Abstract
The movement of leukocytes into tissues is regulated by the local production of chemical mediators collectively referred to as chemoattractants. Although chemoattractants constitute a diverse array of molecules, including proteins, peptides, and lipids, they all appear to signal leukocytes through a related family of seven transmembrane–spanning G protein–coupled receptors. The eosinophil is a potent proinflammatory cell that is attracted into tissues during allergic inflammation, parasitic infection, and certain malignancies. Since the molecular mechanisms controlling eosinophil recruitment are incompletely understood, we performed a degenerate polymerase chain reaction on cDNA isolated from murine eosinophils to identify novel chemoattractant receptors. We report the isolation of a cDNA that encodes a 351–amino acid glycoprotein that is 78% identical to a human gene that has been reported to be a purinoceptor (P2Y7) and a leukotriene B4 (LTB4) receptor (BLTR). Chinese hamster ovary (CHO) cells transfected with this cDNA specifically bound [3H]LTB4 with a dissociation constant of 0.6 ± 0.1 nM. Furthermore, LTB4 induced a dose-dependent intracellular calcium flux in transfected CHO cells. In contrast, [35S]dATP did not specifically bind to these transfectants. This mRNA was expressed at high levels in interleukin 5–exposed eosinophils, elicited peritoneal macrophages and neutrophils, and to a lesser extent interferon γ stimulated macrophages. Low levels of expression were detected in the lung, lymph node, and spleen of unchallenged mice. Western blot analysis detected the mBLTR protein in murine eosinophils and alveolar macrophages as well as human eosinophils. In addition, elevated levels of mBLTR mRNA were found in the lungs of mice in a murine model of allergic pulmonary inflammation in a time course consistent with the influx of eosinophils. Our findings indicate that this murine receptor is an LTB4 receptor that is highly expressed on activated leukocytes, including eosinophils, and may play an important role in mediating eosinophil recruitment into inflammatory foci.
Keywords: chemotactic factor, eosinophil, leukotriene B4, asthma, G protein–coupled receptor
The attraction of leukocytes from the blood into the tissue is a critical component of the host inflammatory response. The eosinophil is a blood-borne leukocyte that is thought to play an important role in host defense against helminthic parasites by exocytosing its cytotoxic granular constituents, such as eosinophil major basic protein and eosinophil peroxidase, in the vicinity of the pathogen (1). However, these cytotoxic proteins are also harmful to host tissue and contribute to the tissue destruction seen in a variety of diseases where eosinophils accumulate, such as asthma and other allergic diseases (2). The regulation of eosinophil recruitment into the tissue is a complex process that is not completely understood but involves adhesion molecules, such as vascular cell adhesion molecule 1, very late activation antigen 4, and chemoattractants (3–5).
A number of eosinophil chemoattractants have been identified, including the proteolytic fragment of complement, C5a, the formylated peptide met-leu-phe (FMLP),1 numerous chemokines, and several lipid mediators (4, 6). The chemokines are a large superfamily of 8–10-kD secreted proteins that play a role in controlling the movement of leukocytes (7, 8). Several chemokines, including eotaxin-1 and -2, RANTES (regulated on activation, normal T cell expressed and secreted), macrophage inflammatory protein (MIP)-1α, and monocyte chemoattractant protein (MCP)-2, -3, and -4, chemoattract unstimulated peripheral blood eosinophils isolated from nonatopic donors, whereas IL-8 is active on activated eosinophils (5, 9–13). The lipid mediator platelet-activating factor (PAF) is a potent chemoattractant for purified human and mouse eosinophils, whereas the metabolite of arachidonic acid, leukotriene B4 (LTB4), is a potent chemoattractant for purified guinea pig eosinophils with relatively little activity on unstimulated human eosinophils (14–18).
Chemoattractants direct the movement of leukocytes by signaling cells through specific surface receptors. The chemoattractant receptors constitute a subfamily of the G protein– coupled seven transmembrane–spanning family of receptors (STR) whose members are related in structure and function and include the receptors for C5a (19), PAF (20), FMLP (21), lipoxin A4 (22), and the chemokine receptors (7, 23). Several of these receptors have been shown to be expressed in eosinophils, including the C5a receptor (C5aR; references 24, 25) and the chemokine receptors CCR3, CCR1, and CXCR2 (26–28). We were interested in isolating novel eosinophil chemoattractant receptors and used degenerate PCR on mouse eosinophil cDNA to isolate a cDNA that is highly expressed in these eosinophils. This gene encodes a 351–amino acid glycoprotein that is 78% identical to a human gene that has been reported to be a purinoceptor (P2Y7; reference 29) and an LTB4 receptor (BLTR; reference 30). We present data demonstrating that this murine gene encodes a functional receptor for LTB4, and in concordance with the human designation (30) and the consensus nomenclature (31), we have proposed the name murine (m) BLTR. Furthermore, we show that mBLTR is highly expressed on eosinophils and in the lungs of mice after allergic inflammation, suggesting that this receptor is important for trafficking eosinophil into tissue.
Materials and Methods
Purification of Mouse Leukocytes.
Murine eosinophils were isolated from the spleens of the CD2 promoter IL-5 transgenic mice (32) by negative selection with anti–Thy-1 (M5/49), anti-B220 (6B2), anti–Lyt-2 (53-6.7), goat anti–rat serum-coated magnetic beads, and a MACS magnet (Miltenyi Biotech, Auburn, CA; reference 33). The resulting eosinophil purity was >90% as determined by microscopic examination of Diff-Quik–stained (Baxter Scientific, McGaw Park, IL) cytocentrifuge preparations; contaminating cells were mononuclear. Peritoneal cells were harvested from healthy mice by peritoneal lavage. Intraperitoneal injections of 9% sodium casein were used to elicit activated cells into the peritoneal cavity. Elicited cells were collected by peritoneal lavage and macrophages and neutrophils purified by Percoll gradients (34). The cellular composition of each fraction was determined by Diff-Quik–stained cytocentrifuge preparations. Bone marrow cells were harvested from the femurs of pathogen-free FVB female mice and were maintained in RPMI with either 30% L cell–conditioned medium (source of M-CSF) and 20% FBS for 2 wk to obtain macrophages or 50% WEHI conditioned medium (source of IL-3) and 10% FBS for 4 wk to obtain mast cells.
Isolation of a Novel mSTR cDNA.
Purified murine eosinophil poly(A)+ RNA was used as a template to generate oligo dT primed first strand cDNA (cDNA synthesis kit; Boehringer Mannheim Corp., Indianapolis, IN). Degenerate oligomers were synthesized based on the well-conserved transmembrane domains (TM) of chemoattractant G protein–coupled STRs: 5′ TM II tac (c/a)(t/g)gctg(a/c)acctg(t/g)c(c/a)(t/g)tggccgac, 5′ TM III gaccg (t/c)tacctggccat(t/c)gtcca(t/c)gccac, 3′ TM III gtgc(a/g)tggac(a/g)- atggccaggta(a/g)cggtc, 3′ TM VII (g/a)taga(t/g)ga(t/g)ggggtt(g/c)- ag(g/a)ca(g/a)c(t/a). PCR was performed using 100 pmol of the oligomer pairs 5′ TM II and 3′ TM III, 5′ TM II and 3′ TM VIII, 5′ TM III and 3′ TM VIII, 1 μl murine eosinophil first stand cDNA, and 0.5 U Taq polymerase (Perkin Elmer, Foster City, CA) in 75 μl with an initial 5 cycles of annealing at 37°C for 60 s, followed by 25 cycles at 50°C for 60 s (denaturation at 95°C for 30 s and extension at 72°C for 90 s). PCR products were ligated into TA cloning vector (Invitrogen Corp., Carlsbad, CA) and sequenced. Several clones from the PCR reaction using the TM II and TM III primers designated C1.2, C1.4, and C1.5 contained an identical, novel ∼200-bp insert with sequence homology to the STRs. A murine eosinophil cDNA library was prepared using the ZAP express cDNA library kit in the pBK-CMV phagemid vector (Stratagene, La Jolla, CA) that contained 1.75 × 106 independent clones (98% recombinants). 106 clones were screened in duplicate with a 32P-Klenow–labeled 200-bp C1.4 insert under conditions of high stringency (50% formamide, 10% dextran sulfate, 5× SSC, 1× Denhardt's solution, 1% SDS, 100 μg/ml denatured herring sperm DNA, and 20 mM Tris at 42°C) and washed at 65°C in 0.1× SSC and 0.1% SDS for 40 min. 92 positive plaques were clearly identified in duplicate and 20 randomly picked for analysis. Sequence analysis revealed that these clones were from the same gene but had variable 5′ ends. One clone, p65b, which represented a class of clones that contained an open reading frame beginning with methionine, had the longest 5′ untranslated region and contained a 3′ poly A tract. Therefore, this clone was selected for further studies.
5′ Rapid Amplification of cDNA Ends.
The 5′ end of the murine p65b gene was isolated using two nested internal 5′ oligomers tagaacaatgggcaacagagacaggg and agaaggtgctgcaggagatgtagt and the 5′ Amplifinder 5′ Rapid Amplification of cDNA Ends (RACE) Kit (CLONTECH Laboratories, Inc., Palo Alto, CA). The RACE products were cloned and sequenced.
Northern Analysis.
RNA was isolated from the organs of normal FVB mice or from fresh lymphomas by lysing the tissue in guanidinium isothiocyanate using a polytron and pelleting the RNA through a 5.7 M CsCl2 cushion. The poly(A)+ fraction was isolated from total RNA by oligo dT cellulose chromatography (Pharmacia Biotech, Inc., Piscataway, NJ). Spontaneously arising lymphomas were obtained from bigenic mice containing a c-myc transgene and p53 null alleles (35). Histologically, these tumors were comprised of nonadherent cells with a high mitotic rate, containing large pleomorphic nuclei, multiple nucleoli, and scanty cytoplasm, and were not infiltrated by normal leukocytes (35). These tumors were Thy-1 positive by Northern blot and described in Elson et al. (35). RNA STAT-60 (Tel-Test “B”, Inc., Friendswood, TX) was used to isolate RNA from mouse leukocytes and cell lines. RNA was fractionated on a 1.2% agarose gel containing 0.7% formaldehyde, transferred to GeneScreen (Dupont-NEN, Boston, MA) overnight using 10× SSC and hybridized with a 32P-dCTP Klenow-labeled random primed p65b cDNA probe and the ribosomal protein (rp) L32 (36) or a 28s RNA probe (37) as a control of RNA loading. RNA was fixed to membrane by first UV cross-linking (Stratalinker; Stratagene, La Jolla, CA). The membranes were hybridized and washed under conditions of high stringency described above.
