Abstract
Dendritic cells (DCs), nature's adjuvant, must mature to sensitize T cells. However, although the maturation process is essential, it is not yet fully understood at the molecular level. In this study, we investigated the course of expression of the unique hypusine-containing protein eukaryotic initiation factor 5A (eIF-5A), which is part of a particular RNA nuclear export pathway, during in vitro generation of human DCs. We show that eIF-5A expression is significantly upregulated during DC maturation. Furthermore, an inhibitor of the hypusine modification, GC7 (N 1-guanyl-1,7-diaminoheptane), prevents CD83 surface expression by apparently interfering with nucleocytoplasmic translocation of the CD83 mRNA and, importantly, significantly inhibits DC-mediated T lymphocyte activation. The data presented suggest that CD83 mRNA is transported from the nucleus to the cytoplasm via a specific nuclear export pathway and that hypusine formation appears to be essential for the maturation of functional DCs. Therefore, pharmacological interference with hypusine formation may provide a new possibility to modulate DC function.
Keywords: dendritic cells, CD83, hypusine, eIF-5A, nuclear export
Introduction
Dendritic cells (DCs) are specialized to sensitize helper and killer T cells and thus act as “nature's adjuvant” in inducing T cell–mediated immunity 1. Immature DCs capture antigens in the periphery but lack full T cell–stimulatory capacity. In the presence of appropriate stimuli (such as microbial products and/or inflammatory cytokines), the DCs then mature. DCs upregulate T cell adhesion and costimulatory molecules as well as selected chemokine receptors that guide DC migration into secondary lymphoid organs for priming of antigen-specific T cells 2. DCs are defined by their potent T cell–stimulatory capacity (e.g., in the allo-MLR) as well as a characteristic morphology (nonadherent cells with motile veils) and phenotype (upregulation of CD86 and de novo expression of CD83; references 3 and 4).
DCs can be generated in vitro either from rare proliferating CD34+ or frequent, nonproliferating CD14+ monocytic precursors 5 6 7 8. The generation of DCs from monocytes under the use of GM-CSF and IL-4 yields homogenous DC progenitors that are well suited for studying the maturation of DCs in vitro 9.
Eukaryotic initiation factor 5A (eIF-5A) is the only cellular protein known to contain the unusual amino acid hypusine, a modification that appears to be required for cell proliferation 10 11. The hypusine modification is a spermidine-dependent posttranslational reaction that is catalyzed by two enzymes. This includes the transfer of the aminobutyl moiety of spermidine to the ε-NH2 group of lysine at position 50 in eIF-5A by deoxyhypusine synthase 12 13 14. The intermediate that is generated is subsequently hydroxylated by deoxyhypusine hydroxylase 15, resulting in the active form of eIF-5A. Although eIF-5A was originally designated as an “initiation factor” 16 17, more recent in vitro and in vivo data have demonstrated that eIF-5A is not an initiator of protein translation 18 19 20. In fact, the subsequent finding that eIF-5A is a cellular cofactor of HIV-1 Rev and HTLV-I Rex transactivator proteins suggested an entirely different eIF-5A activity 21 22 23 24. Both Rev and Rex are nucleocytoplasmic shuttle proteins that mediate the nuclear export of incompletely spliced and unspliced viral mRNAs 25 26. Thus, eIF-5A appears to be part of a specific nuclear export pathway that is exploited by the Rev/Rex class of retroviral RNA transport factors. This notion is further supported by the finding that, in yeast, eIF-5A affects the decay of specific mRNAs that are transported from the nucleus to their cytoplasmic site of degradation 20 27 and, more recently, that eIF-5A is a high-copy suppressor of transport-deficient TATA binding protein mutants 28. Furthermore, the eIF-5A protein itself has been shown to accumulate at the nucleoplasmic site of nuclear pore complexes to interact with the general nuclear export receptor CRM1 and to translocate from the nucleus to the cytoplasm in mammalian cells 29.
Investigation of eIF-5A mRNA levels in human cells revealed that eIF-5A is constitutively expressed in cell lines as well as in various tissues 30. In contrast, the eIF-5A gene appears to be subject to distinct regulation in primary lymphoid cells. In particular, eIF-5A gene expression is constitutively low but inducible with T lymphocyte–specific stimuli in human PBMCs 30. The combined data suggest that the hypusine-containing protein eIF-5A fulfills a specific and presumably essential function during activation and/or proliferation of primary human blood cells.
In this work, we investigated the expression level of the hypusine-containing protein eIF-5A during maturation of primary human DCs. Using an inhibitor of hypusine modification, we are able to show that formation of hypusine is required for the expression of the DC-specific molecule CD83 and the full stimulatory activity of mature DCs, demonstrating a potentially novel approach by which to interfere with DC function.
Materials and Methods
Cell Culture Medium.
Cells were cultured using a standard medium (referred to as 1% human plasma medium), which consisted of RPMI 1640 (BioWhittaker) supplemented with glutamine (300 μg/ml) (BioWhittaker), penicillin/streptomycin (20 μg/ml), 10 mM Hepes, pH 7.5 (Sigma-Aldrich), and 1% heat-inactivated (56°C; 30 min) human plasma from a single AB donor, obtained from the Department of Transfusion Medicine, Erlangen, Germany.
Generation of DCs.
PBMCs (5 × 107) were isolated from buffy coats by sedimentation in Ficoll-hypaque (Amersham Pharmacia Biotech) and seeded onto IgG-coated (10 μg/ml γ-globulin from Cohn fraction; Sigma-Aldrich) 100 mm-culture dishes and incubated at 37°C in 5% CO2. After 1 and 7 h of incubation, nonadherent cell fractions were harvested, and the remaining adherent cells were further cultured in 1% human plasma medium supplemented with the cytokines GM-CSF (800 U/ml) and IL-4 (1,000 U/ml). Fresh medium (5 ml) containing 4,000 U GM-CSF and 5,000 U IL-4 was added to the culture dish at day 3 of this incubation period. On day 4 or 5, nonadherent cells were collected, counted, and transferred into new dishes at a density of 0.3–0.5 × 105 cells/ml. For final DC maturation, 1% human plasma medium was supplemented with TNF-α (25 ng/ml), prostaglandin E2 (PGE2; 1 ng/ml), GM-CSF (400 U/ml), and IL-4 (500 U/ml) 31. In the studies using the hypusine inhibitor GC7 (N 1-guanyl-1,7-diaminoheptane), immature DCs were pretreated at day 4 or 5 with GC7 at a concentration of 1 μM for 10 min before addition of the final maturation medium.
Cytokines and GC7.
