Abstract
A high proportion of tumors arise due to mutation of the p53 tumor suppressor protein. A p53 hotspot mutation at amino acid position 273 from R to H, flanking a peptide epitope that spans residues 264–272, renders cells resistant to killing by human histocompatibility leukocyte antigen (HLA)-A*0201–restricted cytotoxic T lymphocytes (CTLs) specific for this epitope. Acquisition of the R to H mutation at residue 273 of the human p53 protein promotes tumor growth in vivo by selective escape from recognition by p53.264–272 peptide-specific CTLs. Synthetic 27-mer p53 polypeptides covering the antigenic nonamer region 264–272 of p53 were used as proteasome substrates to investigate whether the R to H mutation at the P1′ position of the COOH terminus of the epitope affects proteasome-mediated processing of the protein. Analysis of the generated products by tandem mass spectrometry and the kinetics of polypeptide processing in conjunction with CTL assays demonstrate that the R to H mutation alters proteasomal processing of the p53 protein by inhibiting proteolytic cleavage between residues 272 and 273. This prevents the release of the natural CTL epitope that spans flanking residues 264–272 as well as a putative precursor peptide. These results demonstrate that mutation of p53 not only leads to malignant transformation but may also, in some instances, affect immune surveillance and should be considered in the design of cancer vaccines.
Keywords: p53, tumor antigens, cytotoxic T lymphocytes, antigen processing, proteasomes
Inactivation of the p53 tumor suppressor protein, through mutation or deletion, occurs in the majority of human cancers (1, 2). Often this involves a missense mutation at one of several defined mutational hotspots in the molecule (1–3). The altered p53 protein accumulates to high levels within the cell and has been used as a marker for malignant transformation (1, 2). Previous studies in both murine and human (Hu)1 models have demonstrated the preferential susceptibility of transformed cells to lysis by CTLs that are specific for peptides representative of processed wild-type (WT) p53 protein sequences and are presented on the cell surface by class I MHC molecules (4–9). Using transgenic (Tg) mice that express the HLA-A*0201 (A*0201) molecule, we previously identified two peptide sequences from the Hu WT p53 molecule, p53.149–157 and 264–272, that bind A*0201 and serve as endogenously processed target epitopes for CTL recognition and lysis (4). Importantly, CTLs specific for these p53-derived peptide epitopes are able to recognize and lyse a broad range of p53-overexpressing and A*0201+ Hu tumor cells (4). In contrast, nontransformed cells expressing A*0201 are not killed by the same CTLs, presumably due to insufficient levels of expression of p53 and p53-derived CTL epitopes (4, 8). These peptide epitopes are also able to stimulate A*0201-restricted CTLs from Hu peripheral blood, suggesting the presence of CTL precursors with specificity for p53 that may be potentially mobilized to destroy tumor cells expressing high levels of p53 peptides (10–13).
Peptides that are presented on the cell surface by class I MHC molecules for recognition by CTLs most often are derived from proteolytic processing of cellular proteins by the multicatalytic proteasome complex (14–16). Proteolytic degradation of p53 has been found to be dependent on proteasomal processing (17–20). Proteasome-generated peptide products are subsequently translocated into the endoplasmic reticulum (ER) by the transporters associated with antigen processing, where they are loaded into the peptide-binding groove of nascent class I molecules (21–23). In this report, we demonstrate that proteasomal processing of the natural A*0201-restricted CTL epitope p53.264–272 and of a putative precursor peptide is profoundly affected by a mutational p53 hotspot (R to H) at the COOH-terminal flanking residue 273. As a consequence, target cells that overexpress p53 harboring the 273 R to H mutation are not susceptible to in vitro and in vivo lysis by A*0201- restricted CTLs specific for the flanking p53 epitope, 264– 272. To our knowledge, these experiments demonstrate the first example of a naturally occurring mutation flanking a CTL epitope and affecting the ability of the proteasome to generate a defined MHC class I ligand.
Materials and Methods
Mice.
The derivation of homozygous A2/Kb-Tg mice expressing a chimeric transgene that consists of the α1 and α2 domains of A*0201 and the α3 domain of H-2Kb has been described previously (4, 8). Mice were propagated and maintained under specific pathogen–free conditions. All experimental procedures were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Peptides.
27-mer polypeptides spanning residues 256–282 of Hu WT (TLEDSSGNLLGRNSFEVRVCACPGRDR) and mutant (273 R to H) p53 were synthesized on an automated peptide synthesizer (432A; PE Applied Biosystems, Foster City, CA). Peptides p53.264–272 (LLGRNSFEV) and 260–272 (SSGNLLGRNSFEV) were synthesized by the core facility of TSRI (430A synthesizer; PE Applied Biosystems). Purity of synthetic peptides was ascertained by reverse phase (RP)-HPLC and mass spectrometry (MS). Amino acid residues are given in single letter code.
Cell Lines.
Previously described cell lines and transfectants used in these studies included T1 and T2 cells (24), the naturally A*0201-expressing, p53-deficient osteosarcoma line Saos-2, and the same cells transfected with Hu p53 genes harboring mutations at residue 143 (V to A), 175 (R to H), and 273 (R to H) (4, 25). Murine thymoma lines (EL4) transfected with A*0201/Kb (EA2Kb) or both A*0201/Kb and a Hu p53 gene harboring an R to H mutation at residue 273 (EA2Kb.1p53) have been generated as previously reported (8).
CTL Lines.
The derivation and maintenance of A*0201- restricted CTL lines specific for Hu WT p53.264–272 (CTL A2 264) and p53.149–157 (CTL A2 149) have previously been described (4). CTL lines were used as effector cells at the indicated E/T ratios in 51Cr-release assays (4).
Viral p53 Recombinants.
The derivation of parental canarypox virus and canarypox virus p53 recombinants expressing Hu p53 without (WT) or with mutations at residue 175 (R to H) or 273 (R to H) has been previously described (5). Saos-2 targets were labeled with 51Cr and infected with parental and recombinant canarypox viruses at an infectious dose of 20 PFU/cell. Cells were washed after 1 h and incubated for another 3-h period before responder CTLs were added for a 3-h 51Cr-release assay (4). Recombinant vaccinia viruses (rVV) which contain minigenes encoding either p53.149–157 (rVV-ES149) or p53.264–272 (rVV-ES264) downstream of the ER insertion sequence of E19 (26), and rVV-VPE16 that expresses the gp160 gene of HIV type 1 (27), were provided by Drs. Jack Bennink and Jonathan Yewdell (National Institute of Allergy and Infectious Diseases, Bethesda, MD).
Protection of Tumor Growth in A2/Kb-Tg Mice.
A2/Kb-Tg mice were either nonimmunized or immunized intravenously with 5 × 106 PFU of rVV-ES149, rVV-ES264, or rVV-VPE16. 2 wk later, mice were challenged subcutaneously in the right flank with 5 × 105 EA2Kb or EA2Kb.1p53 tumor cells. The size of tumors was determined by the formula (a2 × b/2), in which a defines the horizontal and b the vertical diameter of the tumor mass as determined by calipers. Three mice were used for each experimental and control group.
Purification of 20S Proteasomes.
20S proteasomes were purified from Hu T1 cells by standard procedures (28–31). Cell pellets were lysed in lysis buffer (80 mM KAc, 5 mM MgAc2, 10 mM Hepes, pH 7.2, and 0.1% Triton X-100), dounce homogenized, and spun at 40,000 g for 20 min. Supernatant was adsorbed to equilibrated DEAE-Sephacel (Amersham Pharmacia Biotech, Piscataway, NJ) for 45 min, unbound protein was removed with wash buffer (80 mM KAc, 5 mM MgAc2, and 10 mM Hepes, pH 7.2), and bound protein was eluted (500 mM KAc, 5 mM MgAc2, and 10 mM Hepes, pH 7.2). The protein containing eluate was concentrated by ultrafiltration, loaded on a 10–40% sucrose gradient in wash buffer, and ultracentrifuged (40,000 rounds/min in an SW40 Ti rotor for 15.5 h; Beckman, Fullerton, CA). Gradient fractions containing 20S proteasome complexes were identified by enzyme assays using fluorogenic substrate peptides, pooled, and concentrated for chromatography on a MonoQ HR 5/5 anion exchange column (Amersham Pharmacia Biotech; eluent A: 100 mM KCl, 5 mM MgCl2, and 10 mM Hepes, pH 7.2; eluent B: 1 M KCl, 5 mM MgCl2, and 10 mM Hepes, pH 7.2; linear gradient). The 20S proteasomes obtained were again identified by enzymatic activity and eluted as single peak upon rechromatography. Purity of 20S proteasome preparations was >90% as assessed by Coomassie-stained PAGE gels. Two-dimensional gel electrophoresis revealed that all detectable proteasomal subunits were present in 20S proteasomes purified from the Hu T1 cell line.