In Vitro Translation.
Transcription of the p65b cDNA plasmid linearized with Xba1 was performed using T3 polymerase to produce a sense cRNA (38). Transcription of the p65b cDNA plasmid linearized with BamH1 was performed using T7 polymerase to produce an antisense cRNA. The cRNA was in vitro translated in the presence of [35S]methionine (Dupont-NEN) using a rabbit reticulocyte lysate (Promega Corp., Madison, WI). Translation was performed in the presence or absence of canine pancreatic microsomal membranes. 5 μl of each translation reaction was analyzed by 4–20% gradient Laemmli SDS-PAGE, treated with Autofluor (National Diagnostics, Atlanta, GA) and exposed to autoradiography.
Cell Culture and Transfection.
Chinese hamster ovary (CHO) cells were grown in F-12/HAM medium supplemented with 10% FCS, 2 mM glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Mediatech, Herndon, VA) at 37°C and 5% CO2. CHO cells were transiently transfected with 10 μg of the p65b cDNA plasmid (which is in the pBK-CMV vector) per 10-cm dish using Lipofectamine (GIBCO BRL, Gaithersburg, MD) and cultured for 72 h before analysis. Mock transfected cells were treated identically without the addition of DNA. CHO cells were stably transfected with 15 μg of the p65b cDNA plasmid linearized with Ssp1 in a 10-cm dish by the calcium phosphate method (38) and selected with G418 (800 μg/ml; Mediatech) for 10 d. 48 clones were selected and 19 analyzed by Northern blot analysis for the expression of the p65b mRNA. Clone A1B2 that highly expressed the p65b mRNA was selected for a second round of cloning and a subclone A1B2.1 was used for further analysis. The mouse cell lines EL4 T cell, MIC B cell, P815 mastocytoma, and the macrophage WEHI and RAW 264.7 as well as the rat basophilic leukemia (RBL) cell line (American Type Tissue Culture Collection, Rockville, MD) were maintained in RPMI supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin, 50 μg/ml streptomycin, 2 mM l-glutamine and 57 μM 2-mercaptoethanol at 37°C, and 5% CO2. RAW 264.7 were stimulated with 100 U/ml of mIFN-γ (R & D Systems, Inc., Minneapolis, MN) in complete medium for 18 h.
Radioligand Binding Assays.
Membrane protein was prepared from p65b transiently transfected CHO cells and mock-transfected CHO cells in buffer A (50 mM Tris, 1 mM EDTA, 1 mM EGTA, pH 7.4, 1 mM benzamidine, 0.1 mM PMSF, 0.01% bacitracin, 0.001% trypsin inhibitor, and 20 μg/ml aprotinin) as previously described (39). 10 μg of membranes were incubated with 1 nM of [5,6,8,9,11,12,14,15-3H(N)]LTB4 (150–200 Ci/mmol) or 20 nM [35S]dATPαS (1,250 Ci/mmol; DuPont-NEN) in the absence or presence of excess cold competitor (1 μM unlabeled LTB4 or 100 μM 2-methylthio-ATP (2-MeSATP). For saturation binding studies, different concentrations of [3H]LTB4 ranging from 0.1 to 2 nM were incubated with 10 μg membrane protein in 500 μl of buffer A on ice for 1 h and terminated by rapid filtration through GF/C glass fiber filters (presoaked in 20 mM sodium pyrophosphate) on a Millipore vacuum manifold. The filters were immediately washed five times with 5 ml ice-cold 50 mM Tris-HCl, pH 7.4, dried and bound radioactivity determined by scintillation counting. Nonspecific binding at each concentration of [3H]LTB4 was determined by coincubation of samples with an excess (1 μM) of unlabeled LTB4 (Calbiochem-Novabiochem, La Jolla, CA). As a control, membrane fractions prepared from mock-transfected CHO cells were treated identically in parallel with transfected cells. For the competitive displacement experiments, increasing concentrations of nonradiolabeled 12(S)- hydroxy-(5Z, 8Z, 10E, 14Z)-eicosa-tetraenoic acid (12(S)-HETE), prostaglandin D2 (PGD2) and LTD4 (Sigma Chemical Co., St. Louis, MO) were incubated with 10 μg membrane protein of transiently transfected CHO cells in the presence of 1 nM of [3H]LTB4. Binding curves and K d and K i calculations were performed using Prism (GraphPad Software Inc., San Diego, CA).
Calcium Flux.
CHO cell clones stably expressing p65b and untransfected CHO cells in F-12/HAM containing 1% FCS and purified murine eosinophils in HBSS containing 0.05% BSA were loaded at 107 cells/ml with 5.0 μM of the acetoxymethyl ester of fura-2 (fura-2 AM; Molecular Probes, Inc., Eugene, OR) for 60 min at 37°C in the dark. Loaded cells were washed twice and resuspended in a buffer containing 145 mM NaCl, 4 mM KCl, 1 mM NaHPO4, 0.8 mM MgCl2, 1.8 mM CaCl2, 25 mM Hepes, and 22 mM glucose. 2 ml of cells (5 × 106 cells/ml) were placed in a continuously stirring cuvette at 37°C in DeltaRAM (Random Access Monochromator) fluorimeter (Photon Technology International, Monmouth Junction, NJ). Changes of cytosolic free calcium were determined after addition of the agonist by monitoring the excitation fluorescence intensity emitted at 510 nm in response to sequential excitation at 340 nm and 380 nm. The data are presented as the relative ratio of fluorescence at 340/ 380 nm.
Chemotaxis.
50 μl of purified mouse eosinophils at 2.5 × 106 cells/ml in HBSS and 0.05% BSA were placed in the top of a 48-well modified Boyden micro-chemotaxis chamber (NeuroProbe, Cabin John, MD). 30 μl of 10-fold serial dilutions of LTB4 in HBSS and 0.05% BSA were placed in the bottom wells of the chamber and were separated from the cells by a 5-μm pore polycarbonate filter. The apparatus was incubated at 37°C and 5% CO2 for 60 min and cells that migrated across the filter and adhered to the bottom side of the filter were stained with Diff-Quik and counted.
Western Blot Analysis.
The peptide MAANTTSPAAPSSPGGM(C) corresponding to the NH2-terminal 17 amino acids of mBLTR plus a COOH-terminal cysteine for coupling was synthesized on an Applied Biosytems Peptide Synthesizer. 2 mg of this peptide was coupled to 2 mg of Imject® maleimide activated keyhole limpet hemocyanin (KLH; Pierce Chemical Co., Rockford, IL). The conjugate was purified by size exclusion chromatography on a NAP-1 column (Pharmacia Biotech, Inc.) and used to immunize two New Zealand white female rabbits. Immune serum from both rabbits was pooled and affinity purified on a Sulfolink® gel (Pierce Chemical Co.) to which the peptide had been coupled via its COOH-terminal cysteine residue to a 12-atom spacer arm attached to the gel. Purified murine eosinophils from IL-5 transgenic mice or purified human eosinophils were washed twice in cold PBS, and lysed in 10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, NP-40 1%, and Triton X-100 1% supplemented with antiproteases (10 μg/ml leupeptin, 1 μg/ml pepstatin A, and 4 μg/ml aprotinin). Murine alveolar macrophages (mMAC) were obtained by bronchoalveolar lavages (BAL; 5 ml PBS and 0.6 mM EDTA) of normal male BALB/c mice and were lysed in the same lysis buffer. Cell lysates were boiled in sample buffer (250 mM Tris-HCl, pH 6.8, 20% glycerol, and 2% 2-mercaptoethanol) for 5 min. Protein extracts were loaded onto a 10% SDS-PAGE gel and electrophoretically separated. Proteins were transferred onto nitrocellulose membrane (Hybond ECL; Amersham Pharmacia Biotechnology Inc., Piscataway, NJ) and blocked overnight at 4°C in TBS, pH 7.6, containing 0.1% Tween-20 (TBST) and 5% nonfat dry milk (blotto). The membrane was sequentially incubated for 1 h at room temperature with the affinity-purified rabbit anti-BLTR (1 μg/ml) in blotto and then with a HPR-conjugated goat anti–rabbit (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) diluted to 1:5,000 in blotto. The membrane was washed repeatedly in TBST between incubations. Blots were developed using ECL kit from Amersham. Controls were performed using preimmune rabbit serum at 1/20,000 (∼0.5 mg/ml) and by preincubating 5 μg of the rabbit anti-BLTR antibody with 100 μg of the immunizing peptide in a volume of 110 μl PBS (∼1,500-fold molar excess of peptide) for 1 h at room temperature before dilution into 10 ml of blotto for the Western blot analysis.
Aspergillus fumigatus Model of Allergic Pulmonary Inflammation.
A mixture of culture filtrate and mycelial extracts of Aspergillus fumigatus (Af ) was provided by Bayer Pharmaceuticals (Spokane, WA) and was diluted to a concentration of 2 mg/ml with normal saline. Mice were anesthetized by Metofane inhalation (methoxyfluorane; Pittman-Moore, Mundelein, IL), and 100 μg (50 μl) of Af antigen or 50 μl of normal saline was applied to the left nostril using a micropipette with the mouse held in the supine position. After instillation, mice were held upright until alert. Mice were immunized three times a week for 3 wk. 12 h after the final sensitizing dose animals were killed. RNA was prepared from lung tissue using a modified version of guanidium isothiocyanate method. In brief, 4 M guanidium isothiocyanate, 25 mM sodium citrate, pH 7.0, 0.5% sarcosyl, and 0.1 M β-mercaptoethanol was used to solubilize lung tissue (10 ml/lung). Phenol/chloroform (1:1) and sodium acetate, pH 4.8, were added and the mixture was passed through a 23-G needle three times. Tubes were spun at 10,000 g for 20 min and RNA was precipitated from aqueous phase with equal volume of isopropanol for 1 h at −20°C. RNA was suspended in the extraction solution and again precipitated with isopropanol. Pellets were washed twice with 75% ethanol, resuspended in dH20, and heated to 65° for 10 min. 5 μg of RNA/lane was run on a low-formaldehyde/1% agarose gel, capillary blotted onto GeneScreen Plus (Dupont-NEN).