Recombinant human (rh)GM-CSF and GC7 32 were obtained from Novartis Research Institute, rhIL-4 from Genzyme, rhTNF-α from Boehringer, and PGE2 from Cayman Chemical.
FACS® Analyses.
For flow cytometry analyses, mAbs recognizing the following antigens were used: CD83 (Immunotech), CD80, CD86, CD13, CD68, MHC class I, MHC class II, and CD95 (Becton Dickinson). The isotype controls IgG1a and IgG2b were obtained from Becton Dickinson and were run in parallel. Cell populations were phenotyped with the panel of mAbs listed above and analyzed on a FACScan™ (Becton Dickinson) as described previously 8. Nonviable cells were gated out on the basis of their light scattering properties.
Western Blot Analyses.
Cells (0.5 × 106) were harvested on days 4, 5, 6, and 7 of the DC generation procedure, washed with PBS, and solubilized in gel loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.1% bromophenol blue). After SDS-PAGE, the separated proteins were transferred onto nitrocellulose membranes, and specific antibodies were used to detect eIF-5A 33, histone H1 (StressGen Biotechnologies), and CD83 as described previously 29.
RNA Analyses.
CD83 and eIF-5A mRNA levels were analyzed by PCR. Total cellular RNA from 106 cells was isolated at different time points of DC maturation using TRIzol™ Reagent (GIBCO BRL). Subsequently, the RNA was reverse transcribed into single-stranded cDNA using AMV reverse transcriptase according to the manufacturer's protocol (Roche Molecular Biochemicals). Serial dilutions of the cDNA preparations were subjected to AmpliTaq® (Perkin-Elmer) DNA polymerase–mediated PCR amplification using primer pairs specific for eIF-5A (5′-GCAGATGACTTGGACTTCGAGACAGG-3′ and 5′-CCTTGATTGCAACAGCTGCCTCCTC-3′) and CD83 (5′-GTTATTGGAGGGTGGTGAAGAGAGG-3′ and 5′-GTGAGGAGTCACTAGCCCTAAATGC-3′). Amplification of the mRNA coding for the ribosomal protein S14 served as an internal control (5′-GGCAGACCGAGATGAATCCTCA-3′ and 5′-CAGGTCCAGGGGTCTTGGTCC-3′). The profile for amplification involved 30 cycles of denaturation at 95°C for 60 s, primer annealing at 55°C for 60 s, and primer extension at 72°C for 90 s. The reaction products were analyzed on ethidium bromide–stained 2% agarose gels.
Northern blot analysis of 5 μg of total cellular RNA derived from either DCs or HeLa cells was performed as described previously 34. CD83-specific transcripts were detected using the radiolabeled synthetic oligonucleotide probe 5′-TCTCCATCCTCTCTTCACCACCCTCCAATAAC-3′. To control for comparable RNA amounts, the filters were stripped and rehybridized using a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe, 5′-CCATGGTGGTGAAGACGCCAGTGGACTCC-3′.
Immunofluorescence Microscopy.
2 d after addition of the final maturation cocktail, cells were harvested and washed with RPMI medium. Subsequently, 0.5 × 106 cells were spun onto polylysine-coated microslides (Menzel-Gläser) for 30 s at 300 rpm using a cytospin3 centrifuge (Shandon), fixed in 2% paraformaldehyde (Merck), and washed three times with PBS and 1% glycerol (Sigma-Aldrich). Blocking was performed with 1% BSA (Sigma-Aldrich). Cells were then incubated with primary anti-CD83 mAb, followed by secondary Cy3-conjugated, affinity-purified goat anti–mouse IgG (Rockland). Reactions were performed for 30 min at ambient temperature. Finally, the slides were washed five times with PBS, and cells were embedded in Moviol (Calbiochem). The samples were analyzed using a ZEISS Axiovert-135 microscope. Images were recorded with a cooled MicroMax CCD camera (Princeton Instruments) and processed using the IPLab spectrum and Adobe Photoshop software.
In Situ Hybridization.
12 h after addition of the final maturation cocktail, 0.5 × 106 untreated or GC7-treated cells were seeded onto adhesion slides (Bio-Rad) and fixed for 15 min with 3% paraformaldehyde. After washing with PBS, cells were permeabilized with 0.5% Triton X-100 for 5 min, washed twice with PBS, and incubated twice for 10 min with 2× SSC. For RNA detection, the following digoxigenin-labeled oligonucleotides from Eurogentec were used: CD83, 5′-TGGTTCTTTCGACGC-3′ and 5′-TGTGGACTTGCCCTG-3′; CD86, 5′-ACTGACAAGACGCGG-3′ and 5′-CAAGTATATGGGCCG-3′. For hybridization, cells were incubated in hybridization solution containing 10 ng/μl of each labeled oligo, 25% deionized formamide, 1 μg/μl Escherichia coli tRNA (Sigma-Aldrich), 2× SSC, 0.5% BSA, and 10% dextran sulfate for 16 h at 42°C in a humidified chamber. Unbound probe was removed by two 30-min washes in 2× SSC and one 15-min wash in 1× SSC. Subsequently, cells were washed with 0.1% Triton X-100 in PBS and blocked with 1% BSA for 30 min. Hybrids were stained for 30 min with primary α-digoxigenin mAb (Roche Molecular Biochemicals). After extensive wash steps in PBS, cells were incubated with the appropriate secondary antibody coupled to Cy3 fluorophore (Biotrends). The cells were then treated with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole; Roche Molecular Biochemicals) in PBS, washed again several times in PBS, and mounted in Mowiol. Samples were analyzed by immunofluorescence microscopy as described above.
Allogeneic MLR.
CD4+ and CD8+ T cells were isolated from buffy coats and stimulated with mature allogeneic DCs, which were either untreated or pretreated with GC7 (end concentration of 1 μM) during DC maturation. T cells (2 × 105 per well) and DCs were cocultivated for 4 d in 200 μl RPMI, supplemented with 5% human serum from a single AB donor in 96-well cell culture dishes. Then, cells were pulsed with [3H]thymidine (1 μCi/well; Amersham Pharmacia Biotech) for 8–16 h. The culture supernatants were harvested onto glass fiber filtermates using an IH-110 harvester (Inotech), and filters were counted in a 1450 microplate counter (Wallac).
Results
The Hypusine Inhibitor GC7 Affects CD83 Surface Expression in DCs.
To investigate eIF-5A expression during DC maturation, we generated human DCs in vitro, following a modified two-step protocol 31. The mature DCs generated by this protocol displayed the specific parameters that are routinely used to define mature DCs. Mature DCs are characterized by the typical morphology of nonadherent cells, a distinct cytofluorographic profile (HLA-DR+++CD86++ CD14−CD83++p55++), and a highly potent stimulatory capacity in allogeneic MLRs at DC/T cell ratios of ≤1:300. Furthermore, these cells stably maintained the DC phenotype when cultured for an additional 1–2 d in the absence of exogenously added cytokines (also known as the “wash-out” or “stability” test; reference 8).
