0 (39) myeloma cells for fusions 2 through 5. Hybridoma supernatants were screened by ELISA for secretion of micrometer/liter Ig and the lack of κ Ig secretion.
Analysis of Hybridoma Ig Rearrangements.
Hybridoma genomic DNA was digested with EcoRI or BamHI, then fractionated on 0.8% agarose gels, blotted to nylon membrane, and hybridized with the RS 0.8 (27) or IVS probes (Fig. 1 A). Vκ-RS rearrangements were identified by genomic Southern blot analysis using the RS probe and/or by PCR amplification using primer A and primer B to yield a PCR product of ∼255 bps. VκJκ-intron-RS rearrangements were identified by genomic Southern blot analysis using both the RS and IVS probes and/or by PCR amplification using primer C (Jκ intron, 5′ CTGACTGCAGGTAGCGTGGTCTTCTAG 3′) and primer B, and amplified for isolation and sequencing using primers A and B. The resulting products were isolated from 1.8% low-melt agarose gels, cycle sequenced directly (Dye Terminator Cycle Sequencing Ready Reaction kit; PE Applied Biosystems, Norwalk, CT) and analyzed using an ABI 377 DNA Sequencer (PE Applied Biosystems). To obtain near full-length sequences of hybridoma 1-2A7, 1-2E11, 2-2H11, 3-15D6, 3-15C4 and 3-17B10 Vκ genes, a consensus FWR1 oligo (amino acids −1 through 8; 5′ GGTGACATTGTGCTGACCCAGTCTCCA 3′) was used with antisense Jκ-intron oligos for PCR amplification, followed by cycle sequencing of the products. For hybrids 1-3E8, 2-15E11, 3-7G5, 4-1D2, 1-10A11 and 1-11A4, Vκ leader specific oligos (Ig Prime kit; Novagen, Madison, WI) were used for amplification.
Cloning and Expression of V(D)J Rearrangements for Analysis of H/κL Chain Pairing.
The H chain V(D)J and the L chain VκJκ-RS rearrangements from hybridoma 2H11 were genomically cloned as previously described (40), with the modifications that λZap (Stratagene, La Jolla, CA) was the cloning vector and the RS and IVS probes were used to screen clones for the VκJκ-RS rearrangement. The 6.5-kb EcoRI fragment containing the V(D)J rearrangement and the 4.0-kb EcoRI–XbaI fragment containing the VκJκ rearrangement were gel isolated and ligated to pRμSal, a Cμ expression vector (41) and pSV2-neo-Cκ, a Cκ expression vector (42) respectively. The H chain from hybridoma 15E11 was cloned by PCR amplification using a leader intron oligo (5′ GAACTGGCAGGACCTGAGGTGAAAATGACA 3′) and an oligo that spanned the XbaI site downstream of JH4 (5′ CAGGCTCCACCAGACCTCTCTAGA 3′). The resulting product was digested with EcoO109I and XbaI and ligated into EcoO109I and XbaI digested 8-1Cμ (40), a Cμ expression vector. The VκJκ-RS rearrangement from hybridoma 15E11 was also cloned by PCR using a leader intron oligo (5′ TGGAATTCCAGGTTCTACTGGAGACATTGT-3′) and an oligo which spanned the XbaI site downstream of Jκ5 (5′ ACGAATTCGTCTAGAAGACCACGCTACCT 3′). The resulting product was digested with EcoRI and subcloned into a shuttle vector containing Vκ21C leader and promoter elements. An XbaI fragment that contained the promoter elements, leader, and the 15E11 VκJκ rearrangement was isolated and cloned into the XbaI site in pSV2-neo-Cκ (42). The 2H11 and 15E11 H and L chain constructs were then cotransfected into SP2/0 myeloma cells and selected for expression of IgMκ as previously described (40).
IgMκ ELISA for Analysis of H/κL Pairing.
Supernatants from 2H11 and 15E11 H and L chain transfectoma clones were assayed for IgMκ expression by ELISA. In brief, goat anti–mouse IgM (Southern Biotechnology Associates, Inc., Birmingham, AL) was diluted in PBS, coated onto 96-well Immulon 2HB plates (Dynex Technologies, Inc., Chantilly, VA) and incubated at room temperature for 3 h. Plates were washed five times with PBS/Tween 20, then incubated with blocking buffer (PBS, 0.5% BSA, 0.4% Tween 20) for 1 h at room temperature. Serial dilution of transfectoma and parental hybridoma supernatants were added and incubated at room temperature for 2 h. Plates were washed and horseradish peroxidase–conjugated goat anti–mouse κ (Southern Biotechnology Associates, Inc.) was added and incubated for 2 h. After a final wash, the chromogenic substrate 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Sigma Chemical Co.) was added in McIlvain's buffer (84 mM Na2PO4/48 mM citrate, pH 4.6) with 0.005% H2O2 and OD 410 nm was read using an automated plate reader (Dynatech, Alexandria, VA). Transfectoma and hybridoma antibody concentrations were estimated by comparison to a TEPC 183 (μκ) standard curve.