Results
Molecular Cloning of a Novel mSTR.
Degenerate primers were designed based on the sequences of the conserved transmembrane domains II, III, and VII of chemoattractant receptors for FMLP, C5a, PAF, Lipoxin A4, and the chemokine receptors CXCR1, CXCR2, CXCR4, and CCR1. RT-PCR using degenerate primers to transmembrane domains II and III and mRNA isolated from murine eosinophils amplified an ∼200-bp product, a size consistent with the known distance between these transmembrane domains. Sequence analysis of this PCR product identified a novel open reading frame with ∼30% homology to other chemoattractant receptors reported at the time. Therefore, this PCR product was used to screen a murine eosinophil cDNA library. 92 out of ∼106 independent recombinant plaques hybridized strongly with this PCR product and 20 were randomly selected for further analysis. Restriction digestion, Southern blot, and sequence analyses revealed that these clones were from the same gene but contained different 5′ ends. Clone p65b represented a class of clones that contained an open reading frame beginning with methionine and contained the most 5′ sequences and was selected for further analysis.
Analysis of the p65b cDNA.
To determine the cap site of this gene, 5′ RACE was performed using eosinophil mRNA and a nested set of internal primers. This analysis revealed 20 additional bases 5′ of the cDNA p65b (Fig. 1 A, underlined). This makes the mature p65b cDNA 1,406 bp in length with a 1,053-bp open reading frame, a 137-bp 5′-untranslated region, and a 198-bp 3′-untranslated region that contains the polyadenylation signal ATTAAA (Fig. 1 A, double underlined). The open reading frame encodes a polypeptide of 351 amino acids with an estimated molecular mass of 38 kD (Fig. 1 A). The primary structure of the p65b open reading frame was 78% identical to a human gene independently entered in the data base as an orphan STR (CMKRL1 [40] and R2 [41]) as a novel purinoceptor (P2Y7; reference 29) and as a receptor for LTB4 called BLTR (30; Fig. 1 B). The amino acid sequence of the p65b open reading frame was also related to other G protein–coupled chemoattractant receptors sharing 32% identity with the murine C5a receptor (mC5aR), 32% identity with the murine IL-8 receptor (mCXCR2), 30% identity to mFMLPR, 29% identity with the murine lipoxin A4 receptor (mLXAR), and 26% identity with the murine platelet-activating factor receptor (mPAFR). Using the p65 cDNA as a probe we screened a human lung library under reduced stringency and isolated six clones that were all identical to this human gene CMLKR/R2/P2Y7 /BLTR. The fact that we isolated only this gene that is 79 and 78% identical in nucleic acid and amino acid sequence, respectively, to p65b, strongly suggests that p65b is the mouse orthologue of this human gene. Based on this and the functional data presented below we have named p65b mBLTR.
Figure 1.

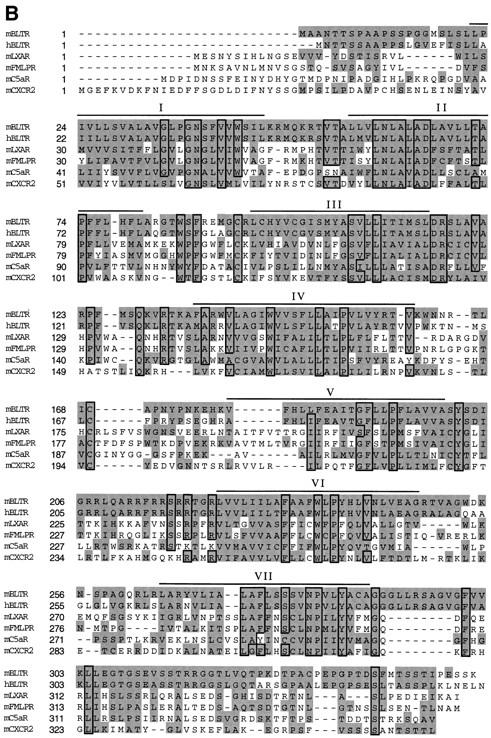
Nucleotide and amino acid sequence of mBLTR cDNA. (A) Nucleotide sequence is numbered on the left and the deduced amino acid sequence is numbered beginning with the predicated initiating methionine on the right. 20 nucleotides obtained from 5′ RACE are underlined and the polyadenylation signal sequence is double underlined. Two putative N-linked glycosylation sites are indicated in bold. These data are available from GenBank/EMBL/DDBJ under accession number AF044030. (B) Amino acid sequence alignment of the mBLTR open reading frame with the chemoattractant receptors: human LTB4 receptor (hBLTR), murine lipoxin A4 receptor (mLXAR), murine formyl-Met-Leu-Phe receptor (mFMLPR), murine C5a receptor (mC5aR), and murine CXCR2. The transmembrane domains predicted from a Kyte-Doolittle hydrophobicity analysis are overlined and labeled as I-VII. The shaded area indicates ≥90% sequence homology and the boxed area indicates regions of identity.
The mBLTR open reading frame exhibits several sequence motifs that are characteristic of G protein–coupled receptors, including seven hydrophobic transmembrane domains, conserved prolines in several of the transmembrane domains, and conserved cysteines in extracellular domains II and III for intramolecular chain disulfide bonding (23). In addition, similar to other chemoattractant receptors, mBLTR has two potential N-linked glycosylation sites (Fig. 1 A, bold), one in the extracellular NH2-terminal segment and the other in the third extracellular domain. The receptor also contains a serine and threonine-rich COOH-terminal intracytoplasmic segment. Analogous regions of other STRs are sites of phosphorylation involved with receptor desensitization and internalization (23). When comparing the amino acid sequence of the mouse and human BLTR, it is striking that the three intracytoplasmic loops are identical across species, while not being conserved across the subfamily of chemoattractant receptors (Fig. 1 B). This suggests that the BLTRs may be coupled to a unique, well-conserved, signaling pathway among the chemoattractant receptors.
In Vitro Translation.
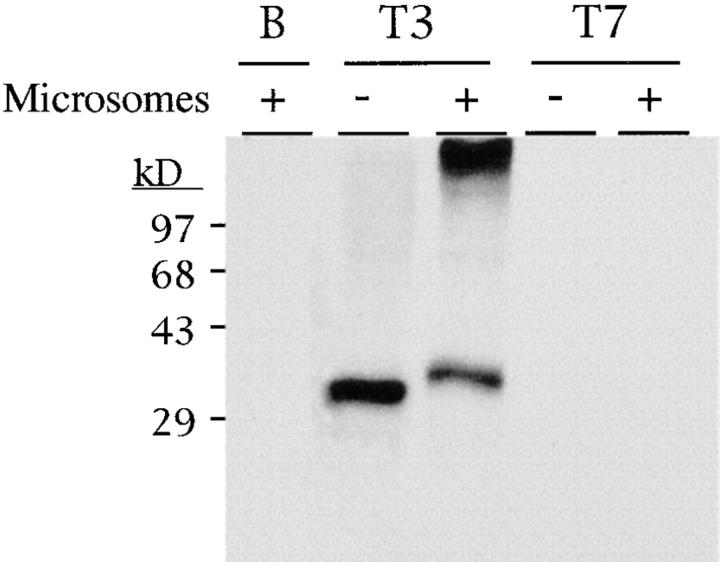
To determine if the p65b cDNA encoded a complete open reading frame, in vitro transcription and translation were performed (Fig. 2). This was relevant because the open reading frame for the human gene reported by one group (41) was 25 amino acids upstream of the one we have proposed for the murine cDNA. In addition, the murine cDNA contains no in frame stop codons 5′ of the proposed initiating methionine. A linearized p65b plasmid was used as a template to synthesize sense (T3) and antisense (T7) cRNA. In vitro translation using a rabbit reticulocyte lysate revealed that the sense cRNA encoded a protein that migrated as an ∼31-kD species on SDS-PAGE. As controls, the translation of antisense cRNA (T3) and no input cRNA (B) did not produce a protein product. To determine if this protein is glycosylated, in vitro translation was performed in the presence of dog pancreatic microsomes (Fig. 2). This revealed an upward shift in mobility on SDS-PAGE of ∼4 kD, indicating that the mBLTR protein is N-linked glycosylated and ∼35 kD. This experiment also told us that the p65b cDNA contained a translatable message. In fact, the upstream methionine proposed by this one group (41) is likely the result of using PCR on genomic DNA because the two other reports identifying this human gene as a cDNA (29, 40) revealed an initiating methionine in a similar position to the one that we found for the murine gene.
Figure 2.
In vitro transcription and translation of the mBLTR cDNA (p65b). Translation in the presence (+) or absence (−) of pancreatic microsomal membranes of p65b cRNA transcribed with T3 polymerase (sense) and T7 polymerase (antisense control). B (blank) translation in the absence of added cRNA. Molecular mass markers in kD are indicated on the left.
Functional Characterization, Specific Binding to LTB4.