We first examined eIF-5A levels by Western blot analysis using total protein extracts from various stages of DC generation. As shown in Fig. 1 B, the eIF-5A protein, migrating at a relative molecular mass of ∼18 kD 35 36, was barely detectable within the initial days of cell culture (lanes 1 and 2). Clearly, a significant increase in eIF-5A protein was detected toward the end, namely at days 6 and 7, of the maturation period (Fig. 1 B, lane 3 and 4). As eIF-5A may be required for functional expression of DC-specific molecules, identical protein blots were subsequently probed with antibodies directed against CD83, a molecule of unknown function that is, nonetheless, one of the best cell surface markers for mature DCs 37. These analyses demonstrated that the time course of CD83 protein expression followed similar kinetics to those of eIF-5A during DC maturation (compare Fig. 1A and Fig. B). This observation could mean that the expression of CD83 may depend on eIF-5A function.
Figure 1.
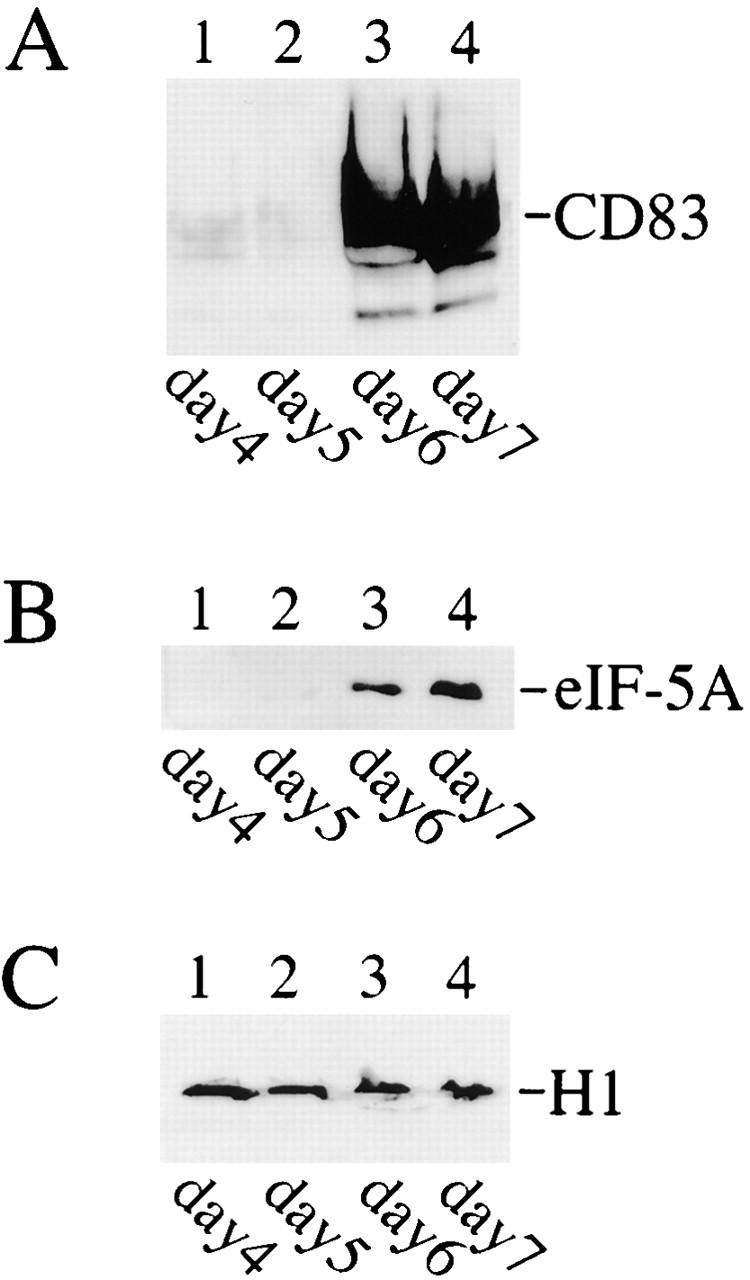
Detection of eIF-5A and CD83 protein expression during in vitro generation of human DCs by Western blot analysis. Cellular protein extracts from different DC generation time points (indicated at the bottom) were resolved by SDS-PAGE, transferred to nylon membranes, and probed with (A) CD83-specific, (B) eIF-5A–specific, or (C) histone H1–specific (loading control) antibodies.
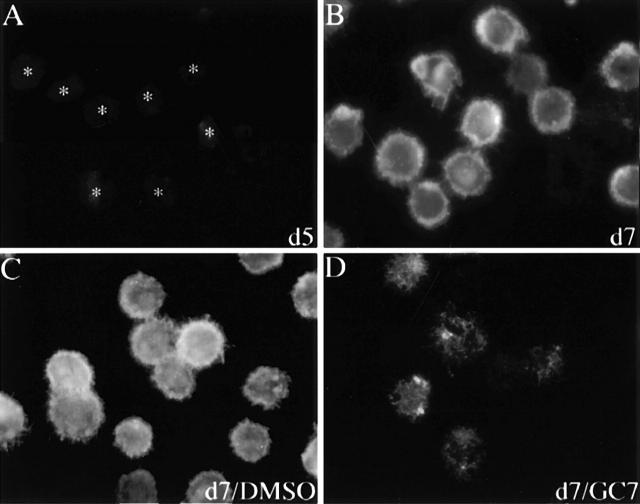
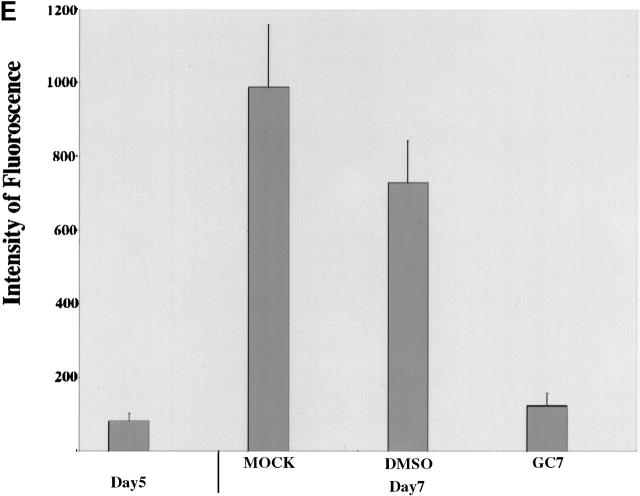
The 154–amino acid eIF-5A protein is unique because it is the only cellular protein known to date to contain the unusual amino acid hypusine 10 11. This enzyme-catalyzed posttranslational modification is essential for eIF-5A function and can be blocked in cell culture with deoxyhypusine synthase inhibitors 38 39 40. In particular, the low-molecular-mass drug GC7 has been shown to be the most specific and potent inhibitor, with a K i value <10 nm 32 38. Therefore, we next investigated the effect of GC7 on CD83 surface expression. As seen in Fig. 2, CD83-specific immunofluorescence analysis revealed that although CD83 surface expression was absent on day 5, it was easily detectable on day 7 of DC maturation (compare panels A and B), reflecting the data obtained in the experiments using total cell protein extracts (Fig. 1 A). In sharp contrast, when GC7 was added to the cell culture medium on day 5 (1 μM), a strong decrease in CD83-specific immunofluorescence was observed at the end of the maturation period (day 7, Fig. 2 D). A control experiment demonstrated that DMSO (solvent for GC7) alone had no significant inhibitory effect on CD83 surface expression (Fig. 2 C). Quantification of the fluorescence signals (Fig. 2 E) confirmed that the presence of the hypusine modification inhibitor GC7 indeed reduced the surface expression of CD83 at least fivefold during the in vitro generation of human DCs.