Results
Strategy for the Isolation of Editing Remnants.
To determine the extent to which RS recombination inactivates functional, in-frame VκJκ joins in the preimmune B cell repertoire, IgM+λ+ splenic B cells were isolated by fluorescence activated cell sorting and their genomic DNA was analyzed by the PCR strategy outlined in Fig. 1 C. This cell sorting strategy should exclude from the template pool cells that are κ+, H chain isotype switched, surface (s)Ig−, or cells of a sIglo, germinal center phenotype. In a second series of experiments, IgMλ secreting hybridomas were isolated and their κ loci analyzed in detail. To simplify these analyses, all B cells analyzed were heterozygous for a targeted deletion of the Jκ–Cκ locus (JCκD/+), in which only a single κ locus and RS allele could rearrange (33). The potential κ gene and RS element rearrangements are depicted in Fig. 1.
Analysis of IgMλ Cells Reveals Frequent Receptor Editing.
Genomic DNA from sorted IgMλ cells was used as template for a PCR using a panspecific Vκ FWR 3 oligonucleotide primer, which recognizes ∼80% of Vκ genes (37), together with an RS-specific primer to amplify VκJκ-intron-RS rearrangements (Fig. 1 C, primers A and B). VκJκ-intron-RS rearrangements containing Vκ genes rearranged to each of the four functional Jκ genes were detected by PCR amplification and Southern blotting (data not shown), but VκJκ5-intron-RS rearrangements were most abundant, in part because their smaller size promoted preferential amplification. Amplified VκJκ5-intron-RS rearrangements were gel-purified and cloned, and a total of 52 clones were sequenced across both the VκJκ and the Jκ-intron-RS joins (Fig. 2). These two different recombination joins, present on each PCR product analyzed, provided markers for uniqueness. PCR products that were identical to one another, or that differed by just one nucleotide, were assumed to represent repeated isolates derived from the same initial template (i.e., derived from a single B cell clone). This represents an underestimate because the single base changes could have reflected real differences and because it was possible that some of the apparent repeats were independent events that happened to have identity in the portions of the genes studied, but not in upstream portions of the V genes. In this sample, at least 37 of the 52 clones represented independent events. Analysis of the VκJκ join sequences allowed an assessment of the potential prior functionality of the VκJκ5 joins just upstream of intron-RS rearrangements. Surprisingly, 15 of the 37 clones (41%) contained VκJκ joins that were in-frame (Fig. 2), and if the apparent repeats were not excluded 23 out of 52 (44%) were in-frame.
Figure 2.
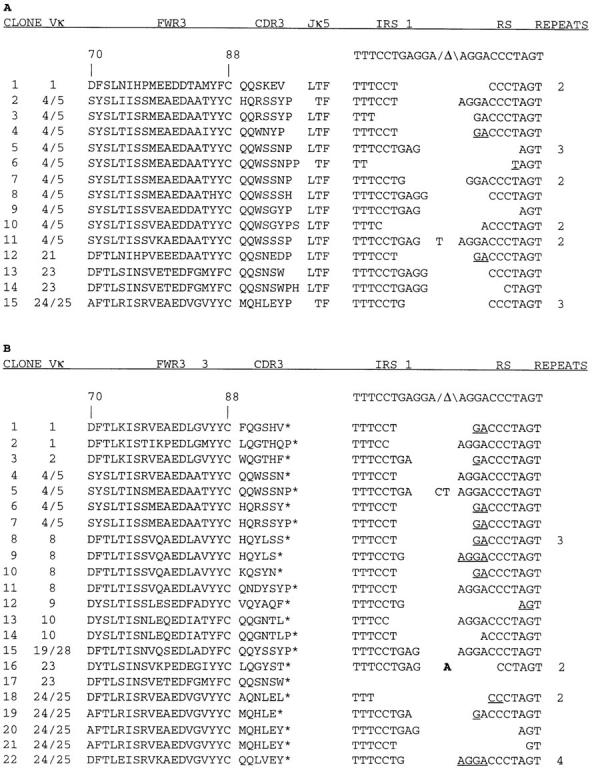
Sequence analysis of (A) productive and (B) nonproductive VκJκ-intron-RS rearrangements from FACS® sorted, IgM+λ+ splenic B cells. Vκ gene family and Jκ gene usage were assigned based on homologies to expressed VκJκ genes (52) or homology searches of Genbank and the Kabat Ig database. Translated Vκ FWR3 (codons 70–88), CDR3, and Jκ5 sequences to the conserved phenylalanine (F) are shown for productive rearrangements, whereas FWR3 and CDR3 sequences are shown for nonproductive rearrangements, with the asterisk (*) adjacent to CDR3 in B denoting an out of frame VκJκ join. The nucleotide sequences of the unrearranged Jκ intronic recombining sequence 1 (IRS1) and RS element (both of which contain a consensus heptamer sequence adjacent to the Δ symbol) are shown above the sequences of the IRS1-RS joins present in each PCR clone. The RS join sequence for clone 17 was not determined. Underlined nucleotides could have been donated by either the IRS1 or the RS sequence, and N region addition (bold) and P-encoded nucleotides are shown between the joins. Repeats denote the number of times a particular sequence was observed.