In Vitro Digestion of Polypeptide Substrates by Purified 20S Proteasomes and Recognition by CTLs.
20 μg of synthetic 27-mer p53 polypeptide substrates were incubated for the indicated periods of time at 37°C with and without 1 μg of purified 20S proteasome in a total volume of 300 μl of assay buffer consisting of 20 mM Hepes-KOH, pH 7.8, 2 mM MgAc2, and 1 mM dithiothreitol (28–31). 51Cr-labeled T2 cells were pulsed for 30 min with degraded 27-mer peptide products at the indicated concentrations in serum-free RPMI 1640 medium supplemented with 5% vol/ vol BSA and Hu β2 microglobulin (β2m) at 10 μg/ml. CTL A2 264 and CTL A2 149 were used as effector cells at an E/T ratio of 20:1 in a 6-h 51Cr-release assay.
Peptide Analysis by MS.
10 μl of 20S proteasome degraded peptide digests were separated by a RP-HPLC SMART-System equipped with a μRPC C2/C18 SC 2.1/10 column (Amersham Pharmacia Biotech) and eluted with a gradient of 15–65% of eluent B (70% acetonitrile in 0.05% TFA) in eluent A (0.05% TFA) in 33 min at a flow rate of 50 μl/min (29–31). Mass analysis of peptides was performed online by a tandem quadrupole mass spectrometer (TSQ 7000; Finnigan MAT, San Jose, CA) equipped with an electrospray ion source. Each scan was acquired over the range mass/charge (m/z) 300–1150 every 2 s (29–31). Peptides were identified by their molecular mass calculated according to the m/z peaks of single or multiple charged ions. Abundant peptides with signal intensities of at least threefold above background were sequenced by tandem MS (MS/MS) after fragmentation of the relevant peptides with argon atoms (30). Peptide sequence was determined from the masses of the fragmented peptide ions.
HPLC Separation of Processed Peptides, Identification by CTLs, and Sequence Analysis of Antigenic Peptides by MS/MS.
50 μl of the bulk 20S proteasome degraded peptide products were separated by RP-HPLC into 1- (50 μl) and 0.5- (25 μl) min fractions (30). 51Cr-labeled T2 target cells were pulsed for 40 min with half of each of the HPLC fractions in serum-free RPMI 1640 medium supplemented with 5% vol/vol BSA and Hu β2m at 10 μg/ml. CTL A2 264 were used as effector cells at an E/T ratio of 20:1 in a 6-h 51Cr-release assay. The antigenic HPLC fractions were dried and resuspended in 50% methanol/1% acetic acid. Antigenic peptides present in the pooled HPLC fractions were identified by coelution of synthetic peptides and sequenced by MS/MS (30). Ions of m/z corresponding to the relevant double protonated peptides were fragmented by argon atoms. Collision-activated dissociation fragments of relevant m/z and derived from the pooled antigenic HPLC fractions were compared with those obtained after argon atom–mediated fragmentation of the corresponding synthetic peptides.
Extraction of Natural Peptides from Class I MHC Molecules.
Adherent Saos-2/143 p53 transfectants were grown at ∼107 cells/ flask. Cells were washed twice with HBSS, and class I MHC– bound peptides were extracted by exposing cells for 1 min in 5 ml of buffer consisting of 0.13 M citric acid and 0.061 M Na2HPO4 at pH 3.0 (32–34). Cells were washed twice in RPMI 1640 and recultured in complete medium. Extracts were spun and the peptide containing supernatant was frozen. This procedure was repeated every other day for 3 wk to collect peptide extracts from the equivalent of 1.2 × 109 Saos-2/143 cells. Extracts were thawed, pooled, and loaded on C-18 spice cartridges (Analtech, Inc., Newark, DE) that had been washed with 4 ml each of methanol and H2O. Cartridges were again washed with H2O (10 ml) and peptides were eluted with 4 ml of 0.1% vol/vol TFA in acetonitrile. The peptide containing eluate was vacuum dried, resuspended in H2O, and cleared of debris by centrifugation. The supernatant was filtered through Centricon-10 (Amicon, Beverly, MA), and the resultant peptide extract was again vacuum dried.
HPLC Separation of Natural Peptide Extracts and Reconstitution of CTL Lysis.
1 ml of resuspended natural peptide extract or 100 pmol of either the p53.264–272 or the p53.260–272 synthetic peptides were separated by RP-HPLC at a flow rate of 50 μl/ min and eluted with a gradient of 20% of eluent B (70% acetonitrile in 0.05% TFA) in eluent A (0.05% TFA) in 32 min, a gradient of 20–48% of eluent B in eluent A in 74 min, and a gradient of 48–95% of eluent B in eluent A in 4 min. HPLC fractions were collected from 32 to 76 min (fractions 1–11: 4 min fractions at 200 μl/fraction) and from 76 min onwards (fractions 12–48: 1 min fractions at 50 μl/fraction). 51Cr-labeled T2 target cells were pulsed for 40 min in serum-free RPMI 1640 medium supplemented with 5% vol/vol BSA and Hu β2m at 10 μg/ml with 100 μl (fractions 4–11) or 25 μl (fractions 12–44) of fractions derived from the HPLC separation of the natural peptide extract. The corresponding volumes of fractions derived from the HPLC separations of synthetic p53.264–272 and p53.260–272 peptides and used to pulse T2 targets were 8 (fractions 4–11) and 2 μl (fractions 12–44). CTL A2 264 were used as effector cells at an E/T of 20:1 in a 5.5-h 51Cr-release assay. Antigenic fractions obtained from the HPLC-separated natural peptide extract were pooled and half of the pooled natural peptides was used for rechromatography by RP-HPLC. HPLC conditions remained almost the same, yet 0.5-min fractions were collected from 56 min onwards (fractions 10–44 at 25 μl/fraction). The amount of synthetic p53.264–272 and p53.260–272 peptides used for the second HPLC separations was 60 pmol. 51Cr-labeled T2 target cells were again pulsed for 40 min under serum-free conditions with 18 μl of fractions 10–32 derived from the rechromatography of the pooled antigenic natural HPLC fractions. The corresponding volumes of fractions derived from the HPLC separations of the synthetic p53.264–272 and p53.260–272 peptides and used to pulse T2 targets were 2 μl (fractions 8, 19, 20, 29) and 0.02 μl, 0.2 μl, and 2 μl (fractions 9–18 and 21–28), respectively. CTL A2 264 were used as effector cells at an E/T of 13:1 in a 6-h 51Cr-release assay.
Results and Discussion
Target Cells Expressing a p53 Mutational Hotspot at Residue 273 Are Not Recognized by CTLs Specific for the Flanking Epitope 264–272.
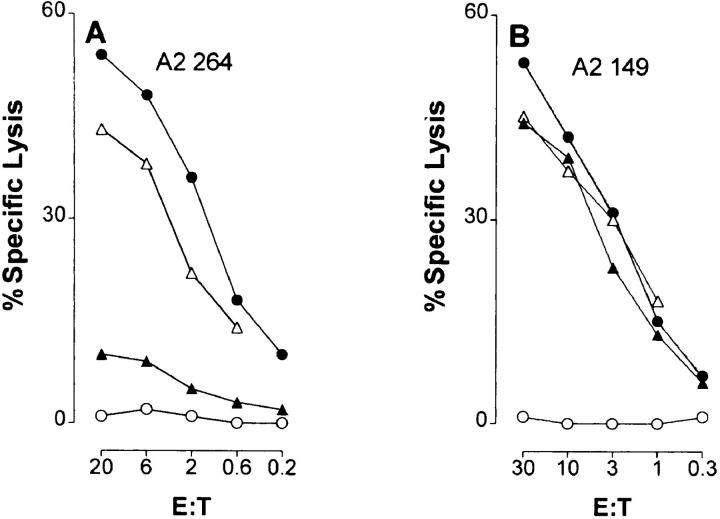
The p53-deficient Saos-2 tumor cell line transfected with a Hu p53 gene expressing the R to H hot spot mutation at amino acid residue 273 was not susceptible to lysis by CTL A2 264 specific for the flanking peptide 264–272 (Fig. 1 A). This contrasted with the successful recognition and lysis of cells transfected with Hu p53 genes expressing mutations at amino acid positions 143 or 175 (Fig. 1 A). All three p53 transfectants were killed by CTLs specific for CTL A2 149, indicating that other epitopes of the protein were being processed and presented by the A*0201 molecule (Fig. 1 B). Furthermore, as demonstrated in Fig. 1, C and D, Saos-2 cells infected with canarypox virus recombinants expressing Hu p53 with or without mutation at residue 175 were recognized by either CTL, whereas cells infected with a canarypox virus p53 recombinant expressing the 273 R to H mutation were lysed only by CTL A2 149 and not by CTL A2 264. Taken together, these results suggested that the R to H mutation at position 273 of the p53 protein may have prevented formation of the 264–272 peptide epitope.