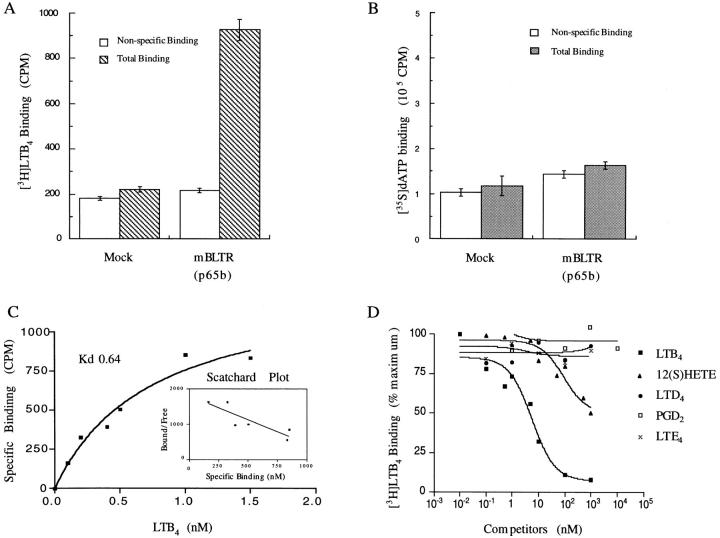
Since the proposed human orthologue of this gene has been shown to be both a receptor for LTB4 (30) and dATP (29), we performed radioligand binding experiments using [3H]LTB4 and [35S]dATP on CHO cells transiently transfected with the mBLTR cDNA p65b. Membrane fractions of transiently transfected and mock-transfected CHO cells were prepared and incubated with 1 nM [3H]LTB4 and 20 nM [35S]dATP (Fig. 3, A and B). To determine nonspecific binding, membrane fractions were also incubated with [3H]LTB4 in the presence of excess (1 μM) nonradiolabeled LTB4 and [35S]ATP in the presence of excess (100 μM) nonradiolabeled 2-MeSATP. Transfected cells specifically bound [3H]LTB4, whereas nontransfected cells did not (n = 4 separate transfections; Fig. 3 A). In contrast, there was no detectable specific binding of [35S]dATP to either mock transfected or mBLTR (p65b)-transfected CHO cells (Fig. 3 B). Saturation binding analysis and Scatchard analysis revealed a single class of specific high affinity binding sites for [3H]LTB4 with a dissociation constant (K d) of 0.64 ± 0.14 nM (mean ± SE of four independent measurements performed in duplicate; Fig. 3 C). To test the specificity of this interaction, we examined the competitive displacement of [3H]LTB4 by different eicosanoids in the arachidonic acid metabolic pathway on membrane fractions of mBLTR (p65b)-transfected CHO cells. LTB4 competed the binding of 1 nM [3H]LTB4 with a K i of 2.1 nM, whereas the structurally related compound 12(S)-HETE competed [3H]LTB4 binding with a K i of 34 nM (Fig. 3 D). LTD4, LTE4, and PGD2 did not significantly compete for [3H]LTB4 binding to the transfected cell membranes (Fig. 3 D). Thus, these data suggest that LTB4 is a specific high affinity ligand for this receptor and hence the proposed designation as murine BLTR.
Figure 3.
Binding of [3H]LTB4 to mBLTR transfected CHO cell membranes. (A) 1 nM [3H]LTB4 and (B) 20 nM [35S]dATP binding to membranes (10 μg) of mBLTR (p65b) transiently transfected CHO cells or mock transfected CHO cells in the absence (Total Binding) and presence (Nonspecific Binding) of unlabeled excess ligand (mean ± SE, n = 4). (C) Binding isotherm and Scatchard analysis of [3H]LTB4 binding to membrane of mBLTR (p65b) transfected CHO cells (mean, n = 4). (D) Inhibition of [3H]LTB4 binding to membrane fractions of mBLTR-transfected CHO cells by various eicosanoids, including 12(S) HETE, LTD4, LTE4, and PGD2.
Functional Characterization, LTB4-induced Calcium Flux.
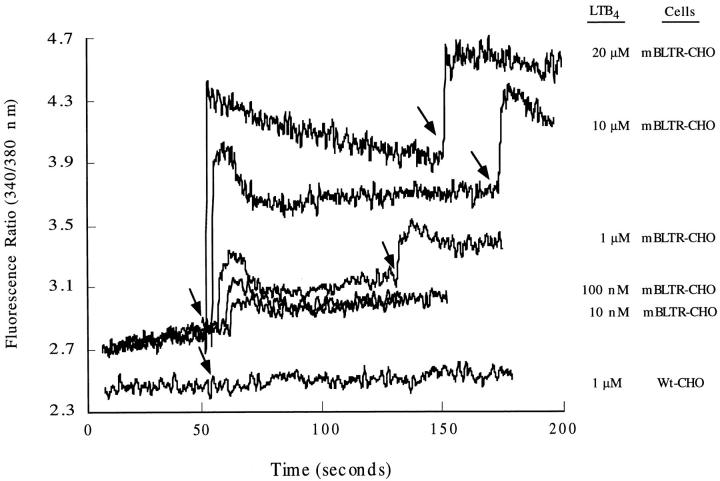
Signaling through G protein–coupled seven transmembrane spanning chemoattractant receptors typically generates a transient rise in intracellular calcium. We have generated stable CHO cell clones that express the mBLTR cDNA p65b and investigated the ability of LTB4 to induce a calcium flux in these cells. As shown in Fig. 4, LTB4 at concentrations ranging from 10 nM to 20 μM induced a rapid calcium flux in fura-2 loaded mBLTR-CHO cells, but not the parental (wild-type) CHO cells. This response was dose dependent with increasing concentrations of LTB4 inducing a greater increase intracellular calcium. These results indicate that LTB4 induces a calcium flux in CHO cells through the cell surface receptor mBLTR. Surprisingly, we did not observe desensitization in these CHO transfectants at concentrations ranging from 10 nM to 20 μM (Fig. 4).
Figure 4.
LTB4-induced calcium flux in mBLTR-CHO transfectants. Fura-2 loaded stable mBLTR-CHO clone (A1B2.1) and the parental wild-type CHO cells (Wt-CHO) were stimulated with LTB4 (concentrations indicated on the right) at the times indicated by the arrows. [Ca2+]i levels were measured as ratio fluorescence of excitation at 340/380 nm over time.
mBLTR mRNA Expression.
Northern blot analysis of mRNA from various mouse tissues revealed that the mature mBLTR transcript migrates as a 1.45-kb mRNA and is quite restricted in its expression pattern (Fig. 5). Low levels of constitutive expression were detected only in the lung, lymph nodes, and spleen. No expression was seen in the thymus, kidney, liver, brain, heart, or testis. Purified eosinophils from IL-5 transgenic mice expressed high levels of mBLTR mRNA constitutively, while bone marrow–derived macrophages, bone marrow–derived mast cells (data not shown), and various mouse cell lines, including the EL4 T cell line, MIC B cell line, P815 mastocytoma line, and the rat RBL basophilic leukemia line did not (Fig. 5 B). The WEHI macrophage line constitutively expressed low levels of mBLTR mRNA and IFN-γ induced its expression in the RAW 264.7 macrophage cell line (Fig. 5 B). Resting peritoneal cells did not express detectable mBLTR mRNA; however, its expression was dramatically induced in both activated macrophages and neutrophils elicited into the peritoneum (Fig. 5 C). Furthermore, mBLTR was highly expressed in T cell lymphomas (Fig. 5 D) that spontaneously arose in c-myc transgenic mice homozygous for p53 null alleles (35), suggesting that T cells can express BLTR. The single RNA species seen above 1.45 kb (Fig. 5, B and C) almost certainly represents the primary unspliced BLTR transcript. The multiple RNA species seen below 1.45 kb in the eosinophil lane in Fig. 5 B are degraded BLTR transcripts. These lower bands were not seen in poly(A)+ eosinophil RNA (Fig. 5 E). Thus, mBLTR mRNA is predominantly expressed in activated leukocytes, including IL-5 exposed eosinophils, elicited peritoneal neutrophils and macrophages, and IFN-γ–simulated macrophages.
Figure 5.
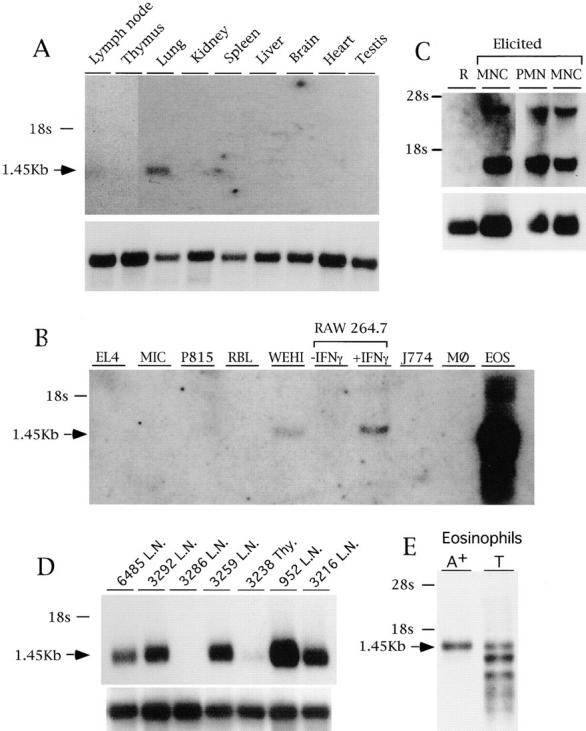
Expression of the mBLTR mRNA. (A) Northern blot analysis of 1.5 μg poly(A)+ RNA isolated from the indicated organs of a healthy FVB mouse. (B) Northern blot analysis of 10 μg total RNA isolated from the EL4 T cell line, MIC B cell line, P815 mastocytoma line, RBL basophilic line, WEHI macrophage line, RAW 264.7 macrophage cell line untreated or treated for 18 h with 200 U/ml IFN-γ, J774 macrophage line, bone marrow derived macrophages (MØ), and eosinophils purified from IL-5 transgenic mice. (C) Northern blot analysis of 10 μg total RNA isolated from leukocytes recovered from peritoneal lavage before (R, resting) or after (elicited) intraperitoneal casein injection and percoll density gradient purification. M, macrophage fraction (76% macrophages, and 24% neutrophils); N, neutrophil fraction (95% neutrophils, 5% macrophages). (D) Northern blot analysis of 1.5 μg poly(A)+ RNA isolated from Thy1 positive fresh lymphomas obtained from lymph nodes (L.N.) or thymus (Thy). Each lane contains RNA isolated from a lymphoma that spontaneously arose in a different bigenic mouse containing a c-myc transgene and p53 null alleles numbered and described in Elson et al. (35). (E) Northern blot analysis of 2 μg of poly(A)+ RNA and 10 μg of total RNA before poly A selection (T) isolated from eosinophils purified from IL-5 transgenic mice. Blots were sequentially probed with the mBLTR (p65b) cDNA (top of each panel) and rpL32 or 28s as a control for RNA loading (bottom of panel).