To verify the analysis of the PCR-amplified VκJκ-intron-RS rearrangements and to increase the sample size, an independent sampling of VκJκ-intron-RS rearrangements was derived from JCκD/+ splenocytes in the form of B cell hybridomas. A total of 133 IgMλ-expressing hybrids were obtained from five separate fusions and their κ locus rearrangements were analyzed. Genomic Southern blot and PCR analysis revealed that at least 74% of the λ+ hybrids (99 out of 133) had inactivated the wild-type κ locus by RS rearrangements (Table 1), a value in accord with previous estimates (31, 34). Two hybridomas apparently had undergone inversional Vκ-RS rearrangements, as they had unique restriction fragments that retained the Cκ locus as revealed by the intron (IVS) probe (data not shown), but scored positive in a Vκ-RS PCR (Fig. 1 B). Approximately 25% (26 out of 99) of the hybridomas with RS rearrangements had Jκ-intron-RS joins (Table 1), as detected with primers B and C (Fig. 1 C). Genomic Southern blot analysis of 18 out of 20 hybrids scoring PCR positive for Jκ- intron-RS rearrangements demonstrated that the RS rearrangements colocalized with EcoRI restriction fragments hybridizing with the IVS probe (data not shown), thus independently confirming the VκJκ-intron-RS rearrangement phenotype. VκJκ-intron-RS rearrangements from individual hybridomas were PCR amplified and directly sequenced, rather than cloned, a procedure that diminishes potential Taq polymerase-generated mutations. Like the VκJκ-intron-RS PCR clones, most of the VκJκ-intron-RS loci from hybridomas used Jκ5, although four hybridomas had rearrangements to upstream Jκs, including one to Jκ2 and three to Jκ4, suggesting that developing B cells do not frequently undergo RS rearrangement until all of the Jκs are rearranged. Sequence analysis over both the VκJκ and intron-RS joins clearly showed that each cell line had a unique sequence at the VκJκ join and that, remarkably, 12 of 20 (60%) of the VκJκ joins were in-frame (Fig. 3 A).
Table 1.
κ Locus Rearrangement Status of IgMλ Hybridomas
| κ locus genotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hybridomas | RS− | Vκ-RS | VκJκ-RS | Productive/ nonproductive | ||||||
| n | ||||||||||
| Fusion 1 | 37 | 12 | 16 | 9/9* | 5/4 | |||||
| Fusion 2 | 44 | 10 | 30 | 4/4 | 2/2 | |||||
| Fusion 3 | 12 | 1 | 6 | 5/4 | 4/0 | |||||
| Fusion 4 | 25 | 9 | 11 | 5/2 | 1/1 | |||||
| Fusion 5 | 15 | 2 | 10 | 3/1 | 0/1 | |||||
| Totals | 133 | 34 | 73 | 26/20 | 12/8 | |||||
RS− denotes hybridomas that lacked a detectable RS rearrangement. Vκ-RS and VκJκ-RS are defined in Fig. 1. Asterisk indicates total number of VκJκ-intron-RS loci that were isolated followed by the number that we were able to PCR amplify with the consensus FWR3 oligo.
Figure 3.