Figure 1.
Cells expressing Hu p53 that contains an R to H mutation at residue 273 are resistant to lysis by A*0201-restricted CTLs specific for the flanking peptide epitope p53.264–272. A*0201-restricted CTL lines specific for peptide epitopes p63.264–272 (A2 264) (A and C) and p53.149–157 (A2 149) (B and D) were tested for cytotoxicity at the indicated E/T ratios in 5-h (A and B) and 3-h (panel C and D) 51Cr-release assays. Targets in A and B were p53 deficient Saos-2 cells (○), and the same cells transfected with Hu p53 genes harboring mutations at residue 143 (V to A) (•), 175 (R to H) (▵), and 273 (R to H) (▴). Targets in C and D were Saos-2 cells (○), the same cells infected with canarypox virus recombinants expressing Hu p53 without (•) or with mutations at residue 175 (R to H) (▵) and 273 (R to H) (▴), and Saos-2 cells infected with the parental canarypox virus (□).
Acquisition of the R to H Mutation at Residue 273 Promotes Tumor Growth In Vivo by Escape from Immune Recognition.
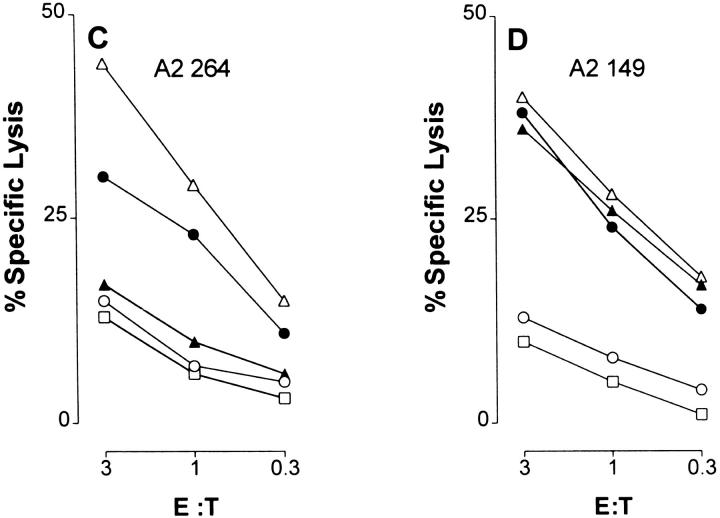
To determine if tumors that express Hu p53 harboring the R to H mutation at residue 273 would be resistant to growth inhibition by A*0201-restricted CTLs specific for p53.264–272, A2/Kb-Tg mice were immunized with rVV strains containing minigenes encoding the sequence of either the Hu p53.149–157 (rVV-ES149) or the Hu p53.264–272 peptide (rVV-ES264). Control mice either received no vaccination or were immunized with rVV encoding the HIV type 1 envelope protein gp160 (rVV-VPE16). 2 wk later, mice were challenged with syngeneic EA2Kb or EA2Kb.1p53 transfectants, the latter of which express the R to H mutation at residue 273 of Hu p53. Preimmunization with rVV-ES149 specifically prevented growth of EA2Kb.1p53 tumors, whereas vaccination with rVV-ES264 did not (Fig. 2). Vaccination with either virus led to equivalent priming of CTLs specific for the respective peptide (data not shown). Tumors that eventually grew out in rVV-ES149 preimmunized mice were found to have lost expression of Hu p53 protein (35). EA2Kb.1p53-derived tumors in untreated or rVV-VPE16–vaccinated A2/Kb-Tg mice progressed with the same growth rate as in rVV-ES264– immunized animals (35). Tumor growth of the parental EA2Kb cells, which express only low levels of murine WT p53 (8), was not prevented by either vaccination (35). These results indicate that tumors harboring the R to H mutation at residue 273 of the Hu p53 protein are able to selectively escape recognition in vivo by CTLs specific for the flanking peptide 264–272.
Figure 2.
Immunization of A2/Kb-Tg mice with the p53.264–272 epitope does not prevent growth of tumor cells expressing both A*0201/Kb and Hu p53 containing an R to H mutation at residue 273. A2/ Kb-Tg mice were immunized intravenously with 5 × 106 PFU of rVV-ES149 (○) or rVV-ES264 (•). 2 wk later (day 0), mice were challenged subcutaneously in the right flank with 5 × 105 EA2Kb.1p53 (273 R to H) tumor cells. Nontreated or rVV-VPE16–vaccinated (encoding the HIV type 1 envelope protein gp160) A2/Kb-Tg mice behaved the same as rVV-ES264–immunized animals; tumors appeared on day 10 after challenge and progressed with the same growth rate. These negative controls are not shown for clarity.
Peptides Derived from 20S Proteasome Degradation of the Synthetic 27-mer Polypeptide p53.256–282 Carrying an R to H Mutation at Residue 273 Are Not Recognized by CTL A2 264.
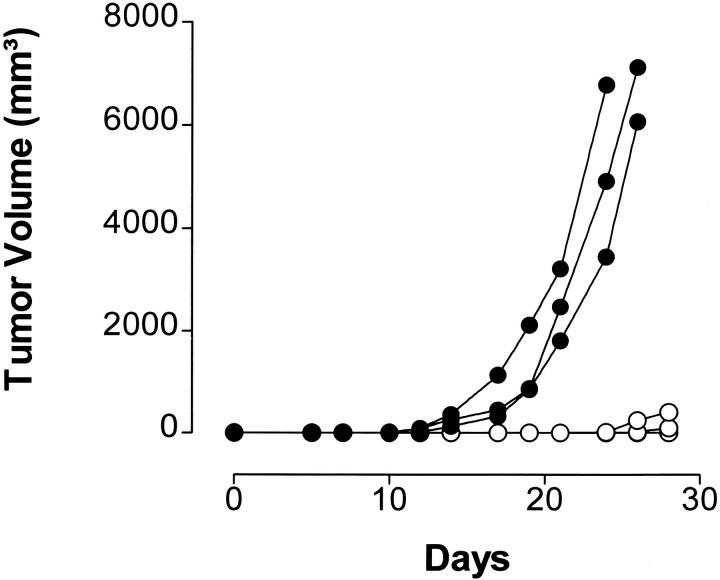
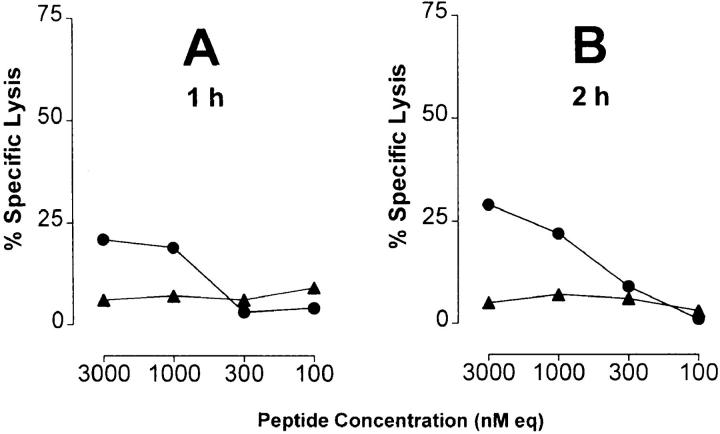
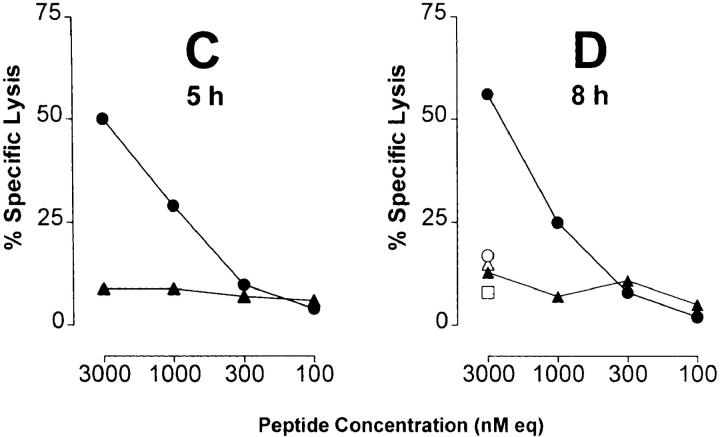
In several reports, it has been demonstrated that the proteasome system represents a major source for the generation of MHC class I ligands (14–16, 28, 36). As demonstrated previously, degradation of p53 is dependent on proteasomal processing (17–20). Accordingly, one mechanism that could interfere with the availability of the 264–272 epitope would be the inability of the proteasome to produce this peptide from the 273 R to H mutant p53. This could arise if the COOH-terminal cleavage site of the 264– 272 epitope was abrogated by the change from R to H at the epitope flanking residue 273. To test this hypothesis, synthetic 27-mer p53 peptides spanning residues 256–282 and containing the 264–272 epitope flanked by either the WT or mutant residue at position 273 were used as substrates for in vitro digestion by purified 20S proteasomes. T2 target cells that express predominantly empty A*0201 molecules (24) were pulsed with the peptide digests and tested for recognition by CTL A2 264. Substantial lysis of T2 targets was observed when cells were pulsed with peptides derived from the 20S proteasome–degraded WT 27-mer, but not with those derived from degraded mutant 27-mer (Fig. 3). Recognition of the synthetic 9-mer p53.264–272 epitope as compared with the WT p53.256–282 peptide digest was 2.4 logs more efficient (2.3–2.7 logs as compared with three independent WT 27-mer peptide digests), suggesting that the 264–272 epitope did not represent an abundant product generated from the WT 27-mer by the 20S proteasome. The selective recognition of WT as opposed to mutant 27-mer peptide digests by CTL A2 264 was dependent on degradation by the 20S proteasome as no lysis was detectable when WT and mutant 27-mer peptides were incubated in the absence of the 20S proteasome. Kinetic studies revealed that WT degradation products recognized by T cells became detectable after 1 and 2 h of incubation of the polypeptide with purified 20S proteasome (Fig. 4, A and B), reached a peak of activity after a period of between 5 and 8 h of proteasomal cleavage (Fig. 4, C and D), and could still be observed after 27 h of proteasome incubation (data not shown). These results indicate that the R to H mutational change at residue 273 affected the ability of the proteasome to release an antigenic peptide from the 27-mer that could be recognized by CTL A2 264.