BLTR Protein Expression on Eosinophils and Alveolar Macrophages.
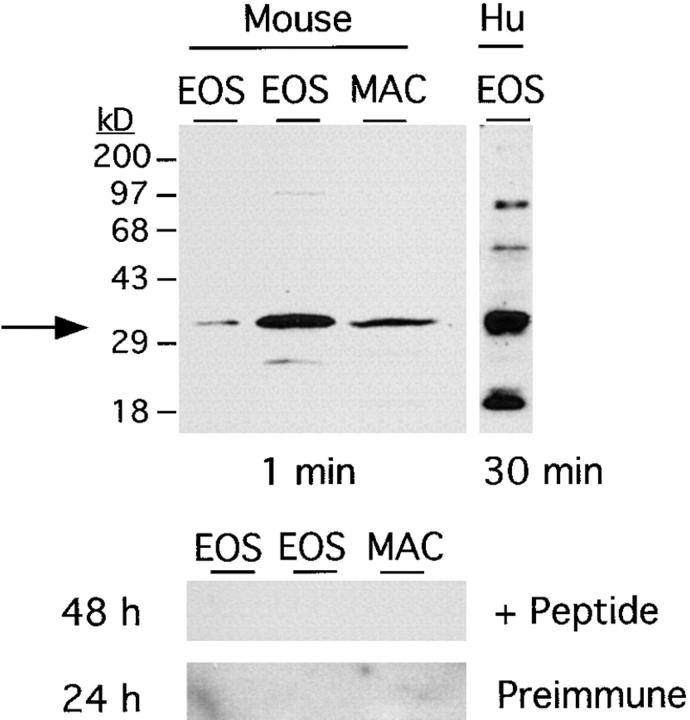
To study the expression of the mBLTR protein, we have generated an affinity-purified rabbit antibody directed against the NH2-terminal 17 amino acids of the predicted open reading frame. This was done by immunizing rabbits with the peptide coupled to KLH and then purifying the immune serum from two rabbits by peptide affinity chromatography. Western blot analysis using this affinity-purified antibody on total cell lysates of eosinophils purified from IL-5 transgenic mice and alveolar macrophages isolated by bronchoalveolar lavage (BAL) from wild-type mice revealed an ∼32.5-kD species in both murine eosinophils and macrophages (Fig. 6). Interestingly, lower levels of a comigrating band were also seen in human eosinophils purified from a nonatopic donor (Fig. 6). This antibody reaction was specific for the immune serum and was not seen with the preimmune serum (Fig. 6). In addition, the BLTR immunoreactivity seen with the immune serum was completely blocked by an excess of the mBLTR immunizing peptide (Fig. 6).
Figure 6.
Expression of the mBLTR protein. Protein extracts of eosinophils obtained from two IL-5 transgenic mice (30 μg/lane), murine alveolar macrophages (20 μg/lane), and human eosinophils purified from a normal donor (30 μg/ lane), were loaded onto a 10% SDS-PAGE gel and electrophoretically separated. Western blot analysis was performed using an affinity-purified rabbit anti– mouse BLTR antibody. The position of BLTR protein is indicated by the arrow and molecular mass markers in kD are indicated to the left of the top blot. The affinity-purified antibody cross-reacts with the human receptor. This antibody reaction was specific for the immune serum and was not seen with the preimmune serum and was completely blocked by excess BLTR peptide (bottom two blots). The exposure times are indicated below the top blots and to the left of the bottom blots. EOS, eosinophils; MAC, alveolar macrophages; Hu, human.
mBLTR Expression in a Murine Model of Allergic Pulmonary Inflammation.
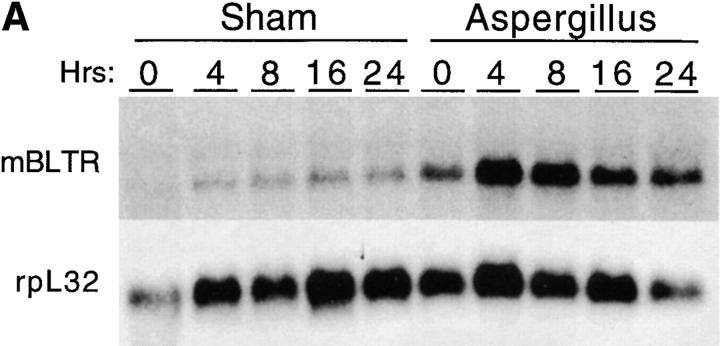
To assess the expression of mBLTR mRNA in the setting of allergic inflammation, we subjected mice to inhalation of the mold allergen Aspergillus fumigatus (Af). We have previously reported that repeated intranasal instillation of Af gives rise to an intense eosinophil-predominant inflammatory infiltrate (42). BAL eosinophil counts are elevated by two to three orders of magnitude in Af-treated mice (105–106 eosinophils/ml) compared with controls (mean 103 eosinophils/ml) and the lungs of mice treated with Af have dense inflammatory infiltrates. The infiltrate contains predominantly eosinophils with a significant number of admixed lymphocytes and occasional histiocyte-like cells in peribronchial and perivascular cuffs (see cover of this issue).
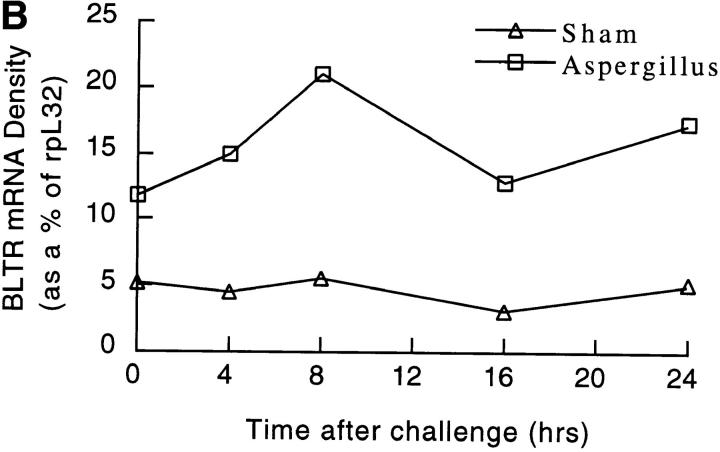
In the setting of repeated allergen inhalation, Af-treated mice develop a baseline level of eosinophils in their BAL and lung biopsies. Superimposed on this background inflammation is a new influx of eosinophils elicited by each successive Af dose. We have found that waves of infiltrating eosinophils in the BAL and tissue peak 12 h after each Af exposure (Oettgen, H.C., personal observation). As can be seen in Fig. 7, levels of mBLTR are elevated (approximately threefold) in the lungs of mice after repeated allergen inhalation (compare time 0 in the saline- and Af-treated mice). In addition, after inhalation of Af in sensitized mice, mBLTR is further upregulated, peaking at ∼8 h (approximately fivefold over saline treated), consistent with the receptor being expressed on influxing eosinophils.
Figure 7.
Expression of mBLTR in lungs of mice in the Af model of allergic pulmonary inflammation. Mice were treated three times a week for a period of three weeks with 50 μg Af intranasally (or normal saline in “sham” animals). (A) Northern blot analysis of 5 μg of total RNA isolated from the lungs of mice at the indicated times after the last Af treatment or saline treatment (sham control). Each lane represents a different mouse. The blot was probed sequentially with the mBLTR cDNA and rpL32 as a control for RNA loading. (B) Quantitation of the Northern blot using a PhosphorImager expressed as the percentage of rpL32 signal for each lane (PhosphorImager signal of BLTR/rpL32 × 100).
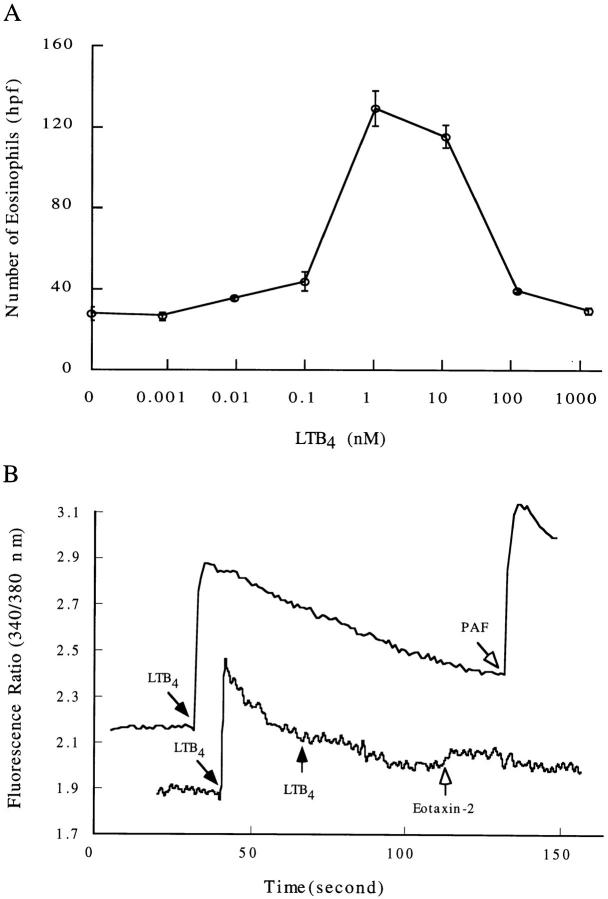
Murine Eosinophils Respond to LTB4.