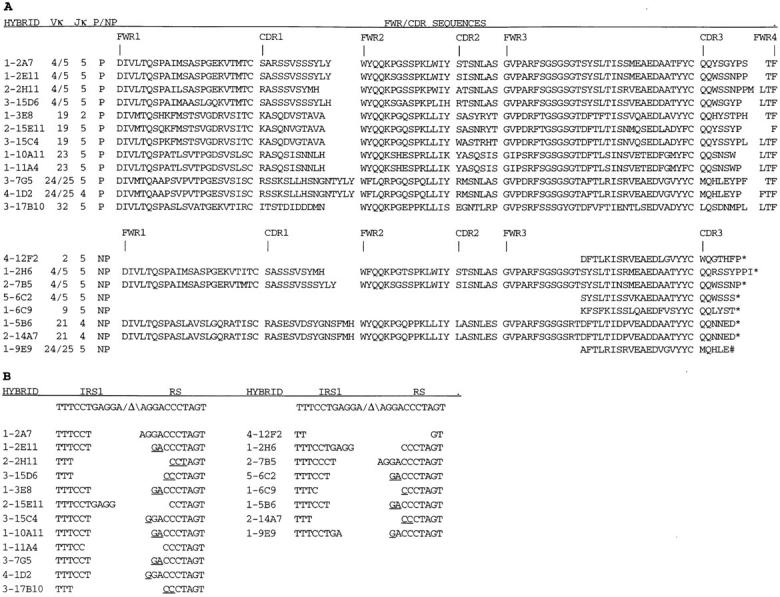
Sequence analysis of VκJκ- intron-RS rearrangements from IgMλ hybridomas. (A) Sequences of the VκJκ rearrangements. The first digit in the hybridoma name indicates the fusion experiment number. Myeloma fusion partners were either NSO-bcl2 (fusion 1) or SP2/0 (fusions 2–5). Vκ gene family and Jκ gene usage were assigned as described in Fig. 2. P and NP denote productive and nonproductive VκJκ rearrangements, respectively. Translated amino acid sequences of Vκ FWR, CDR, and Jκ sequences to the conserved phenylalanine (F) residue are shown for productive rearrangements, and Vκ FWR and CDR sequences, with * denoting an out of frame VκJκ join and # denoting an in-frame stop codon, are shown for nonproductive rearrangements. These sequence data are available from EMBL/Genbank/DDBJ under accession numbers AF087023–AF087034 and AF087460–AF087467. (B) Sequences of the RS rearrangements (as described in Fig. 2).
Diversity of V Gene Usage.
VκJκ-intron-RS rearrangements were clearly diverse because at least 32 different Vκ genes representing 11 of the 19 Vκ families were identified among the 57 independent VκJκ-intron-RS loci analyzed (data not shown). This value of Vκ gene representation in the VκJκ-intron-RS loci analyzed is most likely an underestimate of the diversity because the 5′ PCR primer lies in FWR3 and yields only a short stretch of Vκ gene sequence for interclonal comparison. Despite this limitation, multiple genes were observed within particular Vκ gene families. For example, within the Vκ4/5 family, at least 11 different genes were represented among the 14 in-frame and 7 out-of-frame joins (data not shown). In addition, Vκ genes were sometimes found repeatedly in independent VκJκ-intron-RS rearrangements and there was considerable overlap in usage among hybridoma and PCR clone sequences. 13 of the hybridomas used Vκ genes that were observed in the direct PCR–derived clones, whereas 6 hybridomas expressed distinct Vκ genes that were members of families observed in the PCR clone sample and 1 hybridoma expressed a Vκ32 gene, a Vκ family not seen in the PCR clone samples (Figs. 2 and 3 and data not shown).
Intron/RS Joins.
The sequences of the Jκ-intron-RS joins in both the PCR clones (Fig. 2) and hybridomas (Fig. 3 B) were quite varied and were dominated by deletions at both sides of the joins, as up to nine nucleotides were missing from either the Jκ-intron or RS heptamer-flanking sequences. There did appear to be a bias for a particular join (e.g., clone 4, Fig. 2 A), which was observed to be associated with 13 independent VκJκ rearrangements. Two of the intron-RS joins contained P nucleotides and one contained N-region addition nucleotides, consistent with findings described previously (7, 43).
Rebuilding IgMκ Antibodies for Analysis of H/κL Pairing and Antigen Specificity.
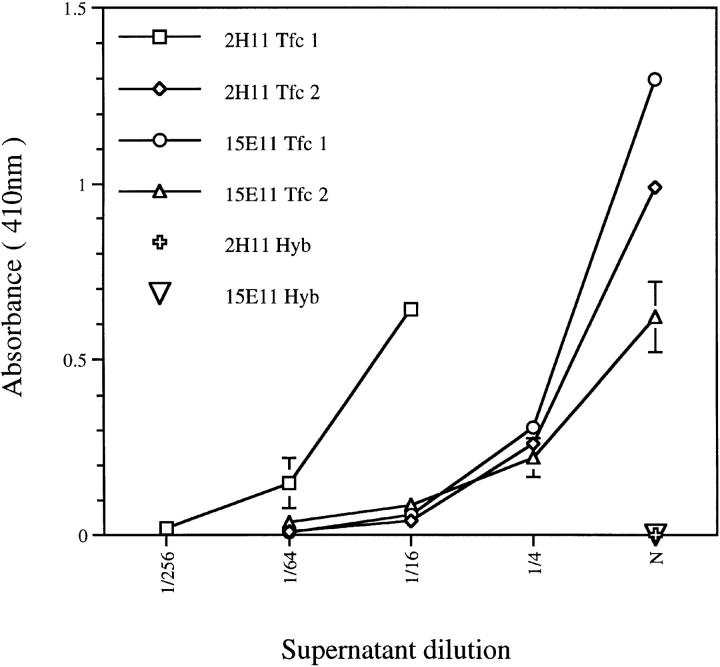
To determine if the high frequency of in-frame VκJκ rearrangements silenced by intron-RS recombination was due to the inability of H chains to pair with their κ L chains, the V(D)J and VκJκ rearrangements from hybridomas 2H11 and 15E11 were cloned into Cμ and Cκ expression vectors, respectively. These H and L chain constructs were cotransfected into SP2/0 myeloma cells to generate transfectoma clones. Analysis of transfectoma supernatants by IgMκ sandwich ELISA revealed that the in-frame κL chains were able to pair with their hybridoma H chains (Fig. 4), suggesting that ongoing RS rearrangement was not due to the inability of H/L chain pairing. The specificity of the μκ transfectoma antibodies remains unknown, however. Attempts in flow cytometry assays to detect recombinant antibody binding to the surfaces of bone marrow cells were unsuccessful (data not shown).