Figure 3.
Peptide products derived from in vitro 20S proteasome degradation of the synthetic 27-mer polypeptide p53.256–282 carrying an R to H mutation at residue 273 are not recognized by CTLs specific for the nonameric p53.264–272 peptide epitope. Synthetic 27-mer polypeptide substrates spanning residues 256–282 of WT (TLEDSSGNLLGRNSFEVRVCACPGRDR) or mutant (273 R to H) p53 were incubated for 24 h with and without purified 20S proteasome. 51Cr-labeled T2 cells were pulsed under serum-free conditions with the indicated concentrations of WT 27-mer products that had been incubated with (•) and without (○) 20S proteasome, mutant 27-mer products that had been incubated with (▴) and without (▵) 20S proteasome, the synthetic p53.264–272 peptide epitope (▪), or no peptide (□). A*0201-restricted CTLs specific for the p53.264–272 peptide epitope were used as effector cells at an E/T ratio of 20:1 in a 6-h 51Cr-release assay. Peptide concentration is given as the equivalent of the input concentration of 27-mer polypeptide substrates before proteasomal degradation (nM eq). Peptide-pulsed T2 targets were not lysed by CTLs specific for the unrelated p53.149–157 peptide (data not shown).
Figure 4.
Kinetics of 20S proteasomal degradation of WT and mutant 27-mer p53 polypeptide substrates. WT and mutant p53.256–282 peptide substrates were incubated for 1 (A), 2 (B), 5 (C), and 8 (D) h with and without purified 20S proteasome. 51Cr-labeled T2 cells were pulsed under serum-free conditions with the indicated concentrations of WT 27-mer products that had been incubated with (•) and without (○) 20S proteasome, and mutant 27-mer products that had been incubated with (▴) and without (▵) 20S proteasome. Material derived from the incubation of 20S proteasome in the absence of any polypeptide substrate served as another negative control (□). CTL A2 264 were used as effector cells at an E/T ratio of 20:1 in a 6-h 51Cr-release assay. Peptide concentration is given as the equivalent of the input concentration of 27-mer polypeptide substrates before proteasomal degradation (nM eq).
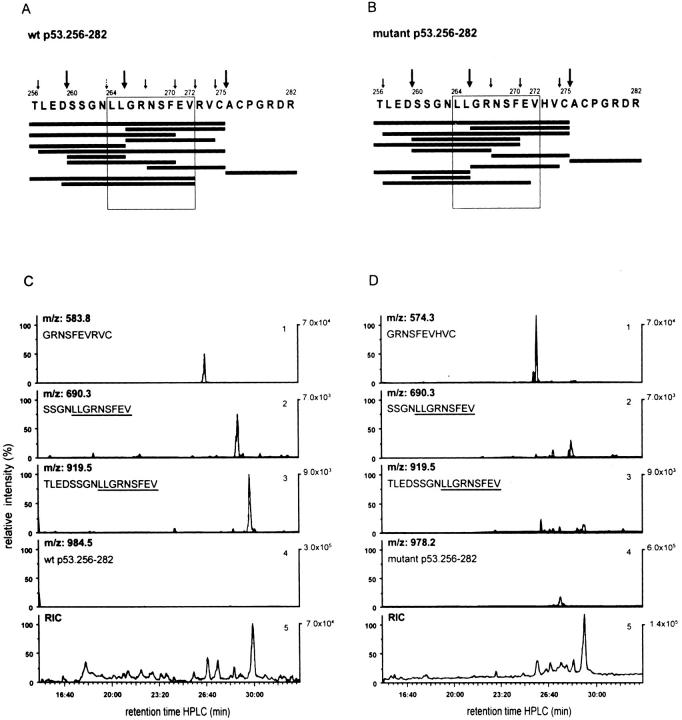
The p53 Mutation at Residue 273 Abrogates the Proteasomal Cleavage Site between p53 Residues 272 and 273.
To prove that the COOH-terminal cleavage site of the 264–272 epitope has indeed been abrogated by the change from R to H at flanking residue 273, the 20S proteasome–degraded (24-h) WT and mutant p53.256–282 peptide products were separated by RP-HPLC, abundant masses were identified online by MS, and the sequence of abundant peptides was confirmed by MS/MS. The majority of the dominant cleavage products generated by the 20S proteasome from the WT and mutant 27-mer polypeptides were comparable in sequence and quantity (Fig. 5, A and B). However, within the range of signal intensities of at least threefold above background, only two peptides, 260–272 and 256–272, both of which contained the COOH-terminal residue 272 of the CTL epitope 264–272, were observed (Fig. 5 A). Notably, the relative signal intensities of peptides 260–272 and 256–272 were found to be 14- and 20-fold higher, respectively, within the WT as opposed to the mutant peptide digests (Fig. 5, C and D, panels 2 and 3). The differential detectability of these peptides within the WT as opposed to the mutant 27-mer peptide digests was not due to differences in the efficiency of proteasomal degradation of either polypeptide. In general, the efficiency of 20S proteasome– induced degradation of either 27-mer peptide was >95% (Fig. 5, C and D, panel 4). Also, comparable amounts (relative signal intensity: twofold more in mutant versus WT) of the peptide 266–275, which did not use the relevant cleavage site between residues 272 and 273, were generated from the WT and mutant 27-mer (Fig. 5, C and D, panel 1).
Figure 5.
The p53 hotspot mutation at residue 273 (R to H) abrogates the proteasomal cleavage site between p53 residues 272 and 273. Bulk peptide products derived after 24 h from 20S proteasome–mediated degradation of the synthetic WT (A and C) and mutant (273 R to H) (B and D) 27-mer polypeptides p53.256–282 covering the A*0201-restricted CTL epitope p53.264–272 (LLGRNSFEV) were separated by RP-HPLC and analyzed online by MS. Abundant peptide products were sequenced by MS/MS. Cleavage products with signal intensities of at least threefold above background and identified by mass (MS) and sequence (MS/MS) are shown in descending order according to their signal intensities (A and B, black bars). The amino acid sequence of WT (A) and mutant (B) peptide substrates with abundant (large arrows) and nonabundant (small arrows) cleavage sites is also presented. The small broken arrow (A) represents the theoretical NH2-terminal cleavage site of the nonameric CTL epitope 264–272. The quantitative comparison of some of the relevant WT (C) and mutant (D) cleavage products is shown by their elution profiles and relative signal intensities as measured by the ion current of double protonated peptide ions. As demonstrated in C and D, peptide 266–275 (GRNSFEV-R/H-VC) represents a dominant product that interferes with the formation of the 264–272 CTL epitope (panel 1). The WT p53 peptides 260–272 (SSGNLLGRNSFEV) and 256–272 (TLEDSSGNLLGRNSFEV) use the COOH-terminal cleavage site of the minimal CTL epitope 264–272 between WT residues 272 and 273 (panels 2 and 3). Panel 4 shows the uncleaved WT and mutant 27-mer substrate peptides left after proteasomal degradation and used for adjusting the scale (percentage of relative intensity). Panel 5 gives the total ion current of the bulk proteasomal degradation products.
