Abstract
We found previously that Id3, which inhibits transcriptional activities of many basic helix-loop-helix transcription factors, blocked T and B cell development but stimulated natural killer (NK) cell development. Here we report that ectopic expression of Id3 and another Id protein, Id2, strongly inhibited the development of primitive CD34+CD38− progenitor cells into CD123high dendritic cell (DC)2 precursors. In contrast, development of CD34+CD38− cells into CD4+CD14+ DC1 precursors and mature DC1 was not affected by ectopic Id2 or Id3 expression. These observations support the notion of a common origin of DC2 precursors, T and B cells. As Id proteins did not block development of NK cells, a model presents itself in which these proteins drive common lymphoid precursors to develop into NK cells by inhibiting their options to develop into T cells, B cells, and pre-DC2.
Keywords: dendritic cells, dendritic cell precursors, basic helix-loop-helix transcription factors, idiotype proteins, lymphoid development
Introduction
It is commonly accepted that mature lymphoid cells are derived from a common lymphoid precursor (CLP). Both in humans 1 and mice 2, the CLP has been described in terms of expression of cell surface proteins. Recent work has provided some insight into the mechanisms of cell fate specification of these CLPs, as several transcription factors have been identified that control differentiation of particular lymphoid lineages. Among these are E proteins, which belong to the family of basic helix-loop-helix (bHLH) transcription factors. Members of this family have been shown to play a role in lineage decisions in many developmental systems in organisms varying from yeast to mammals 3 4. bHLH proteins are characterized by two conserved domains: an HLH domain that mediates protein–protein interactions and a basic domain that confers binding of bHLH dimers to sequences present in target genes. The E proteins control development in concert with the structurally related inhibitor of DNA binding (Id) proteins 5. The four Id proteins, which have different tissue distributions, possess a HLH domain and can interact effectively with bHLH factors, preferentially with E proteins. However, as Id proteins lack a basic motif, complexes of Id and bHLH factors fail to bind to DNA and are therefore transcriptional inactive.
The combined dosage of bHLH and Id proteins is the determining factor in certain cell lineage decisions. The E proteins, E12 and E47, which are different splice products of a single gene, E2A, are essential for B cell development 6 7. They control either directly or indirectly the expression of PAX-5 6, which shuts off the developmental options of lymphoid precursors except to the B cell lineage, thereby consolidating further B cell development 8 9. E2A proteins are also involved in T cell development 10. The thymus of E2A−/− mice is much smaller than that of wild-type mice, and T cell development is partially blocked at the transition of uncommitted into committed T cell precursors 10. Mice deficient for the E protein HEB also have a small thymus and a relative block early in T cell development 11. Bone marrow cells of mice deficient for the bHLH factor HES-1, which belongs to a different class of HLH factors 4, are unable to develop into T cells upon transfer into recombination activating gene (RAG)-1−/− mice 12. Together, these findings indicate that multiple bHLH transcription factors are involved in T cell development. Concordant with findings in the mouse, we documented that ectopic expression of one of the Id proteins, Id3, in human lymphoid precursors blocks T and B cell development. In contrast, NK cell development was promoted by Id3 13 14 15. Recently, it was reported that mice deficient for another member of the Id family, Id2, lack NK cells 16. Together, these findings would be compatible with the idea that Id proteins consolidate NK development by shutting off alternative developmental options of lymphoid precursor cells.