Figure 2.
Effect of GC7 on CD83 cell surface expression. Human DC precursors were subjected to CD83-specific indirect immunofluorescence analysis. (A) No significant CD83 surface expression was detectable in immature DCs (indicated by asterisks) at day 5. (B) Analysis of mature cells at day 7 demonstrated a strong CD83 surface expression. (C) Comparable signals were detected when DMSO was present in the cell cultures. (D) Exposure of precursor cells to the hypusine inhibitor GC7 (1 μM in DMSO) demonstrated a significant inhibitory effect on CD83 cell surface expression. (E) Quantification of the CD83-specific fluorescence signals shown in A–D.
For further characterization of the effect of GC7 on DC precursors, we next extended our studies with GC7 to other cell surface molecules expressed on mature DCs. FACS® analyses confirmed that the cell surface expression of CD83 was significantly reduced (Fig. 3 A). In contrast, CD95 (Fig. 3 C), a molecule involved in programmed cell death 41, the MHC class I and II molecules (Fig. 3D and Fig. E), CD13 (Fig. 3 B), which is a protein constitutively expressed on myeloid cells 42, and CD68 (Fig. 3 H), a molecule that discriminates between DCs and macrophages 43, were not affected by GC7 treatment. Furthermore, expression of the costimulatory molecules CD80 and CD86 was only slightly reduced (Fig. 3F and Fig. G). Importantly, when fully mature DCs were treated with GC7, no alteration in the expression of the cell surface molecules analyzed above was observed, indicating that only immature DC precursors are sensitive to GC7. Finally, GC7 did not induce necrosis or apoptosis as determined by propidium iodide and annexin V staining (data not shown).
Figure 3.
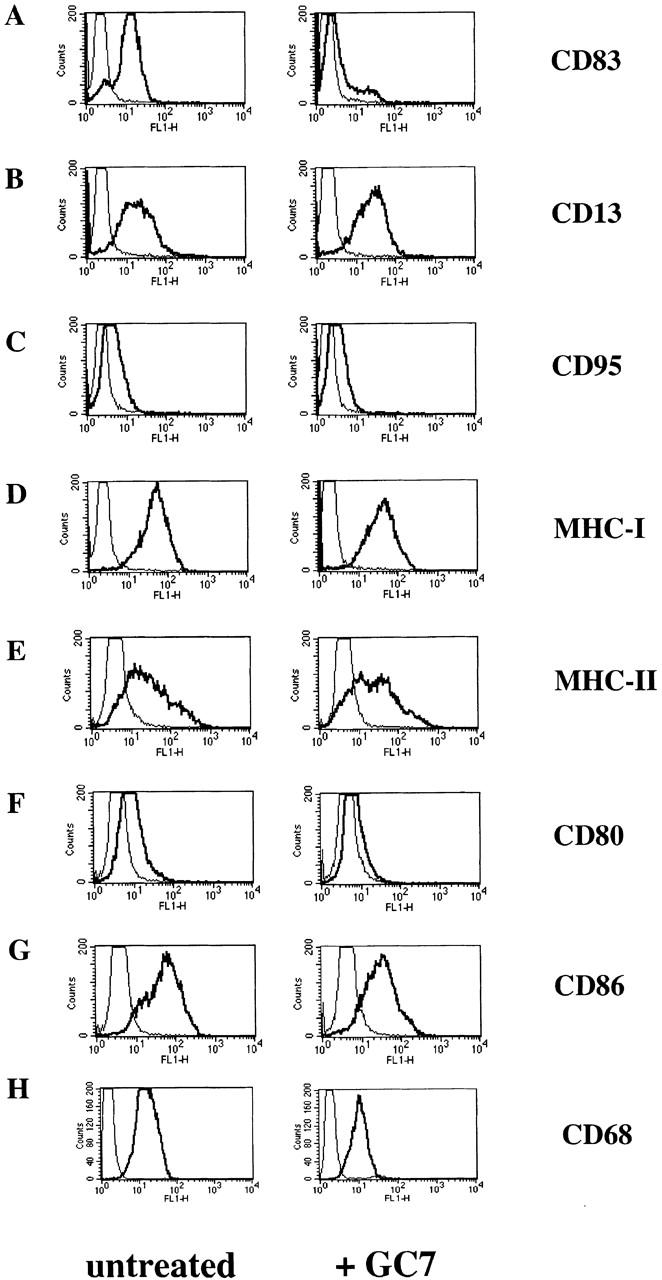
FACS® analyses of untreated or GC7-treated DCs. Left panels show the typical phenotype of untreated mature DCs. Right panels show the effect of GC7 treatment. (A) GC7 strongly reduced CD83 cell surface expression. No significant GC7 effect on cell surface expression was observed in case of CD13 (B), CD95 (C), MHC class I and II molecules (D and E, respectively), and CD68 (H). Only a minimal reduction was observed for CD80 and CD86 cell surface expression (F and G, respectively).
Taken together, the data presented demonstrate that the biosynthesis of the hypusine-containing protein eIF-5A is specifically regulated during DC maturation and that hypusine formation appears to be required for surface expression of the DC marker molecule CD83.
GC7 Affects the Intracellular Distribution of CD83-specific mRNA in DCs.