These results indicate that the p53 R to H mutation at residue 273 retards proteolytic cleavage by the 20S proteasome between residues 272 and 273. However, we could not detect the peptide representing the CTL epitope 264– 272. The proteolytic generation of the 264–272 epitope from the WT 27-mer at a quantity below the threshold of detectability by MS could have been responsible for such lack of detection, even though the bulk WT 27-mer degradation products had been recognized by CTL A2 264. The observation that the 266–275 peptide was an abundant proteasomal cleavage product of both the WT and mutant 27-mer substrate would support this interpretation, as the release of this peptide would interfere with the generation of the 264–272 CTL epitope (Fig. 5). However, it could also be possible that the peptide 260–272, which used the COOH-terminal cleavage site between residues 272 and 273, represents a precursor peptide that is subsequently trimmed in the cytosol or the ER to the size of the optimal antigenic CTL epitope 264–272. Postproteasomal trimming of the NH2 terminus of precursor peptides has been observed previously (31, 37–40).
The p53.264–272 CTL Epitope Is Generated by Proteasomal Degradation of the WT as Opposed to the Mutant 27-mer Polypeptide Substrate.
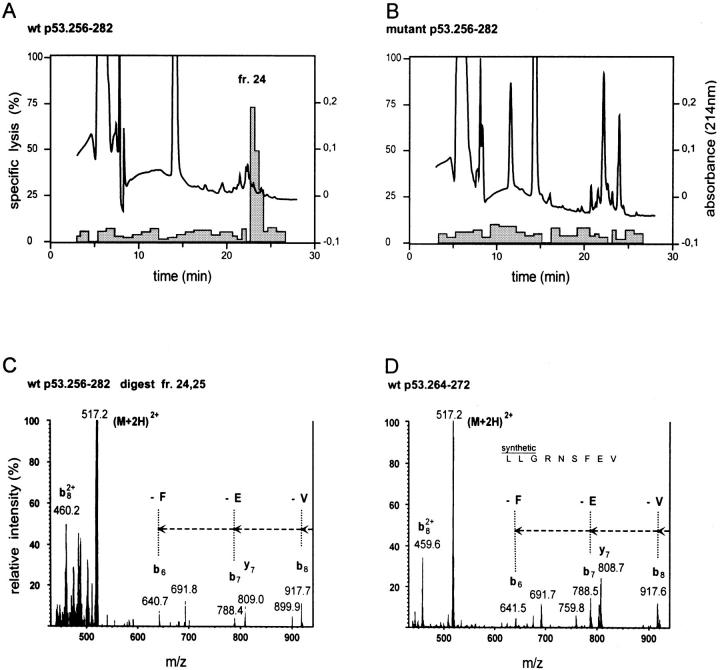
To improve the sensitivity of detection of the 264–272 peptide, the proteasome degraded peptide products (24-h) were fractionated by RP-HPLC. T2 targets were sensitized with the individual HPLC fractions and tested for recognition by CTL A2 264. Only T2 cells that had been pulsed with fractions 24, 25, and 26 of the WT 27-mer peptide digests were recognized by CTL A2 264 (Fig. 6, A and B). The retention time of fraction 24 was matched by that of the synthetic 264–272 peptide (data not shown). WT fractions 24 and 25 were pooled and tested for the presence of the 264–272 peptide by MS/MS. Identical collision-activated dissociation fragments of ions corresponding to the double protonated 264–272 peptide were detected in the pooled WT fractions 24 and 25, and the synthetic 264–272 peptide (Fig. 6, C and D). The relevant collision-activated dissociation fragments were absent in pooled WT fractions 22 and 23, and in pooled mutant fractions 22 and 23 and 24 and 25 (data not shown). These results indicate that the p53.264–272 CTL epitope had been generated directly from the WT 27-mer by the 20S proteasome. Its low abundance is not unique, but a property shared with immunodominant nonameric CTL epitopes detected in 20S proteasome in vitro digests (30, 41– 45). CTL assays of p53-deficient Saos-2 cells infected with rVV expressing minigenes encoding either the 149–157 or the 264–272 peptide without an ER insertion sequence revealed that the 264–272 peptide is efficiently translocated into the ER (Theobald, M., and L.A. Sherman, unpublished observation). These findings demonstrate that the p53 hotspot mutation from R to H at residue 273 prevents the 20S proteasome–mediated COOH-terminal cleavage of the flanking peptide 264–272 and its subsequent presentation by A*0201.
Figure 6.
The p53.264–272 CTL epitope is generated by proteasomal degradation of the WT as opposed to the mutant 27-mer p53.256–282 polypeptide substrate. Bulk 20S proteasome– degraded (24-h) WT and mutant 27-mer peptide products were fractionated by RP-HPLC. 51Cr-labeled T2 target cells were pulsed for 40 min under serum-free conditions with half of each of the WT and mutant HPLC fractions and tested for susceptibility to lysis by CTL A2 264 at an E/T ratio of 20:1 in a 6-h 51Cr-release assay. The HPLC profile (absorbance: —) and the specific lysis (shaded columns) of T2 targets sensitized with individual WT (A) and mutant (B) HPLC fractions is shown. WT fractions 24 and 25 were pooled and tested by MS/MS for detection of the p53.264–272 (LLGRNSFEV) peptide epitope. Ions of m/z = 517.2 corresponding to the double protonated 264–272 9-mer peptide were fragmented by argon atoms. Collision-activated dissociation fragments of m/z 517.2 and derived from the pooled WT fractions 24 and 25 (C) were compared with those obtained after argon atom–mediated fragmentation of the synthetic 264–272 peptide (D). In particular, fragments b8, b7, and b6, lacking the COOH-terminal residues V, E, and F, respectively, were detectable in both the pooled WT fractions 24 and 25 (C) and the synthetic 264–272 9-mer peptide (D).
The more abundant flanking peptide 256–272 eluted in WT fraction 26, which was recognized poorly by CTL A2 264 (Figs. 5 A and 6 A), making it unlikely to be an independent CTL epitope. However, it is of interest that due to the HPLC conditions used, the other abundant flanking peptide 260–272 (Fig. 5 A) was identified by MS/MS to be present in the pooled WT fractions 24 and 25 (data not shown). However, the A*0201-binding affinity of the longer peptide 260–272 was 2.5-fold lower than that of the 9-mer 264–272, and it was found to be recognized 30– 100-fold less efficiently by CTL A2 264 (data not shown). These results favor the view that 260–272 may represent a precursor peptide that is eventually trimmed in the cytosol or the ER to the 9-mer rather than functioning as an independent CTL epitope.
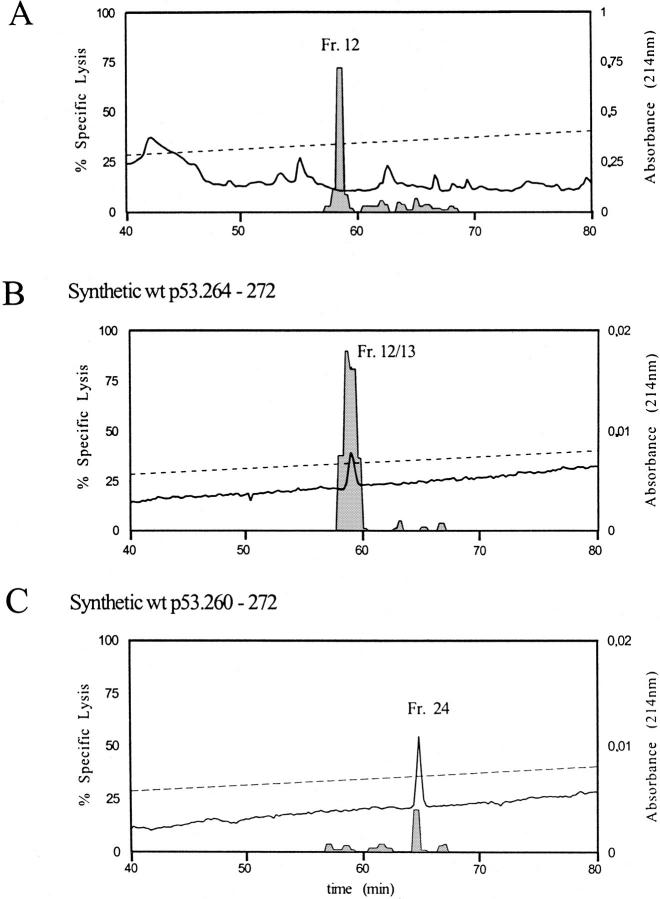
The Hu WT p53.264–272 Peptide Is the Natural Epitope Presented by A*0201 and Recognized by CTL A2 264.