Figure 4.
“Repair” of intron-RS recombination-silenced VκJκ genes by restoration of Cκ exon and surrounding elements reveals that silenced κL chains can pair with their original μ chain partner. The graph shows representative results from a μκ ELISA comparing several IgMκ transfectoma antibodies (Tfc) to their IgMλ parental hybridoma antibodies (Hyb). Antibodies in supernatants were captured on plastic using adsorbed anti-μ chain antibodies and revealed with anti-κ conjugates. Bars indicate the SD determined from antibodies assayed in triplicate. The concentrations of the hybridoma antibodies were at least 10-fold higher than those of the transfectoma antibodies based on comparison to a TEPC 183 (μ, κ) standard curve.
Discussion
In this report we examined the DNA sequences of VκJκ joins located upstream of intronic-RS rearrangements in normal, non-Ig Tg B cells to determine the extent to which RS-mediated recombination silences functionally rearranged κ genes. Nearly half of all VκJκ joins inactivated by RS recombination were in-frame (27 out of 57). This high frequency is clearly incompatible with a strict feedback suppression model of L chain allelic exclusion, which predicts no in-frame VκJκ joins upstream of the RS rearrangements. More strikingly, this high frequency is also significantly higher than 33%, the percentage of in-frame joins expected from random VκJκ rearrangement, indicating that productive VκJκ rearrangements actively induce intron-RS rearrangements. The data also demonstrate a physiological role for the RS element in normal B cell development—the inactivation of functionally rearranged κ genes.
To understand why we conclude that the RS rearrangements were actively induced by functional κ L chains, consider the extreme hypothetical cases of mice in which all κ gene rearrangements result in either autoreactive B cell receptors or nonproductive κ chains (Table 2). If Vκ-to-Jκ and RS rearrangements proceed randomly, albeit with different relative frequencies, then in either case VκJκ joins located upstream of intronic-RS rearrangements should be in-frame at a maximum frequency of one out of three. To significantly exceed this frequency, in-frame VκJκ joins must stimulate the relative rate of (intronic) RS rearrangement. This argument applies to our data because the observed frequency of in-frame joins, 47.4%, is significantly higher than one out of three (P < 0.04, single sample test of a proportion based on a normal approximation). Since it is exceedingly unlikely that the stimulus for increased in-frame rearrangements is mediated by anything other than κ protein, and because κ chains can probably only be perceived by the signaling machinery of B cells through their association with H chains, we conclude that functional κ chains actively stimulate the rate of RS rearrangement based on B cell receptor antigenic specificity. These data also predict that in mice in which the Cκ exon is inactivated, but surrounding cis-acting elements are left intact, VκJκ rearrangement should be extensive, whereas RS rearrangement should be reduced. This is in fact the experimental observation (44).
Table 2.