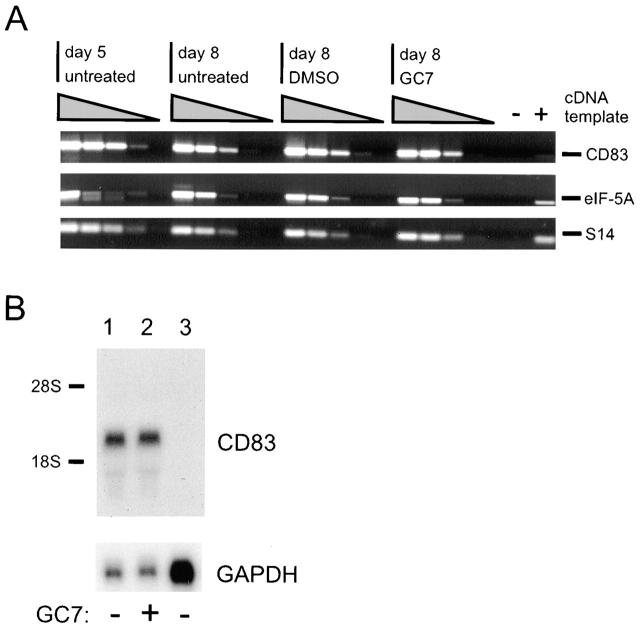
To obtain an insight into how GC7 interferes with CD83 expression, we next prepared total cellular RNA from immature and mature DCs that were cultured in the presence or the absence of GC7. Subsequently, these RNAs were reverse transcribed into single-stranded cDNAs, which were then analyzed by PCR using specific oligonucleotide primer pairs. Amplification of ribosomal protein S14 RNA-derived sequences served as an internal control, and serial 10-fold dilutions of the input templates (cDNAs) indicated linear PCR kinetics. The relative levels of the investigated mRNAs were comparable, irrespective of whether or not GC7 or the solvent control DMSO was present in the cell cultures (Fig. 4 A). This highly sensitive RNA assay also revealed that CD83-specific mRNA is already detectable on day 5 of the DC maturation period and, most importantly, that GC7 treatment did not alter the CD83 mRNA levels. We next subjected total cellular RNA isolated from untreated or GC7-treated (days 5–8) DCs to CD83-specific Northern blot analysis 44, thereby demonstrating again that GC7 does not have a significant influence on the overall CD83 mRNA level in DCs (Fig. 4 B, lanes 1 and 2). As expected, no CD83 messages were present in total RNA derived from HeLa cells (Fig. 4 B, lane 3). Taken together, these data suggested that CD83 surface expression is blocked by GC7 at the posttranscriptional level.
Figure 4.
Detection of eIF-5A– and CD83-specific mRNAs. (A) Total cellular RNA was isolated from untreated, DMSO-treated, and GC7-treated DC precursors at the indicated time points and reverse transcribed. Using specific oligonucleotide primers, serial dilutions of the respective cDNAs were subjected to PCR and subsequently analyzed by agarose gel electrophoresis. Amplification of ribosomal protein S14 RNA-specific sequences served as control for input cDNA amounts. The identities of the various amplification products (indicated at right) were confirmed by DNA sequence determination (not shown). −, negative PCR control; +, positive PCR control. (B) Northern blot analysis of total cellular RNA isolated from DCs that were matured in either the absence (lane 1) or presence (lane 2) of GC7. CD83-specific mRNA species were detected, irrespective of whether or not GC7 was present in the cell cultures. In contrast, no CD83 transcripts were detected in total RNA derived from HeLa cells (lane 3). To control for loading of comparable quantities of RNA, the filters were stripped and rehybridized using a probe specific for GAPDH mRNA.
As the hypusine-containing protein eIF-5A has been shown to be involved in the nucleocytoplasmic transport of HIV-1– and HTLV-I–specific mRNAs 21 22 23 24, it is conceivable that GC7 may affect the intracellular distribution of CD83-specific mRNA. This would mean that eIF-5A activity, which critically depends on the posttranslational hypusine modification of the eIF-5A precursor, mediates efficient nuclear export and thereby, indirectly, the translation of CD83 mRNA.
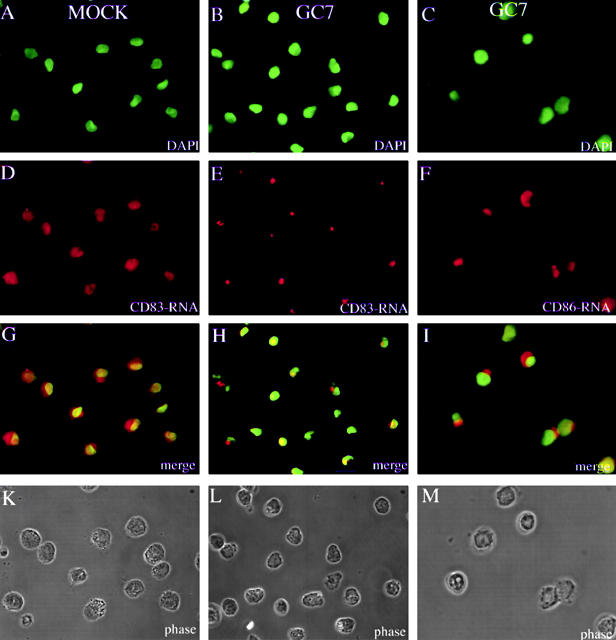
To test this hypothesis, we next directly examined potential GC7-dependent accumulation of CD83 mRNA in the cytoplasmic and nuclear compartment of DCs. As only limited amounts of DCs can be obtained from blood samples, a direct biochemical fractionation of total cellular RNA in nuclear and cytoplasmic subfractions was not feasible. We therefore subjected our in vitro–generated DCs to CD83 mRNA in situ hybridization. The nuclei of fixed DCs (Fig. 5K, Fig. L, and Fig. M) were visualized by DNA staining using DAPI (Fig. 5A, Fig. B, and Fig. C). To prevent the loss of cells through nonadherence in this type of experiment, special adhesion slides were used that affect the typical DC morphology. Inspection of the CD83-specific signals demonstrated that almost comparable levels of CD83 mRNA accumulate in the DC nucleus and cytoplasm (Fig. 5 D). However, in the presence of GC7, the CD83 mRNA appears to be trapped in the nuclear compartment (Fig. 5 E). This becomes particularly obvious when the respective images are merged (compare Fig. 5G and Fig. H). Furthermore, it appears that the CD83 mRNA is not randomly distributed within the nuclei of GC7-treated cells but tends to accumulate at distinct subnuclear sites. This GC7 effect is specific for CD83 mRNAs, since addition of GC7 did not result in nuclear trapping of CD86-specific messages (Fig. 5 F). Please note that these data are in good agreement with the FACS® analyses shown in Fig. 3 G.