Since the mBLTR mRNA and protein was highly expressed in murine eosinophils isolated from IL-5 transgenic mice, and its mRNA was upregulated in the lungs of mice in a murine model of allergic pulmonary inflammation in a time course consistent with eosinophil tissue recruitment, we sought to determine if mBLTR was a functional chemotactic receptor on eosinophils. To do this, we examined the chemotactic and calcium flux responses of mouse eosinophils to different concentrations of LTB4. Eosinophils showed a dose-dependent bell-shaped chemotactic response to LTB4 with an optimum concentration of 1–10 nM (Fig. 8 A). LTB4 also triggered a robust calcium flux in murine eosinophils loaded with fura-2 (Fig. 8 B). In addition, 100 nM LTB4 almost completely desensitized eosinophils to a subsequent 100 nM LTB4 challenge (Fig. 8 B). However, the eosinophils were still able to respond to PAF and human eotaxin-2 indicating that PAF, eotaxin-2, and LTB4 use distinctive receptors on the membranes of eosinophils for signal transduction. Of note, although 100 nM LTB4 induced homologous desensitization in eosinophils (Fig. 8 B), this was not the case for the mBLTR-CHO transfectants (Fig. 4). Although the binding affinity of LTB4 for BLTR is very similar in CHO transfectants and leukocytes, BLTR-mediated signaling appears to have significant differences in CHO cells and eosinophils.
Figure 8.
LTB4-induced chemotaxis and calcium flux of mouse eosinophils. (A) Chemotaxis of murine eosinophils in response to the indicated concentrations of LTB4 (mean ± SE, n = three separate experiments). (B) Calcium flux responses of murine eosinophils. Each tracing represents the [Ca2+]i levels of fura-2-loaded eosinophils measured as relative fluorescence over time. Arrows mark the time of addition of the indicated agonist LTB4 (100 nM), PAF (5 μM), and human eotaxin-2 (1.6 μM).
Discussion
In an effort to identify novel chemoattractant receptors, we performed degenerate PCR based on the sequences of known chemoattractant receptors on cDNA isolated from murine eosinophils and isolated a gene that is 78% identical to a human gene that has been reported to be a purinoceptor (P2Y7; reference 29) and an LTB4 receptor (BLTR; reference 30). We have shown that this murine gene is a functional LTB4 receptor and propose the name mBLTR. Interestingly, mBLTR was highly expressed on murine eosinophils isolated from IL-5 transgenic mice and was upregulated in the lungs of mice after the induction of eosinophilic pulmonary inflammation.
Leukotrienes constitute a class of potent biological mediators of inflammation that have been implicated in a wide variety of human inflammatory disease, including asthma and allergic rhinitis, cystic fibrosis, inflammatory bowel disease, psoriasis, acute respiratory distress syndrome, multiple sclerosis, and rheumatoid arthritis (43–45). Their biosynthesis derives from 5-lipoxygenese-catalyzed oxygenation of arachidonic acid in cells of the myeloid lineage. LTB4 is primarily active on leukocytes causing chemotaxis, adhesion, and activation, whereas the cysteinyl leukotrienes (LTC4, LTD4, and LTE4) are primarily active on smooth muscle, causing bronchoconstriction, capillary leak, and mucus production.
Although the leukotrienes are known to exert their biological actions through stereoselective membrane receptors (31), structural information has been lacking. Radioligand binding studies have identified an LTB4 receptor on leukocytes that has been given the designation BLTR (31). LTB4 can also bind and activate the intranuclear transcription factor peroxisome proliferator-activated receptor (PPAR)α, resulting in the activation of genes that terminate inflammatory processes (46). BLTR has been characterized as a binding site for [3H]LTB4 on human neutrophils with a K d of 10 (47), 0.46 (48), and 1.5 nM (49) and on guinea pig eosinophils with a K d of 1.4 nM (16). Recently, a human BLTR has been molecularly identified as G protein–coupled STR (30). Membrane fractions of COS-7 cells transfected with human BLTR were shown to bind LTB4 with a K d of human 0.154 (30) or 2.1 nM (50). In CHO cells stably expressing this receptor, LTB4 induced a calcium flux, d-myo-inositol-1,4,5-triphosphate (IP3) accumulation, inhibition of adenylate cyclase, and chemotaxis (30). However, this same human receptor was also cloned from a human erythroleukemia cell cDNA library using a chicken P2Y3 cDNA probe, and when it was transfected into COS-7 cells, it resulted in a binding site for dATP and ATP with a K d of 7.3 and 37 nM, respectively (29).
We have isolated a mouse gene that is highly related in sequence to the human gene described above. CHO cells transfected with this murine gene, but not mock-transfected CHO cells, specifically bound LTB4 with a K d of 0.6 ± 0.1 nM. Furthermore, LTB4 induced a dose-dependent intracellular calcium flux in CHO stable transfectants, but not control CHO cells. In contrast, dATP did not specifically bind to these CHO transfectants or CHO cells expressing the human gene (30). However, like Yokomizo et al. (30), we did find that dATP specifically bound to untransfected COS-7 cells (data not shown). In fact, COS-7 were shown to contain an endogenous P2Y2 receptor (51). In addition, a range of nucleotides, including ATP, UTP, ADP, UDP, and dATP, had no effect on phospholipase C or adenyl cyclase activities in 1321N1 human astrocytoma cells expressing an epitope-tagged human BLTR gene (51). These data, together with the low sequence homology of this gene with other P2Y receptors, suggest that the gene we have isolated is a murine LTB4 receptor.
LTB4 was discovered based on its potent chemotactic activity on neutrophils (52) and we have seen high levels of BLTR expression in elicited neutrophils. However, we have also detected the expression of BLTR on other leukocytes where the biological activities of LTB4 are less clear. For example, we have seen BLTR expression in elicited peritoneal macrophages, alveolar macrophages, and IFN-γ–stimulated RAW 264.7 macrophages. Although activated macrophages are a major source of LTB4, BLTR has not been described on monocytes or macrophages. However, LTB4 has been shown to augment IL-6 production by human monocytes (53) and enhance the ability of peritoneal macrophages to phagocytose and kill Salmonella typhimurium and Pseudomonas aeruginosa (54). In addition, mice deficient in 5-lipoxygenase were more susceptible to Klebsiella pneumonia in association with decreased alveolar macrophage phagocytic and bactericidal activities (55). These defects in macrophage function were overcome by the addition of exogenous LTB4, suggesting a functional role for LTB4 in alveolar macrophage host defense (55). We have also detected constitutive expression of BLTR in lymph nodes, and high levels of BLTR expression were seen in T cell lymphomas, suggesting that BLTR can be expressed on T lymphocytes. Although LTB4 receptors have not been described on lymphocytes, LTB4 has been shown to induce IL-2 and IFN-γ production from T cells and to augment NK cell cytotoxicity and sensitivity to IL-2 by upregulating IL-2 receptor expression (56–58). The cloning of BLTR will allow new studies aimed at understanding the role of LTB4 and BLTR in lymphocyte and macrophage biology.
mBLTR was highly expressed in murine eosinophils isolated from IL-5 transgenic mice. Consistent with this, LTB4 was a potent chemoattractant for murine eosinophils, inducing peak chemotaxis at 1–10 nM and inducing a robust calcium flux at 100 nM. LTB4 has been shown to induce guinea pig eosinophil superoxide anion generation and chemotaxis in vitro and in vivo (16, 17, 59). In addition, eosinophils purified from IL-5 transgenic mice, labeled with 111In and injected intravenously into wild-type mice were recruited into the skin by LTB4 (60). In contrast, human eosinophils have been reported to respond poorly to LTB4 when compared with PAF (14, 15, 17). Our results indicate that murine eosinophils from IL-5 transgenic mice respond robustly to LTB4 in manner comparable with PAF. In addition, Western blot analysis revealed high levels of mBLTR protein in these IL-5–exposed murine eosinophils and lower levels of BLTR in human eosinophils isolated from the peripheral blood of a nonatopic donor. It may be that priming with cytokines, such as IL-5, is important for regulating the expression or responsiveness of BLTR on human eosinophils (61).
Specific antagonists of LTB4 have revealed a role of LTB4 in regulating the recruitment of eosinophils into tissues in vivo. In a guinea pig model of antigen-induced pulmonary inflammation, pretreatment of sensitized guinea pigs with a selective LTB4 antagonist before antigen provocation decreased the influx of eosinophils into the lung and BAL fluid but had no effect on the influx of neutrophils (62). Likewise, in a murine model of experimental allergic encephalomyelitis, pretreatment of mice with a selective LTB4 receptor antagonist markedly blocked the recruitment of eosinophils into the spinal cord and completely inhibited the development of paralysis (63). The influx of lymphocytes was unaffected by pretreatment with the LTB4 receptor antagonist, thus revealing an unrecognized role for eosinophils in the pathogenesis of this disease. Furthermore, the specific inhibition of 5-lipoxygenase with zileuton reduced eosinophil influx into the lung tissue and BAL fluid in a murine model of allergic pulmonary inflammation and also prevented airway mucus release in these mice (64). Studies in humans have also implicated leukotrienes in the pathogenesis of allergic airway inflammation. Increased levels of LTB4 and LTC4 were recovered in the BAL fluid after endobronchial antigen challenge compared with prechallenge levels (65, 66). Nocturnal levels of LTB4 and cysteinyl leukotrienes were elevated in the BAL fluid of patients with nocturnal asthma compared with normal controls (67). In these asthmatic subjects, zileuton decreased BAL fluid LTB4 levels, blood eosinophil levels, and improved FEV1 (67). In fact, zileuton is now widely used clinically in the treatment of human asthma (68–70).
Our results clearly demonstrate that LTB4 can be a potent agonist for murine eosinophils and that these cells can express high levels of BLTR. Furthermore, we have seen that levels of BLTR mRNA in the lungs of mice are increased after the induction of eosinophilic pulmonary inflammation in a time course consistent with the influx of eosinophils. Taken together with the above mentioned in vivo studies using specific LTB4 antagonists, these data suggest that LTB4 and BLTR play an important role in the recruitment of eosinophils into tissues. In addition, we have also found that BLTR is expressed in other leukocytes, including alveolar macrophages, IFN-γ stimulated macrophages, elicited peritoneal macrophages and neutrophils, and T cell lymphomas, suggesting a broad role for LTB4 in host defense and inflammation.
In conclusion, while searching for novel chemoattractant receptor genes expressed in murine eosinophils, we have isolated a gene that encodes a functional cell surface receptor for LTB4, mBLTR. The identification of the mBLTR gene will help elucidate the role of the LTB4 pathway in controlling the movement and activation of leukocytes in models of human disease.
Acknowledgments
We are grateful to Dr. Philip Leder in whose laboratory this work began.