Analysis of VκJκ Joins in VκJκ-intron-RS Sequences: Models and Predictions Compared with Experimental Data
| Models | Predicted fraction in-frame | Experimental data | Observed percentage in frame | |||
|---|---|---|---|---|---|---|
| Perfect feedback regulation with functional κ chain preventing λ rearrangement | 0% | 27 out of 57 VκJκ-intron-RS loci | 47.4% (P < 0.04) | |||
| Poor feedback regulation | ≤33% | |||||
| High frequency of Vκ pseudogenes | ≤33% | |||||
| High frequency of H/κ chain mispairing | ≤33% | |||||
| Any combination of the above | ≤33% | |||||
| Extreme model of editing with random RS rearrangements and all κs autoreactive | 33% |
The statistical argument also excludes the possibility that a high frequency of rearrangeable Vκ pseudogenes, L chains that fail to pair with H chains, or a role for positive selection is responsible for our results. Furthermore, complete sequencing of the coding regions from all the in-frame VκJκ rearrangements derived from λ+ hybridomas revealed no stop codons or other obvious defects that would have precluded function (Fig. 3 A). It is also unlikely that frequent aberrant H/L chain pairing is responsible for the high frequency of in-frame VκJκ joins in the VκJκ-intron-RS rearrangements, as demonstrated by the ability of H/κL chains from two hybridomas to pair (Fig. 4). Moreover, there are few examples of L chains that fail to pair with H chains and most experiments suggest that virtually all random H/L pairs can associate (40, 45–48). Finally, if a lack of positive selection of surface Ig was responsible for the high frequency of in-frame joins, this would predict that B cells should frequently express two κ chains, a result that has not been observed.
The receptor editing events documented in this study probably do not represent renewed V(D)J recombination in mature B cells, such as has recently been described in the germinal center (49–51), because the cells analyzed expressed high levels of IgM and λ chain and because they were isolated and, in the case of the hybridomas, stimulated in a manner that should not have induced V(D)J recombination. Another indication that receptor editing in mature B cells is unlikely to explain our results is that the fraction of λ+ cells in newly formed and mature splenic B cells is nearly identical, suggesting that in unmanipulated mice mature κ+ cells rarely give rise to λ+ B cells (44). Overall, it would appear from our data that the RS rearrangements that we studied were actually stimulated, rather than inhibited, by productive κ gene rearrangements, probably as the result of immune tolerance-mediated receptor editing in immature B cells. To definitively test the prediction that the κ chains of the cells that we have analyzed generate autoantibodies in association with the same cell's heavy chain, it will be necessary to generate mice transgenic for these genes.
Acknowledgments
We thank D. Huszar for providing JCκD/JCκD mice; B. Diamond (Albert Einstein College of Medicine, Bronx, NY) for providing the NSO-bcl2 myeloma; S. Sobus for cell sorting; D. Norsworthy for cycle sequence analyses; D. Iklé of the Biostatistics Department for statistical analyses; K. Karjalainen and L. Wysocki for discussions; and M. Hertz, D. Melamed, V. Kouskoff, and other members of the lab for critical reading of the manuscript.
This work was supported by grants from the National Institutes of Health and the Arthritis Foundation.
Abbreviations used in this paper
- FWR
framework region
- JCκD/+
heterozygous κ deficient germline genotype
- RS
recombining sequence
- Tg
transgenic
References
- 1.Coleclough C, Perry RP, Karjalainen K, Weigert M. Aberrant rearrangements contribute significantly to the allelic exclusion of immunoglobulin gene expression. Nature. 1981;290:372–378. doi: 10.1038/290372a0. [DOI] [PubMed] [Google Scholar]
- 2.Nussenzweig MC, Shaw AC, Sinn E, Danner DB, Holmes KL, Morse HC, III, Leder P. Allelic exclusion in transgenic mice that express the membrane form of immunoglobulin μ. Science. 1987;236:816–819. doi: 10.1126/science.3107126. [DOI] [PubMed] [Google Scholar]
- 3.Manz J, Denis K, Witte O, Brinster R, Storb U. Feedback inhibition of immunoglobulin gene rearrangement by membrane μ but not by secreted μ heavy chains. J Exp Med. 1988;168:1363–1381. doi: 10.1084/jem.168.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritchie KA, Brinster RL, Storb U. Allelic exclusion and control of endogenous immunoglobulin gene rearrangement in κ transgenic mice. Nature. 1984;312:517–520. doi: 10.1038/312517a0. [DOI] [PubMed] [Google Scholar]
- 5.Storb, U. 1995. Ig gene expression and regulation in Ig transgenic mice. In Immunoglobulin Genes. Second Edition. T. Honjo and F.W. Alt, editors. Academic Press, San Diego, CA. 345–363.