To identify the natural peptide epitope presented by A*0201 and recognized by CTL A2 264, peptides were extracted from class I MHC molecules of Saos-2/143 p53 transfectants. As the 9-mer (264–272) and 13-mer (260–272) p53 peptides had an almost identical retention time under the HPLC conditions used for purifying the 20S proteasomal peptide digests, a shallower elution gradient was applied for the RP-HPLC fractionation of natural peptide extracts as well as synthetic 9-mer and 13-mer peptides. The pool of natural peptides extracted from Saos-2/143 cells was chromatographed by HPLC and 4-min fractions collected and assayed. To obtain final resolution, antigenic fractions 7 and 8 were pooled and half of the pool was rechromatographed. At the relevant retention time, 0.5 min fractions were collected in order to discriminate between the 264– 272 and the 260–272 peptides. T2 cells sensitized with individual HPLC fractions served as targets for CTL A2 264. As shown in Fig. 7 A, the peak of CTL activity obtained after rechromatography of fractions 7/8 and derived from the natural peptide extract was detected in fraction 12. This HPLC fraction had the same conductivity and retention time as compared with the antigenic fractions (12 and 13) derived from the HPLC separation of the synthetic 264– 272 9-mer peptide (Fig. 7 B). In contrast, the antigenic activity of the synthetic 260–272 13-mer peptide eluted in fraction 24 and its retention time differed from the 264– 272 peptide by 6 min under the conditions used for rechromatography (Fig. 7 C). The lower amount of lysis obtained with fraction 24 of the synthetic 260–272 peptide again indicated less efficient crossrecognition by CTL A2 264. Pulsing T2 targets with a 10-fold higher amount (2 μl) of fraction 24 of the synthetic 260–272 peptide resulted in a substantial increase of lysis (100% specific lysis by CTL A2 26; (data not shown). However, as little as 0.02 μl of either fraction 12 or 13 of the synthetic 264–272 peptide was sufficient to detect substantial lysis (fractions 12 and 13: 50 and 61% specific lysis, respectively) of peptide-pulsed T2 cells by CTL A2 264 (data not shown). No antigenic activity could be observed with fraction 24 after rechromatography of the natural peptide extract (Fig. 7 A). These results indicate that the Hu WT p53.264–272 as opposed to the longer 260–272 peptide is the natural epitope presented by A*0201 and recognized by CTL A2 264. However, as the 13-mer 260–272 (and the 17-mer 256–272) peptide(s) are the more abundant products released from the WT p53 sequence by the 20S proteasome (Fig. 5, A and C), it is possible that these peptides undergo NH2-terminal trimming in the cytosol or ER (37–40) and represent precursor peptides that may thereby contribute substantially to the availability of the low-abundant WT p53 processing product 264–272 for its assembly with nascent A*0201 molecules in the ER.
Figure 7.
The Hu WT p53.264–272 peptide is the natural epitope presented by A*0201 for recognition by CTL A2 264. Natural peptides were extracted from class I molecules of Saos-2/143 p53 transfectants. Synthetic p53.264– 272 and 260–272 peptides and the natural peptide extract were fractionated by RP-HPLC. Individual HPLC fractions were used to sensitize T2 targets and to reconstitute lysis by CTL A2 264. Two antigenic HPLC fractions obtained from the natural peptide extract had an almost identical retention time and conductivity as compared with those antigenic HPLC fractions derived from the synthetic p53.264–272 and p53.260–272 peptides. As 4-min HPLC fractions were collected at the relevant retention time, the peak of CTL activity occurred in almost identical HPLC fractions, although the retention time of the synthetic p53.264–272 and p53.260–272 peptides differed from each other by ∼4 min. To separate either peptide from one another and identify the natural antigenic peptide, half of the pooled antigenic fractions of the HPLC-separated natural peptide extract (A) as well as 60 pmol of synthetic 264–272 (B) and 260– 272 (C) peptides were used for rechromatography by RP-HPLC. HPLC conditions remained almost the same, yet 0.5-min fractions (25 μl/fraction) were collected at the relevant retention time in order to discriminate between the 264–272 and the 260–272 peptides. 51Cr-labeled T2 target cells were pulsed for 40 min under serum-free conditions with individual HPLC fractions (fractions derived from the rechromatography of natural peptides: 18 μl; fractions 8, 19, 20, and 29 derived from synthetic p53 peptides: 2 μl; fractions 9–18 and 21–28 derived from synthetic p53 peptides: 0.02 μl, 0.2 μl, and 2 μl) and tested for susceptibility to lysis by CTL A2 264 at an E/T of 13:1 in a 6-h 51Cr-release assay. The HPLC profile (absorbance: —) (A: first HPLC separation of natural peptide extracts) and the specific lysis (shaded columns) of T2 targets sensitized with individual HPLC fractions obtained from the rechromatography of natural peptides (A), 2 μl (fractions 8, 19, 20, and 29), and 0.2 μl (fractions 9–18 and 21–28) of HPLC fractions obtained from the synthetic p53.264–272 (B) and p53.260–272 (C) peptides is shown.
Several recent studies have focused on the mutational alteration within a peptide epitope to demonstrate a way by which viruses can escape recognition by epitope-specific CTLs (46–50). This is generally believed to involve peptide binding to the MHC class I molecule, or recognition by the TCR (46–50). However, in a recent report it was shown that a mutation within a viral peptide epitope affected the production of precursor peptides by proteasomal degradation through the introduction of a dominant new proteasomal cleavage site within the viral epitope (31). Based on studies that evaluated the effect of synthetically altering residues flanking a peptide epitope on its processing, it would be predicted that point mutation outside of the epitope could also interfere with its presentation (30, 36, 44, 51–56). To our knowledge, our studies represent the first report of such a naturally occurring mutation outside the epitope that alters CTL recognition of a flanking peptide. In fact, the single mutation described here interferes not only with the proteasomal processing of putative precursor peptides, but also with the generation of the optimal CTL epitope itself. It is at present unclear why the R to H mutation interferes with the COOH-terminal proteasomal cleavage. It is possible that a change in charge and size of the flanking residue influences the COOH-terminal cleavage site used by the proteasome. However, as the possibility that a particular site will be preferred for proteasomal cleavage is dependent on amino acid sequences within the epitope (31, 51), as well as on flanking residues both up- and downstream (30, 36, 44, 51–56), it is likely that the rules by which ligand generation is governed are more complex and not yet understood.
Treatment of cells with IFN-γ has been reported to alter the processing kinetics, quantity, and quality of MHC class I–bound peptide ligands by affecting the expression both of the IFN-γ–inducible proteasomal subunits (LMP2, LMP7, and MECL-1) and the IFN-γ–inducible PA28-α/β activator complex (11S regulator) of the 20S proteasome (28, 29, 41, 43, 57–69). Exposing Saos-2/273 (R to H) p53 transfectants to IFN-γ (10 ng/ml for 20 h) resulted in an only partial reconstitution of lysis by CTL A2 264 (20% specific lysis of nonpretreated Saos-2/273 cells versus 31% specific lysis of IFN-γ pretreated Saos-2/273 targets at an E/T of 20:1) (Häussler, A., and M. Theobald, unpublished observation). The molecular basis of this IFN-γ–mediated partial reconstitution of Saos-2/273 killing by CTL A2 264 on the level of both the proteasomal subunit composition (LMP2, LMP7, and MECL-1) and the PA28 activator (11S regulator) expression is currently under investigation.
Based on these results, it may be concluded not only that the p53 mutation at residue 273 is associated with malignant transformation, but that it can also affect CTL recognition in vitro and in vivo of tumor cells carrying this mutation. It is tempting to speculate that cells harboring this mutation may have a competitive edge for growth in A*0201+ individuals by evading CTL recognition (70). Several other reports have shown that vaccination of mice with the intact p53 protein expressed in a viral vector or with p53 peptides pulsed onto dendritic cells can prevent growth in vivo of tumors expressing high levels of p53 (5, 7). Furthermore, recent studies have demonstrated the ability of p53.264–272 epitope–specific Hu CTL to lyse squamous cancer cells (6). Thus, knowledge not only of the antigenic epitopes of p53, but also of the biological role of their sequence context and its modulation by frequently arising mutations may explain disease progression, and may assist in the design of efficacious cancer vaccines.