To obtain further support for the hypothesis that E and Id proteins control lymphopoiesis, we sought to test the effect of ectopic expression of Id2 and Id3 on development of another lymphoid cell, the lymphoid dendritic cell (DC). A considerable body of evidence supports the notion that DCs can originate either from myeloid or lymphoid precursors. Cell transfer studies suggest that murine thymic DCs, which express CD8α, are related to lymphoid cells, as CD8α1 DCs can be generated from thymic precursors that are almost entirely devoid of myeloid potential 17 18. Recently, we characterized a cell type present in the medulla of the human thymus with the phenotype CD3−CD4+CD45RA+ expressing high levels of IL-3Rα (CD123). These cells rapidly differentiate into DCs in vitro upon incubation with IL-3 or IL-3 and CD40 ligand (CD40L 19). Interestingly, the thymic CD123high DC precursors are very similar to the recently described pre-DC2 (pDC2) present in the T cell areas of human tonsils 20 and in peripheral blood of adults and neonates 21 22. The CD123high pDC2 are also capable of secreting high levels of IFN-α and -β after infection with HSV-1 or influenza virus 23 24. Therefore, they appear to be identical to a cell type described previously as the natural IFN-producing cell (NIPC [25, 26]). A couple of characteristics of the thymic CD123high pDC2 make it highly likely that they are of lymphoid origin. They express the lymphoid cell–associated markers CD2, CD5, and CD7, which are preferentially expressed on lymphocytes, and lack the myeloid cell–associated markers CD11c, CD13, and CD33 19. Importantly, the thymic DC precursors as well as the tonsillar pDC2 expressed high levels of pre-TCR-α (pTα) transcripts 19, strongly suggesting a close developmental relationship of these pDC2 with T cells.
In this study, we describe a novel assay for development of CD123high pDC2. Using this assay we demonstrate that ectopic expression of Id2 or Id3 into CD34+CD38− fetal liver precursors inhibited generation of CD123high pDC2 but did not affect development of CD34+ cells into DCs with GM-CSF and TNF-α. Id3 also inhibited the generation of CD123high pDC2 from thymic CD34+ cells. As Id overexpression inhibits T cell, B cell, and CD123high pDC2 development but not NK development, our data suggest that Id proteins allow NK development by inhibiting alternative developmental options of lymphoid precursors.
Materials and Methods
Construction of the Vectors and Transduction of Target Cells.
Preparation of the internal ribosomal entry site (IRES)-green fluorescent protein (GFP) and Id3-IRES-GFP constructs have been described previously 14. As a control we constructed into the LZRS-linker-IRES-GFP vector a form of murine Id3 with a mutation in the HLH domain (R72P; provided by Dr. G. Kato, Johns Hopkins University, Baltimore, MD) that has lost the capacity to dimerize with bHLH factors 27. The HLH domains of human and murine Id3 are identical. The coding sequence of human Id2 was cut from the pSG5 Id2 vector (a gift of Dr. R. de Groot, University of Utrecht, Utrecht, Netherlands) with Not1 and was ligated in the Not1 site of polylinker from our plasmid LZRS-linker-IRES-GFP. Selected clones were checked for the correct orientation of the Id2 coding sequence by sequencing. Helper-free recombinant retroviruses were produced after transfection into a 293T-based amphotropic retroviral packaging cell line, Phoenix 28.
Retroviral Transduction.
The transduction procedure using recombinant human fibronectin fragments was performed by a method described previously 13 14. The sorted CD34+CD38− fetal liver or the CD34+CD1a− thymocytes were cultured overnight in the presence of 10 ng/ml human IL-7 and 20 ng/ml stem cell factor (SCF; both from R&D Systems). After overnight culture, the cells were transduced by incubating them for 6 h with virus supernatant in non-tissue culture–treated Falcon petri dishes (3-cm diameter; Becton Dickinson) coated with 30 mg/ml recombinant human fibronectin fragment CH-296 (RetroNectin™; Takara Shuzo Co. 29). After transduction, the cells were washed twice in medium. The transduction efficiency was tested by determining the percentage of GFP+ cells 2 d after transduction.
Antibodies.