- 6.Gollahon KA, Hagman J, Brinster RL, Storb U. Ig λ–producing B cells do not show feedback inhibition of gene rearrangement. J Immunol. 1988;141:2771–2780. [PubMed] [Google Scholar]
- 7.Harada K, Yamagishi H. Lack of feedback inhibition of Vκ gene rearrangement by productively rearranged alleles. J Exp Med. 1991;173:409–415. doi: 10.1084/jem.173.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doglio L, Kim JY, Bozek G, Storb U. Expression of λ and κ genes can occur in all B cells and is initiated around the same pre-B-cell developmental stage. Dev Immunol. 1994;4:13–26. doi: 10.1155/1994/87352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghia P, Gratwohl A, Signer E, Winkler TH, Melchers F, Rolink AG. Immature B cells from human and mouse bone marrow can change their surface light chain expression. Eur J Immunol. 1995;25:3108–3114. doi: 10.1002/eji.1830251118. [DOI] [PubMed] [Google Scholar]
- 10.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radic MZ, Erikson J, Litwin S, Weigert M. B lymphocytes may escape tolerance by revising their antigen receptors. J Exp Med. 1993;177:1165–1173. doi: 10.1084/jem.177.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Radic MZ, Erikson J, Camper SA, Litwin S, Hardy RR, Weigert M. Deletion and editing of B cells that express antibodies to DNA. J Immunol. 1994;152:1970–1982. [PubMed] [Google Scholar]
- 14.Prak EL, Trounstine M, Huszar D, Weigert M. Light chain editing in κ-deficient animals: a potential mechanism of B cell tolerance. J Exp Med. 1994;180:1805–1815. doi: 10.1084/jem.180.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang J, Jackson M, Teyton L, Brunmark A, Kane K, Nemazee D. B cells are exquisitely sensitive to central tolerance and receptor editing induced by ultralow affinity, membrane-bound antigen. J Exp Med. 1996;184:1685–1697. doi: 10.1084/jem.184.5.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertz M, Nemazee D. BCR ligation induces receptor editing in IgM+IgD− bone marrow B cells in vitro. Immunity. 1997;6:429–436. doi: 10.1016/s1074-7613(00)80286-1. [DOI] [PubMed] [Google Scholar]
- 17.Melamed D, Nemazee D. Self-antigen does not accelerate immature B cell apoptosis, but stimulates receptor editing as a consequence of developmental arrest. Proc Natl Acad Sci USA. 1997;94:9267–9272. doi: 10.1073/pnas.94.17.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melamed D, Benschop RJ, Cambier JC, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor editing. Cell. 1998;92:173–182. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Prak EL, Weigert M. Editing disease-associated autoantibodies. Immunity. 1997;6:97–105. doi: 10.1016/s1074-7613(00)80673-1. [DOI] [PubMed] [Google Scholar]
- 20.Pelanda R, Schwers S, Sonoda E, Torres RM, Nemazee D, Rajewsky K. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- 21.Nemazee, D. 1995. B lymphocyte tolerance in the mouse. In Immunoglobulin Genes. Second Edition. T. Honjo and F.W. Alt, editors. Academic Press, Inc., San Diego, CA. 365–378.
- 22.Feddersen RM, Van Ness BG. Corrective recombination of mouse immunoglobulin kappa alleles in Abelson murine leukemia virus-transformed pre-B cells. Mol Cell Biol. 1990;10:569–576. doi: 10.1128/mcb.10.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertz M, Nemazee D. Receptor editing and commitment in B lymphocytes. Curr Opin Immunol. 1998;10:208–213. doi: 10.1016/s0952-7915(98)80250-1. [DOI] [PubMed] [Google Scholar]
- 24.Siminovitch KA, Bakhshi A, Goldman P, Korsmeyer SJ. A uniform deleting element mediates the loss of κ genes in human B cells. Nature. 1985;316:260–262. doi: 10.1038/316260a0. [DOI] [PubMed] [Google Scholar]
- 25.Muller B, Stappert H, Reth M. A physical map and analysis of the murine Cκ-RS region show the presence of a conserved element. Eur J Immunol. 1990;20:1409–1411. doi: 10.1002/eji.1830200631. [DOI] [PubMed] [Google Scholar]
- 26.Daitch LE, Moore MW, Persiani DM, Durdik JM, Selsing E. Transcription and recombination of the murine RS element. J Immunol. 1992;149:832–840. [PubMed] [Google Scholar]
- 27.Durdik J, Moore MW, Selsing E. Novel κ light-chain gene rearrangements in mouse λ light chain-producing B lymphocytes. Nature. 1984;307:749–752. doi: 10.1038/307749a0. [DOI] [PubMed] [Google Scholar]
- 28.Moore MW, Durdik J, Persiani DM, Selsing E. Deletions of κ chain constant region genes in mouse λ chain–producing B cells involve intrachromosomal DNA recombinations similar to V-J joining. Proc Natl Acad Sci USA. 1985;82:6211–6215. doi: 10.1073/pnas.82.18.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selsing, E., and L.E. Daitch. 1995. Immunoglobulin λ genes. In Immunoglobulin Genes. Second Edition. T. Honjo and F.W. Alt, editors. Academic Press, San Diego, CA. 193–203.