Acknowledgments
We thank Jack Bennink and Jonathan Yewdell for providing recombinant vaccinia viruses, and Ilse Drung (Humboldt University) for excellent technical assistance.
Supported by grants from the Deutsche Forschungsgemeinschaft to M. Theobald (SFB 432 A3), T. Ruppert and U.H. Koszinowski (SFB 469), and P.-M. Kloetzel (DFG KL 427/9-2); the “Stiftung Rheinland-Pfalz für Innovation” to M. Theobald; the European Community to U.H. Koszinowski (PL 960505) and P.-M. Kloetzel (BMH 4-2-CT-96-0447); and the National Institutes of Health to L.A. Sherman (CA-57855 and CA-25803). M. Theobald is a fellow of the Stipendienprogramm Infektionsbiologie provided by the German Cancer Research Center (DKFZ) and funded by the German Ministry for Education and Research (BMBF).
Abbreviations used in this paper
- A*0201
HLA-A*0201
- β2m
β2 microglobulin
- ER
endoplasmic reticulum
- Hu
human
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- m/z
mass/charge
- RP
reversed phase
- rVV
recombinant vaccinia virus
- Tg
transgenic
- WT
wild type
References
- 1.Levine AJ, Momand J, Finlay CA. The p53 tumor suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in p53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 4.Theobald M, Biggs J, Dittmer D, Levine AJ, Sherman LA. Targeting p53 as a general tumor antigen. Proc Natl Acad Sci USA. 1995;92:11993–11997. doi: 10.1073/pnas.92.26.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth J, Dittmer D, Rea D, Tartaglia J, Paoletti E, Levine AJ. p53 as a target for cancer vaccines: recombinant canarypox virus vectors expressing p53 protect mice against lethal tumor challenge. Proc Natl Acad Sci USA. 1996;93:4781–4786. doi: 10.1073/pnas.93.10.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Röpke M, Hald J, Guldberg P, Zeuthen J, Norgaard L, Fugger L, Svejgaard A, Van Der Burg S, Nijman HW, Melief CJM, Claesson MH. Spontaneous human squamous cell carcinomas are killed by a human cytotoxic T lymphocyte clone recognizing a wild-type p53-derived peptide. Proc Natl Acad Sci USA. 1996;93:14704–14707. doi: 10.1073/pnas.93.25.14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayordomo JI, Loftus DJ, Sakamoto H, De Cesare CM, Appasamy PM, Lotze MT, Storkus WJ, Appella E, DeLeo AB. Therapy of murine tumors with p53 wild-type and mutant sequence peptide-based vaccines. J Exp Med. 1996;183:1357–1365. doi: 10.1084/jem.183.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theobald M, Biggs J, Hernandez J, Lustgarten J, Labadie C, Sherman LA. Tolerance to p53 by A2.1-restricted cytotoxic T lymphocytes. J Exp Med. 1997;185:833–841. doi: 10.1084/jem.185.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vierboom MPM, Nijman HW, Offringa R, van der Voort EIH, van Hall T, van den Broek L, Fleuren GJ, Kenemans P, Kast WM, Melief CJM. Tumor eradication by wild-type p53-specific cytotoxic T lymphocytes. J Exp Med. 1997;186:695–704. doi: 10.1084/jem.186.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houbiers JGA, Nijman HW, van der Burg SH, Drijfthout JW, Kenemans P, van de Velde CJ, Brand A, Momburg F, Kast WM, Melief CJM. In vitro induction of human cytotoxic T lymphocyte responses against peptides of mutant and wild-type p53. Eur J Immunol. 1993;23:2072–2077. doi: 10.1002/eji.1830230905. [DOI] [PubMed] [Google Scholar]
- 11.Nijman HW, Houbiers JGA, van der Burg SH, Vierboom MPM, Kenemans P, Kast WM, Melief CJM. Characterization of cytotoxic T lymphocyte epitopes of a self-protein, p53, and a non–self-protein, influenza matrix: relationship between major histocompatibility complex peptide binding affinity and immune responsiveness to peptides. J Immunother. 1993;14:121–126. [PubMed] [Google Scholar]
- 12.Nijman HW, van der Burg SH, Vierboom MPM, Houbiers JGA, Kast WM, Melief CJM. p53, a potential target for tumor-directed T cells. Immunol Lett. 1994;40:171–178. doi: 10.1016/0165-2478(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 13.Röpke M, Regner M, Claesson MH. T-cell mediated cytotoxicity against p53-protein derived peptides in bulk and limiting dilution cultures of healthy donors. Scand J Immunol. 1995;42:98–103. doi: 10.1111/j.1365-3083.1995.tb03631.x. [DOI] [PubMed] [Google Scholar]
- 14.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 15.Groettrup M, Soza A, Kuckelkorn U, Kloetzel P-M. Peptide antigen production by the proteasome: complexity provides efficiency. Immunol Today. 1996;17:429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 16.Lehner PJ, Cresswell P. Processing and delivery of peptides presented by MHC class I molecules. Curr Opin Immunol. 1996;8:59–67. doi: 10.1016/s0952-7915(96)80106-3. [DOI] [PubMed] [Google Scholar]
- 17.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 18.Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 19.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 20.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher TN, Kantesaria DV, Heemels MT, Ashton-Rickardt PG, Shepherd JC, Früh K, Yang Y, Peterson PA, Tonegawa S, Ploegh HL. Peptide length and sequence specificity of the mouse TAP1/TAP2 translocator. J Exp Med. 1994;179:533–540. doi: 10.1084/jem.179.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momburg F, Roelse J, Hämmerling GJ, Neefjes JJ. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med. 1994;179:1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Endert PM, Tampe R, Meyer TH, Tisch R, Bach JF, McDevitt HO. A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity. 1994;1:491–500. doi: 10.1016/1074-7613(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 24.DeMars R, Rudersdorf R, Chang C, Petersen J, Strandtmann J, Korn N, Sidwell B, Orr HT. Mutations that impair a posttranscriptional step in expression of HLA-A and -B antigens. Proc Natl Acad Sci USA. 1985;82:8183–8187. doi: 10.1073/pnas.82.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, Levine AJ. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 26.Anton LC, Yewdell JW, Bennink JR. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J Immunol. 1997;158:2535–2542. [PubMed] [Google Scholar]
- 27.Earl PL, Hugin AW, Moss B. Removal of cryptic poxvirus transcription termination signals from the human immunodeficiency virus type 1 envelope gene enhances expression and immunogenicity of a recombinant vaccinia virus. J Virol. 1990;64:2448–2451. doi: 10.1128/jvi.64.5.2448-2451.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuckelkorn U, Frentzel S, Kraft R, Kostka S, Groettrup M, Kloetzel P-M. Incorporation of major histocompatibility complex–encoded subunits LMP2 and LMP7 changes the quality of the 20S proteasome polypeptide processing products independent of interferon-γ. Eur J Immunol. 1995;25:2605–2611. doi: 10.1002/eji.1830250930. [DOI] [PubMed] [Google Scholar]
- 29.Dick TP, Ruppert T, Groettrup M, Kloetzel P-M, Kuehn L, Koszinowski UH, Stefanovic S, Schild H, Rammensee H-G. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell. 1996;86:253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 30.Eggers M, Boes-Fabian B, Ruppert T, Kloetzel P-M, Koszinowski UH. The cleavage preference of the proteasome governs the yield of antigenic peptides. J Exp Med. 1995;182:1865–1870. doi: 10.1084/jem.182.6.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ossendorp F, Eggers M, Neisig A, Ruppert T, Groettrup M, Sijts A, Mengede E, Kloetzel P-M, Neefjes J, Koszinowski U, Melief C. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- 32.Storkus WJ, Zeh HJ, III, Maeurer MJ, Salter RD, Lotze MT. Identification of melanoma peptides recognized by class I restricted tumor infiltrating T lymphocytes. J Immunol. 1993;151:3719–3727. [PubMed] [Google Scholar]
- 33.Itoh T, Storkus WJ, Gorelik E, Lotze MT. Partial purification of murine tumor-associated peptide epitopes common to histologically distinct tumors, melanoma and sarcoma, that are presented by H-2Kb molecules and recognized by CD8+tumor-infiltrating lymphocytes. J Immunol. 1994;153:1202–1215. [PubMed] [Google Scholar]
- 34.Lustgarten J, Theobald M, Labadie C, LaFace D, Peterson P, Disis ML, Cheever MA, Sherman LA. Identification of Her-2/Neu CTL epitopes using double transgenic mice expressing HLA-A2.1 and human CD8. Hum Immunol. 1997;52:109–118. doi: 10.1016/S0198-8859(96)00292-3. [DOI] [PubMed] [Google Scholar]