Figure 5.
Subcellular localization of CD83 and CD86 mRNA in DC precursor cells. Images belonging to the same experiment are aligned in columns. Mock-treated (CD83: A, D, G, and K) or GC7-treated (CD83: B, E, H, and L; CD86: C, F, I, and M) DC precursors were subjected to CD83 mRNA– or CD86 mRNA–specific in situ hybridization. Nuclei were labeled by DNA staining using DAPI (A, B, and C). mRNAs were visualized with digoxigenin-labeled oligonucleotide probes, followed by primary α-digoxigenin and appropriate secondary Cy3-coupled antibodies (D, E, and F). Comparison of the merged images shows equal distribution of CD83 mRNA between the nucleus and the cytoplasm in mock-treated DCs (G). In contrast, GC7 treatment of DCs results in nuclear accumulation of CD83 mRNA (H). As shown in panel I, GC7 treatment of DCs does not result in nuclear trapping of CD86 mRNA. Corresponding phase contrast images are shown in K, L, and M.
These data suggest that CD83 mRNA is transported from the DC nucleus to the cytoplasm by exploitation of a specific pathway. Moreover, interference with this pathway results in nuclear trapping of CD83 mRNA and therefore in decreased CD83 protein synthesis.
GC7 Inhibits the Ability of DCs to Induce Allogeneic T Cell Proliferation.
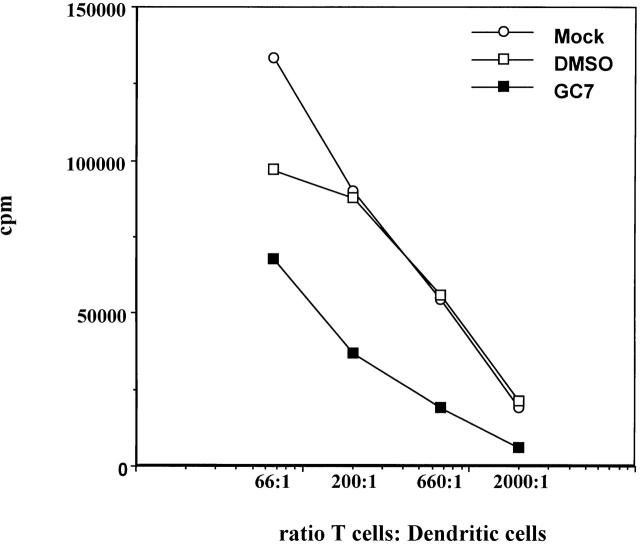
The most distinctive functional characteristic of DCs is their ability to induce a potent T cell response 45. Therefore, we studied whether or not the observed GC7 effect on DC surface molecules, and in particular the prominent inhibition of CD83, also affected the ability of DCs to induce T cell proliferation in an allogeneic MLR. As depicted in Fig. 6, mock- or DMSO-treated DCs displayed comparable T cell proliferation induction rates in this experiment. Clearly, GC7 impaired the ability of DCs to induce a significant T cell response in the MLR. Although these experiments do not show that CD83 downregulation alone is responsible for reduced stimulation, the data indicate that hypusine formation is required for DC function.
Figure 6.
Effect of GC7 on the ability of DCs to induce allogeneic T cell proliferation. Mature DCs derived from untreated (○) or DMSO-treated (□) precursors induce a strong allostimulatory reaction in the primary allogeneic MLR. In contrast, cells derived from GC7-treated precursors demonstrate a reduced allostimulatory capacity (▪).
Discussion
Functional, mature DCs are derived from circulating precursor cells after a period of maturation 4 46. During this period, many activities at the level of gene regulation take place within the precursor cells which ultimately result in formation of mature DCs 47. In addition to their functional qualities, DCs are also characterized by the expression of a specific array of marker molecules. These include the accessory/costimulatory gene products CD40, CD80, and CD86 as well as MHC class I and II 1 3. In particular, the presence of the CD83 molecule is a well characterized marker for fully mature DCs, as CD83 can not be detected on immature DC precursors 37. The functional significance of CD83 is, however, completely unknown.
In this study, we show that CD83 expression is upregulated during DC maturation and is closely mirrored by the expression pattern detected for the hypusine-containing protein eIF-5A. More importantly, our experiments show that GC7, a highly potent inhibitor of the unique hypusine modification in eIF-5A, significantly inhibits CD83 surface expression in these cells. As previously published data have demonstrated that eIF-5A plays a functional role in HIV-1 Rev- and HTLV-I Rex-mediated nuclear export of retroviral mRNAs 21 22 23 24, it was tempting to speculate that GC7 may also interfere with the translocation of the CD83 mRNA across the nuclear envelope in DCs. The notion of a potential GC7 effect on intracellular CD83 mRNA transport was further supported by the recent finding that eIF-5A binds the export receptor CRM1 and accumulates at nuclear pore–associated intranuclear filaments, the site where initial docking of export substrates to the nuclear pore complex is believed to occur 29. Thus, the inhibition of nucleocytoplasmic translocation of CD83 mRNA by GC7 should indeed result in the observed downregulation of CD83 cell surface expression. In fact, examination of the intracellular distribution of CD83 mRNA in our precursor cells demonstrated a marked redistribution of these transcripts between the subcellular compartments upon addition of GC7. GC7 clearly prevented efficient cytoplasmic accumulation of CD83 mRNA, as the messages remained trapped in the cell nucleus, an effect that was not observed for CD86 mRNA. Taken together, these data suggest that hypusine formation is required for efficient nuclear export of CD83 mRNA. Thus, CD83 mRNA exploits a specific pathway for its transport from the nuclear site of RNA transcription and processing to the site of translation in the cytoplasm.
Competition experiments in Xenopus oocytes suggested that different classes of RNA (e.g., mRNA, rRNA, U-rich snRNA, and tRNA) are exported from the nucleus by specific export factors 48. General nuclear export of mRNA appears to be mediated by a class of abundant RNA binding proteins termed heterogeneous nuclear ribonucleoproteins (hnRNPs; for review see references 49 and 50). hnRNPs associate with poly(A)+ RNA in the nucleus as well as in the cytoplasm, and some subsets of hnRNPs have been shown to shuttle between these cellular compartments 51. However, little is known about the regulation of nuclear export of specific groups of mRNAs. A series of recent studies in which the metabolism of early response gene (ERG) mRNA subsets was investigated provided new evidence that specifically regulated mRNA export pathways may indeed exist in mammalian cells (for review see reference 52). ERG mRNAs encode functionally important proteins such as protooncoproteins, cytokines, and lymphokines that are characterized by a short half-life due to instability sequences, termed AU-rich elements (AREs). ARE sequences are commonly found in the 3′-untranslated region of these short-lived mRNAs 53. However, binding of the HuR (also called HuA) nucleocytoplasmic shuttling protein to ARE sequences significantly increases the stability of ERG mRNAs 54 55 56 57 and may subsequently lead to elevated ERG mRNA levels in the cytoplasm. Thus, differences in mRNA nuclear export rates can have pronounced effects on the expression of specific genes, providing cells with an effective mechanism to quickly respond to environmental signals. The data presented in this study also suggest that CD83 mRNA, which does not contain obvious ARE-rich regions 44, can accumulate in the cytoplasm by virtue of a similar mRNA transport mechanism during DC differentiation. Such a mechanism would assure the timely and efficient expression of critical DC proteins. The existence of such a mechanism would indeed be beneficial in the case of CD83. As a result of the specific surface expression on mature DCs only, CD83 is considered to be an important protein for DC function, although its precise activity in DCs remains elusive.