FITC-labeled antibodies specific for CD3, CD5, CD7, CD8, CD14, CD19, CD33, CD40, CD45RA, and CD56, PE-labeled antibodies against CD2, CD4, CD13, CD80, CD123, and HLA-DR, and an unlabeled, nonblocking, anti-CD123 antibody were purchased from Becton Dickinson. Anti-CD1a–PE, anti-CD54–PE, anti-CD83–PE, and anti–TCR-γ/δ–PE were obtained from Beckman Coulter/Immunotech, and anti-CD11c–PE was purchased from Biosource International. Anti-CD4–Tricolor was obtained from Dako and anti–TCR-α/β–Tricolor from Beckman Coulter/Immunotech.
Isolation of CD34+ Cells from Fetal Liver and Postnatal Thymus.
Fetal liver was obtained from elective therapeutic abortions. Gestational age was determined by crown-rump length and ranged from 14 to 17 wk. The use of this tissue was approved by the medical ethical committee of the Netherlands Cancer Institute and was contingent on informed consent. Human fetal liver cells were isolated by gentle disruption of the tissue by mechanical means, followed by density gradient centrifugation over Ficoll-Hypaque (Lymphoprep; Nycomed Pharma). The CD34+ cells were isolated from these samples by immunomagnetic cell sorting, using a CD34 separation kit (varioMACS; Miltenyi Biotec) and were further purified by sorting with a FACStarPLUS™ (Becton Dickinson). Postnatal thymus samples were from children undergoing open-heart surgery. CD34+ cells, enriched by immunomagnetic cell sorting, were labeled with anti-CD34–FITC (HPCA-2; Becton Dickinson) and anti-CD1a–PE (Beckman Coulter/Immunotech) antibodies, and the CD34+CD1a− cells were purified by FACS® sorting. The purity of the populations used in this study was >99%.
Monolayer Cell Culture.
The monolayer cell culture assays for differentiation of CD123high pDC2 were carried out with the murine bone marrow stromal cell line S17 (a gift from Dr. Ken Dorshkind, UCLA, Los Angeles, CA). 2 d before their use in coculture experiments, S17 cells were plated in 24-well tissue culture plates (15 × 104 cells/well in 1 ml medium) or in 96-well flat-bottomed plates (3–5 × 103 cells/well in 100 μl medium). Monolayer cultures were initiated by seeding 5–10 × 104 progenitor cells/ml to the wells seeded with S17 cells. After incubation in Yssel's medium 30 supplemented with 5% FCS (BioWhittaker), the cells were harvested, stained with FITC-, PE-, or Tricolor-labeled human specific antibodies, and analyzed by flow cytometry for cell surface phenotype.
Generation of DCs in IL-3- and CD40L-supported Cultures of Thymic DC Precursors.
CD123high cells were sorted from cocultures of CD34+ cells with S17 cells. The sorted cells were cultured in a 96-well flat-bottomed plate in the presence of 10 ng/ml IL-3 with or without 104 CD40L-transfected mouse fibroblasts (provided by Dr. J. Banchereau, Schering-Plough Corporation, Dardilly, France). The L cells were cultured as a monolayer and had been irradiated with 104 rads before use in the DC differentiation assay. Culture medium consisted of Yssel's medium with 5% normal human serum.
IFN-α Production by CD123high pDC2.
CD123highCD45RA+ cells were purified by FACS® sorting and resuspended at a concentration of 106 cells/ml in Yssel's medium supplemented with 2% human serum. 200 μl of this cell suspension were mixed with 10 PFU/well HSV-1 of a 96-well round-bottomed plate (Costar) and incubated for 24 h at 37°C in a 5% CO2 atmosphere. The IFN-α levels were measured with an ELISA (Biosource International) according to the instructions of the manufacturer.
Generation of DCs from CD34+ Precursor Cells in Media Containing GM-CSF and TNF-α.
Development of CD34+ fetal liver cells and thymocytes to DCs was studied in an in vitro culture system described previously 31. 10,000 progenitor cells were cultured in 200 μl Yssel's medium in the presence 10 ng/ml SCF (R&D Systems), 50 ng/ml GM-CSF (a gift of Dr. R. Kastelein, DNAX, Palo Alto, CA), and 50 U/ml TNF-α (R&D Systems) per well in 96-well microtiter plates (Costar). Appearance of DCs was monitored by analyzing the phenotype by flow cytometry.