- 35.Sherman LA, Theobald M, Morgan D, Hernandez J, Bacik I, Yewdell J, Bennink J, Biggs J. Strategies for tumor elimination by cytotoxic T lymphocytes. Crit Rev Immunol. 1998;18:47–54. doi: 10.1615/critrevimmunol.v18.i1-2.60. [DOI] [PubMed] [Google Scholar]
- 36.Shastri N, Serwold T, Gonzalez F. Presentation of endogenous peptide/MHC class I complexes is profoundly influenced by specific C-terminal flanking residues. J Immunol. 1995;155:4339–4346. [PubMed] [Google Scholar]
- 37.Roelse J, Gromme M, Momburg F, Hämmerling G, Neefjes J. Trimming of TAP-translocated peptides in the endoplasmic reticulum and in the cytosol during recycling. J Exp Med. 1994;180:1591–1597. doi: 10.1084/jem.180.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder HL, Yewdell JW, Bennink JR. Trimming of antigenic peptides in an early secretory compartment. J Exp Med. 1994;180:2389–2394. doi: 10.1084/jem.180.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elliott T, Willis A, Cerundolo V, Townsend A. Processing of major histocompatibility class I–restricted antigens in the endoplasmic reticulum. J Exp Med. 1995;181:1481–1491. doi: 10.1084/jem.181.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craiu A, Akopian T, Goldberg A, Rock KL. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I–presented peptide. Proc Natl Acad Sci USA. 1997;94:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boes B, Hengel H, Ruppert T, Multhaup G, Koszinowski UH, Kloetzel P-M. Interferon γ stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villanueva MS, Fischer P, Feen K, Pamer EG. Efficiency of MHC class I antigen processing: a quantitative analysis. Immunity. 1994;1:479–489. doi: 10.1016/1074-7613(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 43.Groettrup M, Ruppert T, Kuehn L, Seeger M, Standera S, Koszinowski U, Kloetzel P-M. The interferon-gamma-inducible 11S regulator (PA28) and the LMP2/ LMP7 subunits govern the peptide production by the 20S proteasome in vitro. J Biol Chem. 1995;270:23808–23815. doi: 10.1074/jbc.270.40.23808. [DOI] [PubMed] [Google Scholar]
- 44.Niedermann G, Butz S, Ihlenfeldt HG, Grimm R, Lucchiari M, Hoschützky H, Jung G, Maier B, Eichmann K. Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity. 1995;2:289–299. doi: 10.1016/1074-7613(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 45.Niedermann G, King G, Butz S, Birsner U, Grimm R, Shabanowitz J, Hunt DF, Eichmann K. The proteolytic fragments generated by vertebrate proteasomes: structural relationships to major histocompatibility complex class I binding peptide. Proc Natl Acad Sci USA. 1996;93:8572–8577. doi: 10.1073/pnas.93.16.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koup RA. Virus escape from CTL recognition. J Exp Med. 1994;180:779–782. doi: 10.1084/jem.180.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Campos-Lima PO, Levitsky V, Brooks J, Lee SP, Hu LF, Rickinson AB, Masucci MG. T cell responses and virus evolution: loss of HLA A11-restricted CTL epitopes in Epstein-Barr virus isolates from highly A11-positive populations by selective mutation of anchor residues. J Exp Med. 1994;179:1297–1305. doi: 10.1084/jem.179.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goulder PJ, Sewell AK, Lalloo DG, Price DA, Whelan JA, Evans J, Taylor GP, Luzzi G, Giangrande P, Phillips RE, McMichael AJ. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A*0201 are influenced by epitope mutation. J Exp Med. 1997;185:1423–1433. doi: 10.1084/jem.185.8.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moskophidis D, Zinkernagel RM. Immunobiology of cytotoxic T-cell escape mutants of lymphocytic choriomeningitis virus. J Virol. 1995;69:2187–2193. doi: 10.1128/jvi.69.4.2187-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahn YS, Hahn CS, Braciale VL, Braciale TJ, Rice CM. CD8+T cell recognition of an endogenously processed epitope is regulated primarily by residues within the epitope. J Exp Med. 1992;176:1335–1341. doi: 10.1084/jem.176.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenlohr LC, Yewdell JW, Bennink JR. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J Exp Med. 1992;175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Del Val M, Schlicht H-J, Ruppert T, Reddehase MJ, Koszinowski UH. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66:1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 54.Bergmann CC, Tong L, Cua R, Sensintaffar J, Stohlman S. Differential effects of flanking residues on presentation of epitopes from chimeric peptides. J Virol. 1994;68:5306–5310. doi: 10.1128/jvi.68.8.5306-5310.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergmann CC, Yao Q, Ho C-K, Buckwold SL. Flanking residues alter antigenicity and immunogenicity of multi-unit CTL epitopes. J Immunol. 1996;157:3242–3249. [PubMed] [Google Scholar]
- 56.Yellen-Shaw AJ, Wherry EJ, Dubois GC, Eisenlohr LC. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J Immunol. 1997;158:3227–3234. [PubMed] [Google Scholar]
- 57.Arnold D, Driscoll P, Androlewicz M, Hughes E, Cresswell P, Spiess TA. Proteasome subunits encoded in the MHC are not generally required for the processing of peptides bound by MHC class I molecules. Nature. 1992;360:171–173. doi: 10.1038/360171a0. [DOI] [PubMed] [Google Scholar]
- 58.Momburg F, Ortiz-Navarrete V, Neefjes J, Goulmy E, Vandewal Y, Spits H, Powis SJ, Butcher GW, Howard JC, Walden P, Hämmerling GJ. The proteasome subunits encoded by the major histocompatibility complex are not essential for antigen presentation. Nature. 1992;360:174–177. doi: 10.1038/360174a0. [DOI] [PubMed] [Google Scholar]
- 59.Driscoll P, Brown M, Finley D, Monaco JJ. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365:252–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 60.Gaczynska M, Rock KL, Spiess TA, Goldberg AL. γ-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 61.Gaczynska M, Rock KL, Spiess TA, Goldberg AL. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex–encoded genes for LMP-2 and LMP-7. Proc Natl Acad Sci USA. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yewdell JW, Lapham C, Bacik I, Spiess TA, Bennink JR. MHC-encoded proteasome subunits LMP-2 and LMP-7 are not required for efficient antigen presentation. J Immunol. 1994;152:1163–1170. [PubMed] [Google Scholar]
- 63.Van Kaer L, Ashton-Rickardt PG, Eichelberger M, Gaczynska M, Nagashima K, Rock KL, Goldberg AL, Doherty PC, Tonegawa S. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 64.Fehling HJ, Swat W, Laplace C, Kühn R, Rajewsky K, Müller U, von Boehmer H. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 65.Cerundolo V, Kelly A, Elliott T, Trowsdale J, Townsend A. Genes encoded in the major histocompatibility complex affecting the generation of peptides for TAP transport. Eur J Immunol. 1995;25:554–562. doi: 10.1002/eji.1830250238. [DOI] [PubMed] [Google Scholar]
- 66.Sibille C, Gould KG, Willard-Gallo K, Thomson S, Rivett AJ, Powis SJ, Butcher GW, De Baetselier P. LMP2+ proteasomes are required for the presentation of specific antigens to cytotoxic T lymphocytes. Curr Biol. 1995;5:923–930. doi: 10.1016/s0960-9822(95)00182-5. [DOI] [PubMed] [Google Scholar]
- 67.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel P-M. A third interferon-γ–induced subunit exchange in the 20S proteasome. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 68.Groettrup M, Standera S, Stohwasser R, Kloetzel PM. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc Natl Acad Sci USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niedermann G, Grimm R, Geier E, Maurer M, Realini C, Gartmann C, Soll J, Omura S, Rechsteiner MC, Baumeister W, Eichmann K. Potential immunocompetence of proteolytic fragments produced by proteasomes before evolution of the vertebrate immune system. J Exp Med. 1997;185:209–220. doi: 10.1084/jem.186.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiedenfeld EA, Fernandez-Vina M, Berzofsky JA, Carbone DP. Evidence for selection against human lung cancers bearing p53 missense mutations which occur within the HLA A*0201 peptide consensus motif. Cancer Res. 1994;54:1175–1177. [PubMed] [Google Scholar]