- 30.Arakawa H, Shimizu T, Takeda S. Re-evaluation of the probabilities for productive arrangements on the κ and λ loci. Int Immunol. 1996;8:91–99. doi: 10.1093/intimm/8.1.91. [DOI] [PubMed] [Google Scholar]
- 31.Nadel B, Cazenave PA, Sanchez P. Murine lambda gene rearrangements: the stochastic model prevails over the ordered model. EMBO (Eur Mol Biol Org) J. 1990;9:435–440. doi: 10.1002/j.1460-2075.1990.tb08128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu T, Iwasato T, Yamagishi H. Deletions of immunoglobulin Cκ region characterized by the circular excision products in mouse splenocytes. J Exp Med. 1991;173:1065–1072. doi: 10.1084/jem.173.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Trounstine M, Kurahara C, Young F, Kuo CC, Xu Y, Loring JF, Alt FW, Huszar D. B cell development in mice that lack one or both immunoglobulin κ light chain genes. EMBO (Eur Mol Biol Organ) J. 1993;12:821–830. doi: 10.1002/j.1460-2075.1993.tb05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda S, Zou YR, Bluethmann H, Kitamura D, Muller U, Rajewsky K. Deletion of the immunoglobulin κ chain intron enhancer abolishes κ chain gene rearrangement in cis but not λ chain gene rearrangement in trans. EMBO (Eur Mol Biol Organ) J. 1993;12:2329–2336. doi: 10.1002/j.1460-2075.1993.tb05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Ig κ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 36.Gorman JR, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt FW. The Igκ 3′ enhancer influences the ratio of Igκ versus Igλ B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 37.Schlissel MS, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 38.Ray S, Diamond B. Generation of a fusion partner to sample the repertoire of splenic B cells destined for apoptosis. Proc Natl Acad Sci USA. 1994;91:5548–5551. doi: 10.1073/pnas.91.12.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearney JF, Radbruch A, Liesegang B, Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- 40.Retter MW, Eisenberg RA, Cohen PL, Clarke SH. Sm and DNA binding by dual reactive B cells requires distinct VH, Vκ, and VHCDR3 structures. J Immunol. 1995;155:2248–2257. [PubMed] [Google Scholar]
- 41.Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 42.Retter MW, Cohen PL, Eisenberg RA, Clarke SH. Both Sm and DNA are selecting antigens in the anti-Sm B cell response in autoimmune MRL/lprmice. J Immunol. 1996;156:1296–1306. [PubMed] [Google Scholar]
- 43.Graninger WB, Goldman PL, Morton CC, O'Brien SJ, Korsmeyer SJ. The κ-deleting element. Germline and rearranged, duplicated and dispersed forms. J Exp Med. 1988;167:488–501. doi: 10.1084/jem.167.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou YR, Takeda S, Rajewsky K. Gene targeting in the Igκ locus: efficient generation of λ chain-expressing B cells, independent of gene rearrangements in Igκ. EMBO (Eur Mol Biol Organ) J. 1993;12:811–820. doi: 10.1002/j.1460-2075.1993.tb05721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamel PA, Klein MH, Smith-Gill SJ, Dorrington KJ. Relative noncovalent association constant between immunoglobulin H and L chains is unrelated to their expression or antigen-binding activity. J Immunol. 1987;139:3012–3020. [PubMed] [Google Scholar]
- 46.Kaushik A, Schulze DH, Bonilla FA, Bona C, Kelsoe G. Stochastic pairing of heavy-chain and κ light-chain variable gene families occurs in polyclonally activated B cells. Proc Natl Acad Sci USA. 1990;87:4932–4936. doi: 10.1073/pnas.87.13.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Lau WB, Heije K, Neefjes JJ, Oosterwegel M, Rozemuller E, Bast BJ. Absence of preferential homologous H/L chain association in hybrid hybridomas. J Immunol. 1991;146:906–914. [PubMed] [Google Scholar]
- 48.Radic MZ, Mascelli MA, Erikson J, Shan H, Weigert M. Ig H and L chain contributions to autoimmune specificities. J Immunol. 1991;146:176–182. [PubMed] [Google Scholar]
- 49.Papavasiliou F, Casellas R, Suh H, Qin XF, Besmer E, Pelanda R, Nemazee D, Rajewsky K, Nussenzweig MC. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science. 1997;278:298–301. doi: 10.1126/science.278.5336.298. [DOI] [PubMed] [Google Scholar]
- 50.Han S, Dillon SR, Zheng B, Shimoda M, Schlissel MS, Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–305. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- 51.Hikida M, Ohmori H. Rearrangement of λ-light chain genes in mature B cells in vitro and in vivo. Function of reexpressed RAG gene products. J Exp Med. 1998;187:795–799. doi: 10.1084/jem.187.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strohal R, Helmberg A, Kroemer G, Kofler R. Mouse Vκ gene classification by nucleic acid sequence similarity. Immunogenetics. 1989;30:475–493. doi: 10.1007/BF02421180. [DOI] [PMC free article] [PubMed] [Google Scholar]