This work was supported by grants to A.D. Luster from the National Institutes of Health (NIH), the Benjamin Jacobson Family/Cancer Research Institute, the Culpeper Medical Foundation, and Defense Advanced Research Projects Agency. E.A. Garcia-Zepeda was supported by an NIH Fogarty Fellowship.
Abbreviations used in this paper: Af
Aspergillus fumigatus
- BAL
bronchoalveolar lavage
- BLTR
leukotriene B4 receptor
- CHO
Chinese hamster ovary cells
- FBS
fetal bovine serum
- FMLP
formyl-met-leu-phe
- LT
leukotriene
- m
murine
- NS
normal saline
- PAF
platelet-activating factor
- PGD2
prostaglandin D2
- RBL
rat basophilic leukemia
- STR
seven transmembrane–spanning receptor
- TM
transmembrane domain
References
- 1.Weller PF. The immunobiology of eosinophils. N Eng J Med. 1991;324:1110–1118. doi: 10.1056/NEJM199104183241607. [DOI] [PubMed] [Google Scholar]
- 2.Bousquett J, Chanez P, Lacoste JY, Barneon G, Ghavanian N, Enander I, Venge P, Ahlstedt S, Simony-Lafontaine J, et al. Eosinophilic inflammation in asthma. N Eng J Med. 1990;323:1033–1039. doi: 10.1056/NEJM199010113231505. [DOI] [PubMed] [Google Scholar]
- 3.Bochner BS, Schleimer RP. The role of adhesion molecules in human eosinophil and basophil recruitment. J Allergy Clin Immunol. 1994;94:427–428. doi: 10.1016/0091-6749(94)90195-3. [DOI] [PubMed] [Google Scholar]
- 4.Resnick MB, Weller PF. Mechanisms of eosinophil recruitment. Am J Respir Cell Mol Biol. 1993;8:349–355. doi: 10.1165/ajrcmb/8.4.349. [DOI] [PubMed] [Google Scholar]
- 5.Baggiolini M, Dahinden CA. CC chemokines in allergic inflammation. Immunol Today. 1994;15:127–133. doi: 10.1016/0167-5699(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 6.Kitayama J, Carr MW, Roth SJ, Buccola J, Springer TA. Contrasting responses to multiple chemotactic stimuli in transendothelial migration. J Immunol. 1997;158:2340–2349. [PubMed] [Google Scholar]
- 7.Luster AD. Chemokines: chemotactic cytokines that mediate inflammation. N Eng J Med. 1998;388:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 8.Rollins BJ. Chemokines. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- 9.Kita H, Gleich GJ. Chemokines active on eosinophils: potential roles in allergic inflammation. J Exp Med. 1996;183:2421–2426. doi: 10.1084/jem.183.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Zepeda EA, Rothenberg ME, Ownbey RT, Celestin J, Leder P, Luster AD. Human eotaxin is specific chemoattractant for eosinophil cells and provides a new mechanism to explain tissue eosinophilia. Nat Med. 1996;4:449–456. doi: 10.1038/nm0496-449. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Zepeda EA, Combardiere C, Rothenberg ME, Sarafi M, Hamid Q, Murphy PM, Luster AD. Human monocyte chemoattractant protein (MCP)-4: a novel CC chemokine with activities on monocytes, eosinophils, and basophils, induced in allergic and non-allergic inflammation, that signals through the CC chemokine receptors CCR-2 and CCR-3. J Immunol. 1996;157:5613–5626. [PubMed] [Google Scholar]
- 12.Forrsmann U, Uguccioni M, Loetscher P, Dahinden CA, Langen H, Thelen M, Baggiolini M. Eotaxin-2, a novel CC chemokine that is selective for the chemokine receptor CCR3, and acts like eotaxin on human eosinophil and basophil leukocytes. J Exp Med. 1997;185:2171–2176. doi: 10.1084/jem.185.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweizer RC, Welmer JAM, Raaijmakers P, Zanen J, Lammers WJ, Koenderman L. RANTES- and interleukin-8-induced responses in normal eosinophils: effects of priming with interleukin-5. Blood. 1994;83:3697–3704. [PubMed] [Google Scholar]
- 14.Wardlaw AJ, Moqbel R, Cromwell O, Kay AB. Platelet-activating factor: a potent chemotactic and chemokinetic factor for human eosinophils. J Clin Invest. 1986;78:1701–1706. doi: 10.1172/JCI112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morita E, Schroder J-M, Christophers E. Differential sensitivities of purified human eosinophils and neutrophils to defined chemotaxins. Scand J Immunol. 1989;29:709–716. doi: 10.1111/j.1365-3083.1989.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 16.Ng CF, Sun FF, Taylor BM, Wolin MS, Wong PY-K. Functional properties of guinea pig eosinophil leukotriene B4receptor. J Immunol. 1991;147:3096–3103. [PubMed] [Google Scholar]
- 17.Sun FF, Crittenden NJ, Czuk CI, Taylor BM, Stout BK, Johnson HG. Biochemical and functional differences between eosinophils from animal species and man. J Leuk Biol. 1991;50:140–150. doi: 10.1002/jlb.50.2.140. [DOI] [PubMed] [Google Scholar]
- 18.Rothenberg ME, Luster AD, Lilly CM, Drazen JM, Leder P. Constitutive and allergen-induced expression of eotaxin mRNA in the guinea pig lung. J Exp Med. 1995;181:1211–1216. doi: 10.1084/jem.181.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerard NP, Gerard C. The chemotactic receptor for human C5a anaphylatoxin. Nature. 1991;349:614–617. doi: 10.1038/349614a0. [DOI] [PubMed] [Google Scholar]
- 20.Honda Z-I, Nakamura M, Miki I, Minami M, Watanabe T, Seyama Y, Okado H, Tok H, Ito K, Miyamato T, Shimizu T. Cloning and functional expression of platelet-activating factor receptor from guinea-pig lung. Nature. 1991;349:342–346. doi: 10.1038/349342a0. [DOI] [PubMed] [Google Scholar]
- 21.Ye RD, Cavanagh SL, Quehenberger O, Prossnitz ER, Cochrane CG. Isolation of a cDNA that encodes a novel granulocyte N-formyl peptide receptor. Biochem Biophys Res Commun. 1992;184:582–589. doi: 10.1016/0006-291x(92)90629-y. [DOI] [PubMed] [Google Scholar]
- 22.Fiore S, Maddox JF, Perez HD, Serhan CN. Identification of a human cDNA encoding a functional high affinity lipoxin A4Receptor. J Exp Med. 1994;180:253–260. doi: 10.1084/jem.180.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 24.Gerard NP, Hodges MK, Drazen JM, Weller PF, Gerard C. Characterization of a receptor for C5a anaphylatoxin on human eosinophils. J Biol Chem. 1989;264:1760–1766. [PubMed] [Google Scholar]
- 25.Elsner J, Oppermann M, Kapp A. Detection of C5a receptors on human eosinophils and inhibition of eosinophil effector functions by anti-C5a receptor (CD88) antibodies. Eur J Immunol. 1996;26:1560–1564. doi: 10.1002/eji.1830260723. [DOI] [PubMed] [Google Scholar]
- 26.Post TW, Bozic CR, Rothenberg ME, Luster AD, Gerard N, Gerard C. Molecular cloning of two murine eosinophil beta chemokine receptors. J Immunol. 1995;155:5299–5305. [PubMed] [Google Scholar]
- 27.Gao J-L, Sen AI, Tiffany L, Yoshi O, Rothenberg ME, Murphy PM, Luster AD. Identification of a mouse eosinophil-selective receptor for the CC chemokine eotaxin. Biochem Biophys Res Commun. 1996;223:679–684. doi: 10.1006/bbrc.1996.0955. [DOI] [PubMed] [Google Scholar]
- 28.Heath H, Qin S, Rao P, Wu L, LaRosa G, Kassam N, Ponath PD, Mackay CR. Chemokine receptor usage by human eosinophils: the importance of CCR3 demonstrated using an antagonistic monoclonal antibody. J Clin Invest. 1997;99:178–184. doi: 10.1172/JCI119145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akbar G, Dasari V, Webb T, Ayyanathan K, Pillarisetti K, Sandhu A, Athwal R, Daniel J, Ashby B, Barnard E, Kunapuli S. Molecular cloning of a novel P2 purinoceptor from human erythroleukemia cells. J Biol Chem. 1996;271:18363–18367. doi: 10.1074/jbc.271.31.18363. [DOI] [PubMed] [Google Scholar]
- 30.Yokomizo T, Lzumi T, Chang K, Takuwa Y, Shimizu T. A G-protein-coupled receptor for leukotriene B4that mediate chemotaxis. Nature. 1997;387:620–624. doi: 10.1038/42506. [DOI] [PubMed] [Google Scholar]
- 31.Alexander SPH, Peters JA. Receptors and ion channel nomenclature. Trends Pharmacol Sci. 1997;45(Suppl.):50–51. [Google Scholar]
- 32.Dent LA, Strath M, Mellor AL, Sanderson CJ. Eosinophilia in transgenic mice expressing interleukin 5. J Exp Med. 1990;172:1425–1431. doi: 10.1084/jem.172.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothenberg ME, Luster AD, Leder P. Murine eotaxin: an eosinophil chemoattractant inducible in endothelial cells and in IL-4-induced tumor suppression. Proc Natl Acad Sci USA. 1995;92:8960–8964. doi: 10.1073/pnas.92.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Laning J, Devi S, Mak J, Schall TJ, Dorf ME. Biologic activities of the murine β-chemokine TCA3. J Immunol. 1994;153:4616–4624. [PubMed] [Google Scholar]
- 35.Elson A, Deng C, Campos-Torres J, Donehower LA, Leder P. The MMTV/c-myc transgene and p53 null alleles collaborate to induce T-cell lymphomas, but not mammary carcinomas in transgenic mice. Oncogene. 1995;11:181–190. [PubMed] [Google Scholar]
- 36.Shen MM, Leder P. Leukemia inhibitory factor is expressed by the preimplantation uterus and selectively blocks primitive ectoderm formation in vitro. Proc Natl Acad Sci USA. 1992;89:8240–8244. doi: 10.1073/pnas.89.17.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rich BE, Steitz JA. Human acidic ribosomal phosphoproteins P0, P1, and P2: analysis of cDNA clones, in vitro synthesis, and assembly. Mol Cell Biol. 1987;7:4065–474. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 10.32–10.33.