Reverse Transcription PCR Assays and Sequencing of the PCR Product.
RNA was isolated from FACS®-sorted CD123high cells using TRIzol reagent (GIBCO BRL) according to the manufacturer's instructions and reverse transcribed using a poly-dT oligonucleotide (Promega) and 400 U Moloney MuLV reverse transcriptase (GIBCO BRL) at 42°C for 1 h. PCR assays were carried out in 50 μl reaction volumes using 1 μl cDNA template, 2 mM MgCl2, 0.25 mM of each dNTP, 1 μM of each primer, and 3 U Taq polymerase (GIBCO BRL) in 1× buffer (10 mM Tris-HCl, pH 8.5, 50 mM KCl). Reaction conditions were as follows: a 5-min denaturation step at 94°C was followed by 30 cycles of 1 min at 94°C, 1 min at 65°C, and 2 min at 72°C. The PCR products were separated on 1.2% agarose gels, stained with ethidium bromide, and analyzed by videodensitrometry using the Eagle Eye still video system and Eagle Sight Software (Stratagene).
The hypoxanthine ribosyltransferase (HPRT) and pTα primers that were used were: HPRT sense, 5′-TATGGACAGGACTGAACGTCTTGC-3′; HPRT antisense, 5′-GACACAAACATGATTCAAATCCCTGA-3′; pTα sense, 5′-GTCCA-GCCCTACCCACAGGTGT-3′; pTα antisense, 5′-CGGGAA-TTCGACGTCCCTGGCTGTAGAAGCCTCTC-3′. These primers amplify an oligonucleotide spanning position 122 to 420 of the pTα sequence (sequence data are available from EMBL/GenBank/DDBJ under accession no. U36759; the ATG is at position 82). To confirm the identity of the pTα PCR product, the PCR fragments were isolated from the 1.5% agarose 1× Tris-HCl/acetate/EDTA (TAE) gel and subjected to DNA sequencing analysis using either primer used for PCR reaction. As a positive control, a retroviral vector containing the full-length pTα cDNA sequence was used. Sequence reactions were performed with Big Dye Terminator chemistry according to the manufacturer's protocol and analyzed on an ABI model 3700 sequencer (Applied Biosystems).
Results
CD34+ Cells Develop into CD123high pDC2 upon Coculture with the Murine Stromal Cell Line S17.
The murine stromal cell line S17 has been shown previously to induce development of CD19+ B cells from neonatal cord blood and fetal liver CD34+ cells 32 33. In this study we observed that coculture of CD34+CD38− fetal liver cells with S17 cells resulted in the appearance of IL-3Rα (CD123)high cells that coexpressed CD45RA within 4–5 d. The proportion of these cells gradually decreases upon further culture (results not shown). The percentage of CD123high CD45RA+ cells recovered from cultures of CD34+CD38− fetal liver cells with S17 cells were variable; these could be as low as 1% and as high as 15% depending on the donor. On the average, CD34+CD38− cells expand twofold in the 4–5-d S17 cultures. Thus, starting with 105 CD34+CD38− cells, between 2,000 and 30,000 CD123highCD45RA+ cells can be recovered. Fig. 1 shows a typical experiment. The starting CD34+CD38− population (99% pure upon reanalysis) did not express CD45RA and only low levels of CD123. After 4 d of coculture with S17 cells, the CD123highCD45RA+ cells constituted 8% of the cells. To investigate the phenotype of these cells in more detail, we sorted the CD123+ cells to 97% purity (Fig. 1 B) and analyzed the phenotype. All sorted cells expressed CD45RA. They coexpressed CD4, lacked CD11c, and weakly expressed the lymphoid cell–associated markers CD2, CD5, and CD7. They were negative for CD1a, which is expressed on Langerhans type DCs, CD3, and CD8, and for the NK marker CD56. In other experiments we found that these cells also did not express the costimulatory molecule CD80 and the mature DC marker CD83 (data not shown). Expression of the myeloid marker CD13 and CD54 (intracellular adhesion molecule [ICAM]-1) was weak. The phenotypes of the S17-generated CD123high cells, the thymic CD123high pDC2, and peripheral pDC2 are very similar 19 20 34.