Clearly, the inhibitory effect of GC7 on DC activity can not be attributed exclusively to the lack of CD83 expression. We cannot rule out the possibility that GC7 may also affect the metabolism of additional mRNAs in DCs, which have not yet been identified. Nevertheless, interference with eIF-5A function provides a new method to investigate DC function at the molecular level that may ultimately result in the identification of additional functionally important DC proteins. Furthermore, the pharmacological interference with hypusine formation may also provide a novel approach to modulate DC activity.
As DCs are able to induce both immunity and tolerance, they represent a very promising cell type for future immune therapy studies. The elucidation of the mode of action of DC-specific molecules, like CD83 on the molecular level, will not only be important to understand the biology of DC but also be advantageous for the development of new therapeutic strategies.
Acknowledgments
We thank Dr. Sarah L. Thomas for critical comments on the manuscript.
This work received financial support from the Deutsche Forschungsgemeinschaft (SFB 466) and Wilhelm-Sander Stiftung (96.042.2).
Footnotes
M. Kruse and O. Rosorius contributed equally to this study.
D. Bevec's current address is Axxima Pharmaceuticals AG, Am Klopferspitz 19, D-82152 Martinsried, Germany.
Abbreviations used in this paper: AREs, AU-rich elements; DCs, dendritic cells; eIF-5A, eukaryotic initiation factor 5A; ERG, early response gene; hnRNPs, heterogeneous nuclear ribonucleoproteins.
References
- Steinman R.M. The dendritic cell system and its role in immunogenicity. Annu. Rev. Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- Dieu M.C., Vanbervliet B., Vicari A., Bridon J.M., Oldham E., Ait Yahia S., Briere F., Zlotnik A., Lebecque S., Caux C. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 1998;188:373–386. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Steinman R.M., Witmer M.D. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc. Natl. Acad. Sci. USA. 1978;75:5132–5136. doi: 10.1073/pnas.75.10.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caux C., Dezutter Dambuyant C., Schmitt D., Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani N., Gruner S., Brang D., Kampgen E., Lenz A., Trockenbacher B., Konwalinka G., Fritsch P.O., Steinman R.M., Schuler G. Proliferating dendritic cell progenitors in human blood. J. Exp. Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani N., Reider D., Heuer M., Ebner S., Kampgen E., Eibl B., Niederwieser D., Schuler G. Generation of mature dendritic cells from human blood. An improved method with special regard to clinical applicability. J. Immunol. Methods. 1996;196:137–151. doi: 10.1016/0022-1759(96)00078-6. [DOI] [PubMed] [Google Scholar]
- Cella M., Sallusto F., Lanzavecchia A. Origin, maturation and antigen presenting function of dendritic cells. Curr. Opin. Immunol. 1997;9:10–16. doi: 10.1016/s0952-7915(97)80153-7. [DOI] [PubMed] [Google Scholar]
- Park M.H., Wolff E.C., Folk J.E. Hypusineits post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors. 1993;4:95–104. [PubMed] [Google Scholar]
- Park M.H., Wolff E.C., Folk J.E. Is hypusine essential for eukaryotic cell proliferation? Trends Biochem. Sci. 1993;18:475–479. doi: 10.1016/0968-0004(93)90010-k. [DOI] [PubMed] [Google Scholar]
- Joe Y.A., Wolff E.C., Park M.H. Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins. J. Biol. Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- Klier H., Csonga R., Steinkasserer A., Wöhl T., Lottspeich F., Eder J. Purification and characterization of human deoxyhypusine synthase from HeLa cells. FEBS Lett. 1995;364:207–210. doi: 10.1016/0014-5793(95)00394-o. [DOI] [PubMed] [Google Scholar]
- Yan Y.P., Tao Y., Chen K.Y. Molecular cloning and functional expression of human deoxyhypusine synthase cDNA based on expressed sequence tag information. Biochem. J. 1996;315:429–434. doi: 10.1042/bj3150429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonga R., Ettmayer P., Auer M., Eckerskorn C., Eder J., Klier H. Evaluation of the metal ion requirement of the human deoxyhypusine hydroxylase from HeLa cells using a novel enzyme assay. FEBS Lett. 1996;380:209–214. doi: 10.1016/0014-5793(96)00020-8. [DOI] [PubMed] [Google Scholar]
- Benne R., Hershey J.W. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Kemper W.M., Berry K.W., Merrick W.C. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J. Biol. Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- Kang H.A., Schwelberger H.G., Hershey J.W.B. Effect of initiation factor eIF-5A depletion on cell proliferation and protein synthesis. In: Brown A.J.P., Tuite M.F., McCarthy J.E.G., editors. Protein Synthesis and Targeting in Yeast. NATO Series HCell Biology. Springer-Verlag; Berlin, Germany: 1993. pp. 123–129. [Google Scholar]
- Kang H.A., Hershey J.W.B. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae . J. Biol. Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- Zuk D., Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl M., Himmelspach M., Bahr G.M., Hammerschmid F., Jaksche H., Wolff B., Aschauer H., Farrington G. K., Probst H., Bevec D. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J. Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevec D., Jaksche H., Oft M., Wöhl T., Himmelspach M., Pacher A., Schebesta M., Koettnitz K., Dobrovnik M., Csonga R. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science. 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- Katahira J., Ishizaki T., Sakai H., Adachi A., Yamamoto K., Shida H. Effects of translation initiation factor eIF-5A on the functioning of human T-cell leukemia virus type I Rex and human immunodeficiency virus Rev inhibited trans dominantly by a Rex mutant deficient in RNA binding. J. Virol. 1995;69:3125–3133. doi: 10.1128/jvi.69.5.3125-3133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfgang C., Rosorius O., Hofer L., Jaksche H., Hauber J., Bevec D. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc. Natl. Acad. Sci. USA. 1999;96:6229–6234. doi: 10.1073/pnas.96.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.R., Greene W.C. Molecular biology of the type I human T-cell leukemia virus (HTLV-I) and adult T-cell leukemia. J. Clin. Invest. 1991;87:761–766. doi: 10.1172/JCI115078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V.W., Malim M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Johnson A.W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse H., Buratowski R.M., Silver P.A., Buratowski S. The importin/karyopherin kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. USA. 1999;96:12542–12547. doi: 10.1073/pnas.96.22.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosorius O., Reichart B., Krätzer F., Heger P., Dabauvalle M.C., Hauber J. Nuclear pore localization and nucleocytoplasmic transport of eIF-5Aevidence for direct interaction with the export receptor CRM1. J. Cell Sci. 1999;112:2369–2380. doi: 10.1242/jcs.112.14.2369. [DOI] [PubMed] [Google Scholar]