- 39.Simon J, Webb TE, King BF, Burnstock G, Barnard EA. Characterization of a recombinant P2Ypurinoceptor. Eur J Pharm. 1995;291:281–289. doi: 10.1016/0922-4106(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 40.Owman C, Nilsson C, Lolait SJ. Cloning of cDNA encoding a putative chemoattractant receptor. Genomics. 1996;37:187–194. doi: 10.1006/geno.1996.0541. [DOI] [PubMed] [Google Scholar]
- 41.Raport CJ, Schweickart VL, Chantry D, Eddy RL, Shows TB, Godiska R, Gray PW. New members of the chemokine receptor gene family. J Leuk Biol. 1996;59:18–23. doi: 10.1002/jlb.59.1.18. [DOI] [PubMed] [Google Scholar]
- 42.Mehlhop PD, van de Rijn M, Goldberg AB, Brewer JP, Kurup VP, Martin TR, Oettgen HC. Allergen- induced bronchial hyperreactivity and eosinophilic inflammation occur in the absence of IgE in a mouse model of asthma. Proc Natl Acad Sci USA. 1997;94:1344–1349. doi: 10.1073/pnas.94.4.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuelsson B, Dahlen S-E, Lindgren JA, Rouzer CA, Serhan CN. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 44.Lewis RA, Austen KF, Soberman RJ. Leukotrienes and other products of the 5-lipoxygenase pathway. N Eng J Med. 1990;323:645–655. doi: 10.1056/NEJM199009063231006. [DOI] [PubMed] [Google Scholar]
- 45.Henderson WR., Jr The role of leukotrienes in inflammation. Ann Intern Med. 1994;121:684–697. doi: 10.7326/0003-4819-121-9-199411010-00010. [DOI] [PubMed] [Google Scholar]
- 46.Devchand P, Keller H, Peters J, Vazquez M, Gonzalez F, Wahli W. The PPARα-leukotriene B4pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 47.Goldman D, Goetzl E. Specific binding of leukotriene B4to receptors on human polymorphonuclear leukocytes. J Immunol. 1982;129:1600–1604. [PubMed] [Google Scholar]
- 48.Lin A, Ruppel P, Gorman R. Leukotriene B4binding to human neutrophils. Prostaglandins. 1984;28:837–849. doi: 10.1016/0090-6980(84)90038-8. [DOI] [PubMed] [Google Scholar]
- 49.Bomalaski JS, Mong S. Binding of leukotriene B4 and its analogs to human polymorphonuclear leukocyte membrane receptors. Prostaglandins. 1987;33:855–867. doi: 10.1016/0090-6980(87)90114-6. [DOI] [PubMed] [Google Scholar]
- 50.Owman C, Sabirsh A, Boketoft A, Olde B. Leukotriene B4is the functional ligand binding to and activating the cloned chemoattractant receptor, CMKRL1. Biochem Biophys Res Commun. 1997;240:162–166. doi: 10.1006/bbrc.1997.7628. [DOI] [PubMed] [Google Scholar]
- 51.Herold CL, Li Q, Schachter JB, Harden TK, Nicholas RA. Lack of nucleotide-promoted second messenger signaling responses in 1321N1 cells expressing the proposed P2Y receptor, P2Y7 . Biochem Biophys Res Commun. 1997;235:717–721. doi: 10.1006/bbrc.1997.6884. [DOI] [PubMed] [Google Scholar]
- 52.Ford-Hutchinson AW, Bray MA, Doig MV, Shipley ME, Smith M. Leukotriene B, a potent chemokinetic and aggregating substance released from polymorphonuclear leukocytes. Nature. 1980;286:264–265. doi: 10.1038/286264a0. [DOI] [PubMed] [Google Scholar]
- 53.Rola-Pleszcynski M, Stankova J. Leukotriene B4 enhances interleukin 6 (IL-6) production and IL-6 messenger RNA accumulation in human monocytes in vitro: transcriptional and posttranscriptional mechanisms. Blood. 1992;80:1004–1011. [PubMed] [Google Scholar]
- 54.Demitsu T, Katayama H, Saito-Taki T, Yaoita H, Nakano M. Phagocytosis and bactericidal action of mouse peritoneal macrophages treated with leukotriene B4. Int J Immunopharmacol. 1989;11:801–808. doi: 10.1016/0192-0561(89)90134-3. [DOI] [PubMed] [Google Scholar]
- 55.Bailie MB, Standiford TJ, Laichalk LL, Coffey MJ, Strieter R, Peters-Golden M. Leukotriene-deficient mice manifest enhanced lethality from klebsiella pneumonia in association with decreased alveolar macrophage phagocytic and bactericidal activities. J Immunol. 1996;157:5221–5224. [PubMed] [Google Scholar]
- 56.Stankova J, Gagnon N, Rola-Pleszczynski M. Leukotriene B4 augments interleukin-2 receptor-beta (IL-2R beta) expression and IL-2R beta-mediated cytotoxic response in human peripheral blood lymphocytes. Immunol. 1992;76:258–263. [PMC free article] [PubMed] [Google Scholar]
- 57.Rola-Pleszczynski M, Chavaillaz PA, Lemaire I. Stimulation of interleukin 2 and interferon gamma production by leukotriene B4in human lymphocyte cultures. Prostaglandins Leukot Med. 1986;23:207–210. doi: 10.1016/0262-1746(86)90187-3. [DOI] [PubMed] [Google Scholar]
- 58.Rola-Pleszczynski M, Gagnon L, Siros P. Leukotriene B4augments human natural cytotoxic cell activity. Biochem Biophys Res Commun. 1983;113:531–537. doi: 10.1016/0006-291x(83)91758-8. [DOI] [PubMed] [Google Scholar]
- 59.Faccioli LH, Nourshargh S, Moqbel R, Williams FM, Sehmi R, Kay AB. The accumulation of 111In-eosinophils induced by inflammatory mediators, in vivo. . Immunol. 1991;73:222–227. [PMC free article] [PubMed] [Google Scholar]
- 60.Teixeira MM, Wells TNC, Lukacs NW, Proudfoot AEI, Kunkel SL, Williams TJ, Hellewell PG. Chemokine-induced eosinophil recruitment: evidence of a role for endogenous eotaxin in an in vivo allergy model in mouse skin. J Clin Invest. 1997;100:1657–1666. doi: 10.1172/JCI119690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sehmi R, Wardlaw AJ, Cromwell O, Kurihara K, Waltmann P, Kay AB. Interleukin-5 selectively enhances the chemotactic response to eosinophils obtained from normal but not eosinophilic subjects. Blood. 1992;79:2952–2959. [PubMed] [Google Scholar]
- 62.Richards IM, Griffin RL, Oostveen JA, Morris J, Wishka DG, Dunn CJ. Effect of the selective leukotriene B4 antagonist U-75302 on antigen-induced bronchopulmonary eosinophilia in sensitized guinea pigs. Am Rev Respir Dis. 1989;140:1712–1716. doi: 10.1164/ajrccm/140.6.1712. [DOI] [PubMed] [Google Scholar]
- 63.Gladue R, Carroll L, Milici A, Scampoli D, Stukenbrok H, Pettipher E, Salter E, Contillo S, Showell H. Inhibition of leukotriene B4-receptor interaction suppresses eosinophil infiltration and disease pathology in a murine model of experimental allergic encephalomyelitis. J Exp Med. 1996;183:1893–1898. doi: 10.1084/jem.183.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henderson WR, Lewis DB, Albert RK, Zhang Y, Lamm WJE, Chiang GKS, Jones F, Eriksen P, Tien Y, Jonas M, Chi EY. The importance of leukotrienes in airway inflammation in a mouse model of asthma. J Exp Med. 1996;184:1483–1494. doi: 10.1084/jem.184.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wenzel SE, Larsen GL, Johnston NF, Voelkel NF, Westcott JY. Elevated levels of leukotriene C4 in bronchoalveolar lavage fluid from atopic asthmatics after endobronchial allergen challenge. Am Rev Respir Dis. 1990;142:112–119. doi: 10.1164/ajrccm/142.1.112. [DOI] [PubMed] [Google Scholar]
- 66.Wenzel SE, Westcott JY, Larson GL. Bronchoalveolar lavage fluid mediator levels 5 minutes after allergen challenge in atopic subjects with asthma: relationship to the development of late phase responses. J Allergy Clin Immunol. 1991;87:540–548. doi: 10.1016/0091-6749(91)90013-e. [DOI] [PubMed] [Google Scholar]
- 67.Wenzel SE, Trudeau JB, Kaminsky DA, Cohn J, Martin RJ, Westcott JY. Effect of 5-lipoxygenase inhibition on bronchoconstriction and airway inflammation in nocturnal asthma. Am J Respir Crit Care Med. 1995;152:897–905. doi: 10.1164/ajrccm.152.3.7663802. [DOI] [PubMed] [Google Scholar]
- 68.Israel E, Dermarkarian R, Rosenberg M, Sperling R, Taylor G, Rubin P, Drazen JM. The effects of a 5-lipoxygenase inhibitor on asthma induced by cold, dry air. N Eng J Med. 1990;323:1740–1744. doi: 10.1056/NEJM199012203232505. [DOI] [PubMed] [Google Scholar]
- 69.Israel E, Rubin P, Kemp JP, Grossman J, Pierson W, Siegel SC, Tinkelman D, Murray JJ, Busse W, Segal AT, et al. The effects of inhibition of 5-lipoxygenase by zileuton in mild-to-moderate asthma. Ann Intern Med. 1993;119:1059–1066. doi: 10.7326/0003-4819-119-11-199312010-00001. [DOI] [PubMed] [Google Scholar]
- 70.Liu MC, Dube LM, Lancaster J, Group ZS. Acute and chronic effects of a 5-lipoxygenase inhibitor in asthma: a 6-month randomized multicenter trial. J Allergy Clin Immunol. 1996;98:859–871. doi: 10.1016/s0091-6749(96)80002-9. [DOI] [PubMed] [Google Scholar]