Figure 1.
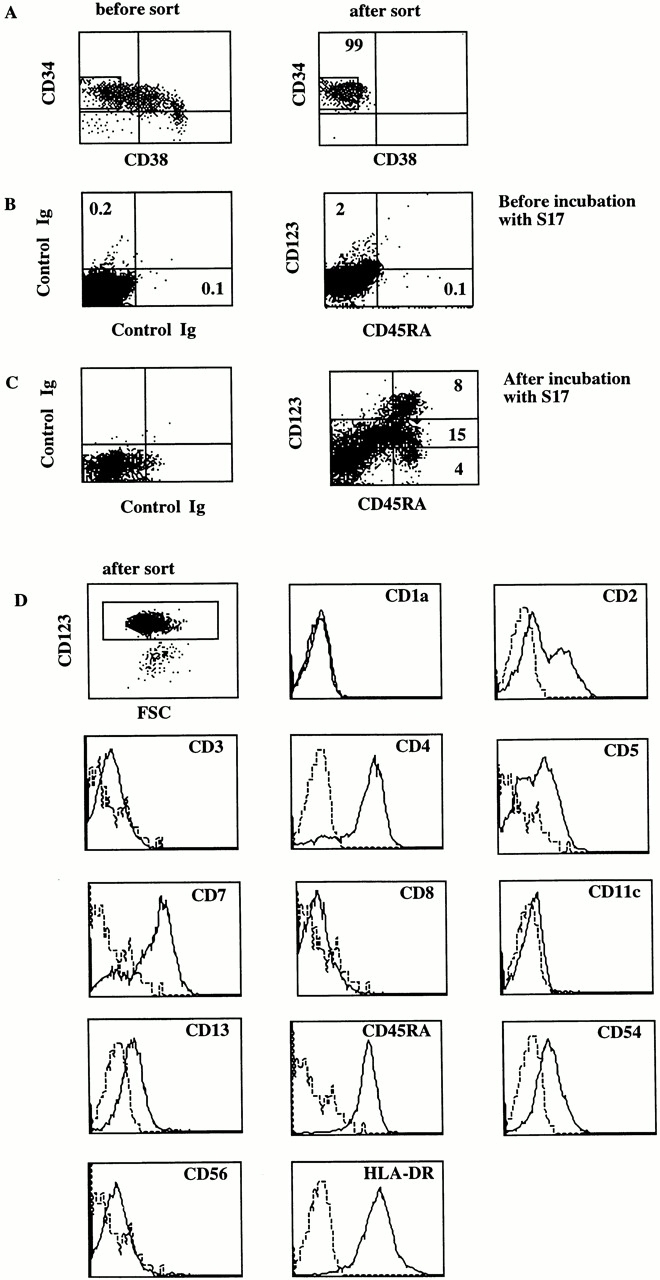
CD34+CD38− fetal liver cells cocultured with S17 cells develop into cells that contain CD123high pDC2. Fetal liver cells were first enriched for CD34+ cells using the MACS. The resulting cell population was sorted and was cocultured with S17 cells. After 5 d of culture the cell surface phenotype of sorted CD123high cells was analyzed. (A) Phenotypes of MACS enriched CD34+ cells before and after the sort. (B) CD123 and CD45RA expression of the sorted CD34+ cells at the start of the coculture with S17 cells. (C) CD45RA and CD123 expression after 5 d coculture with S17 cells. (D) The phenotype of CD123high cells, sorted from the S17 cocultures. The sorted cells were stained with Tricolor- and PE-labeled antibodies. Tricolor-labeled CD5, CD7, and CD56 antibodies were used, and the other antibodies were labeled with PE. The histograms of the Tricolor- and PE-labeled Igs are slightly different because of the different labels. FSC, forward side scatter.