- Bevec D., Klier H., Holter W., Tschachler E., Valent P., Lottspeich F., Baumruker T., Hauber J. Induced gene expression of the hypusine-containing protein eukaryotic initiation factor 5A in activated human T lymphocytes. Proc. Natl. Acad. Sci. USA. 1994;91:10829–10833. doi: 10.1073/pnas.91.23.10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonuleit H., Kuhn U., Muller G., Steinbrink K., Paragnik L., Schmitt E., Knop J., Enk A.H. Pro-inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum-free conditions. Eur. J. Immunol. 1997;27:3135–3142. doi: 10.1002/eji.1830271209. [DOI] [PubMed] [Google Scholar]
- Jakus J., Wolff E.C., Park M.H., Folk J.E. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. J. Biol. Chem. 1993;268:13151–13159. [PubMed] [Google Scholar]
- Schatz O., Oft M., Dascher C., Schebesta M., Rosorius O., Jaksche H., Dobrovnik M., Bevec D., Hauber J. Interaction of the HIV-1 rev cofactor eukaryotic initiation factor 5A with ribosomal protein L5. Proc. Natl. Acad. Sci. USA. 1998;95:1607–1612. doi: 10.1073/pnas.95.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent P., Bevec D., Maurer D., Besemer J., Di Padova F., Butterfield J.H., Speiser W., Majdic O., Lechner K., Bettelheim P. Interleukin 4 promotes expression of mast cell ICAM-1 antigen. Proc. Natl. Acad. Sci. USA. 1991;88:3339–3342. doi: 10.1073/pnas.88.8.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper H.L., Park M.H., Folk J.E. Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell. 1982;29:791–797. doi: 10.1016/0092-8674(82)90441-x. [DOI] [PubMed] [Google Scholar]
- Smit-McBride Z., Schnier J., Kaufman R.J., Hershey J.W.B. Protein synthesis initiation factor eIF-4D. J. Biol. Chem. 1989;264:18527–18530. [PubMed] [Google Scholar]
- Zhou L.J., Tedder T.F. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc. Natl. Acad. Sci. USA. 1996;93:2588–2592. doi: 10.1073/pnas.93.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.H., Wolff E.C., Lee Y.B., Folk J.E. Antiproliferative effects of inhibitors of deoxyhypusine synthase. Inhibition of growth of Chinese hamster ovary cells by guanyl diamines. J. Biol. Chem. 1994;269:27827–27832. [PubMed] [Google Scholar]
- Shi X.P., Yin K.C., Ahern A., Davis J., Stern A.M., Waxman L. Effects of N1- guanyl-1, 7-diaminoheptanean inhibitor of deoxyhypusine synthase, on the growth of tumorigenic cell lines in culture. Biochim. Biophys. Acta. 1996;1310:119–126. doi: 10.1016/0167-4889(95)00165-4. [DOI] [PubMed] [Google Scholar]
- Chen Z.P., Yan Y.P., Ding Q.J., Knapp S., Potenza J.A., Schugar H.J., Chen K.Y. Effects of inhibitors of deoxyhypusine synthase on the differentiation of mouse neuroblastoma and erythroleukemia cells. Cancer Lett. 1996;105:233–239. doi: 10.1016/0304-3835(96)04287-5. [DOI] [PubMed] [Google Scholar]
- Itoh N., Yonehara S., Ishii A., Yonehara M., Mizushima S., Sameshima M., Hase A., Seto Y., Nagata S. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- Rosenzwajg M., Tailleux L., Gluckman J.C. CD13/N-aminopeptidase is involved in the development of dendritic cells and macrophages from cord blood CD34(+) cells. Blood. 2000;95:453–460. [PubMed] [Google Scholar]
- Betjes M.G., Haks M.C., Tuk C.W., Beelen R.H. Monoclonal antibody EBM11 (anti-CD68) discriminates between dendritic cells and macrophages after short-term culture. Immunobiology. 1991;183:79–87. doi: 10.1016/S0171-2985(11)80187-7. [DOI] [PubMed] [Google Scholar]
- Zhou L.J., Schwarting R., Smith H.M., Tedder T.F. A novel cell-surface molecule expressed by human interdigitating reticulum cells, Langerhans cells, and activated lymphocytes is a new member of the Ig superfamily. J. Immunol. 1992;149:735–742. [PubMed] [Google Scholar]
- Thomas R., Davis L.S., Lipsky P.E. Comparative accessory cell function of human peripheral blood dendritic cells and monocytes. J. Immunol. 1993;151:6840–6852. [PubMed] [Google Scholar]
- Van Voorhis W.C., Valinsky J., Hoffman E., Luban J., Hair L.S., Steinman R.M. Relative efficacy of human monocytes and dendritic cells as accessory cells for T cell replication. J. Exp. Med. 1983;158:174–191. doi: 10.1084/jem.158.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno M., Martino M., Sutherland C.L., Gold M.R., Ricciardi Castagnoli P. Dendritic cell survival and maturation are regulated by different signaling pathways. J. Exp. Med. 1998;188:2175–2180. doi: 10.1084/jem.188.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens W.C., Izaurralde E., Mattaj I.W. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Mattaj I.W. RNA Export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Dreyfuss G. Nuclear export of proteins and RNAs. Curr. Opin. Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Dreyfuss G. hnRNP proteinslocalization and transport between the nucleus and the cytoplasm. Trends Cell Biol. 1993;3:151–155. doi: 10.1016/0962-8924(93)90135-n. [DOI] [PubMed] [Google Scholar]
- Keene J.D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Shyu A.B. AU-rich elementscharacterization and importance in mRNA degradation. Trends. Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Atasoy U., Watson J., Patel D., Keene J.D. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- Fan X.C., Steitz J.A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.C., Steitz J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Watson J., Keene J.D., Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]