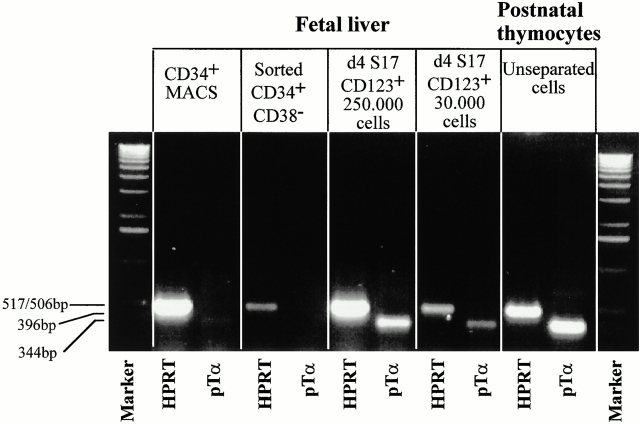
We have found previously that thymic CD123high pDC2 express pTα transcripts 19. However, we could not completely rule out the possibility that a contaminating pre-T cell population was responsible for the pTα signal. To check whether these transcripts are also present in S17-generated CD123high pDC2, we purified CD123high cells from the S17 cultures and analyzed the presence of pTα mRNA by reverse transcription (RT)-PCR. Fig. 2 shows that the pTα PCR product had the predicted size. The PCR products obtained from the CD123high cells and the thymocytes were recovered from the agarose gel and sequenced. As a positive control, we also sequenced a retroviral vector containing the full-length pTα cDNA. The sequences of the two PCR samples were identical to that of the corresponding stretch in the pTα cDNA that was sequenced in parallel, confirming that the PCR samples were derived from pTα transcripts. These data demonstrate that S17-generated pDC2 express pTα mRNA, whereas the freshly isolated CD34+ cells did not express pTα transcripts.
Figure 2.
CD123high cells generated from CD34+ cells express pTα mRNA. Purified CD34+CD38− fetal liver cells were incubated with S17 cells. 5 d later the CD123high cells were sorted (99% pure upon reanalysis) and analyzed for the presence of HPRT and pTα transcripts by reverse transcription PCR. The pTα PCR products were then recovered and sequenced.
It has been reported that pDC2 produce high levels of IFN-α 23 24. To determine whether the S17-generated CD123high cells are able to secrete IFN-α, CD34+CD38− fetal liver cells were sorted and cocultured with S17 cells for 4 d. After this incubation, the CD123highCD4+ cells were sorted and stimulated with 10 PFU/cell gamma irradiation–inactivated HSV-1 as indicated in Materials and Methods. We also determined IFN-α secretion by S17-cocultured cells that expressed no or low levels of CD123. The S17-generated CD123high pDC2 secreted IFN-α upon stimulation with the HSV-1 virus in two separate experiments (7,500 pg/ml and 13,000 pg/ml per 106 cells), whereas the CD123− population secreted much lower levels of IFN-α (350 pg/ml and 25 pg/ml, respectively).
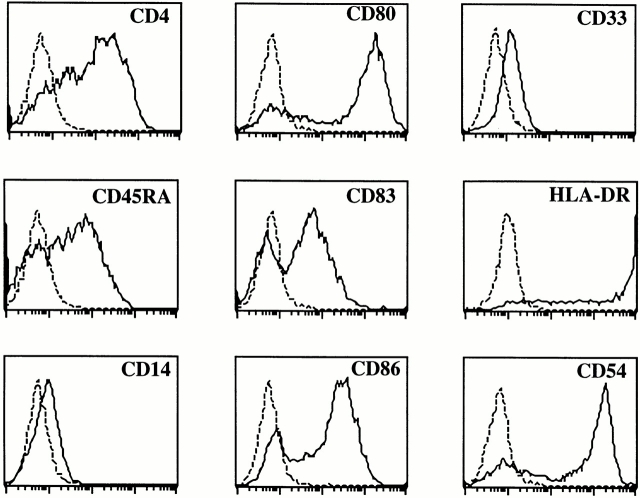
To determine the capacity of the S17-generated CD123high cells to develop into mature DCs, we sorted the CD123highCD4+ cells from the S17-cocultured cells using a nonblocking anti-CD123 antibody and stimulated these cells with IL-3 with or without CD40L-expressing L cells. After 4 d of incubation, the cells acquired the veiled morphology typical for DCs (results not shown). As expected, the IL-3/CD40L–stimulated cells expressed high levels of HLA-DR, the costimulatory molecules CD80 and CD86, and the mature DC marker CD83 (Fig. 3). The expression of HLA-DR and CD54 increased considerably compared with that on the in vitro–generated pDC2 (Fig. 1). The differentiated cells only very weakly expressed the myeloid marker CD33, which is highly expressed on DC1. The phenotype of these DCs is similar to that obtained after differentiation of tonsillar and thymic CD123high pDC2 19 20. Although not shown, these mature DC2 very efficiently stimulated proliferation of allogeneic naive CD4+ CD45RA+ peripheral blood T cells confirming that these cells are antigen-presenting DCs.
Figure 3.
CD123high cells generated from CD34+ cells develop into mature DC2 upon incubation with CD40L and IL-3. Purified CD34+CD38− fetal liver cells were incubated with S17 cells. 5 d later the CD123high cells were sorted (99% pure upon reanalysis) and incubated with CD40L-expressing mouse L cells and IL-3.
CD34+CD1a− but Not CD34+CD1a+ Fetal Thymic Precursors Are Able to Develop into CD123high pDC2.
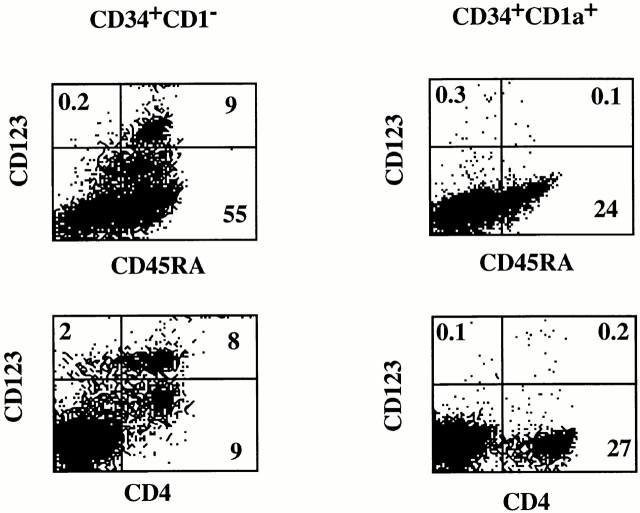
The phenotypic similarity of the S17-generated CD123high cells with freshly isolated CD123high pDC2 and the fact that these cells differentiated into DCs upon culture with IL-3/CD40L indicate that the S17 coculture system can be used as an assay for CD123high pDC2 precursor activity of CD34+ cells. Studies in the mouse have demonstrated that thymic DCs develop from thymic precursors 35. Previous work has shown that CD34+CD1a− thymocytes contain bipotential T/NK cell precursors 36 and are able to develop into DCs upon culture in GM-CSF–containing media 31 37. Upon upregulation of CD1a, the capacity of CD34+ thymocytes to develop into NK cells and DCs is lost. To determine whether CD34+ thymic populations can develop into CD123high pDC2, we sorted CD34+ populations on the basis of CD1a expression and tested these populations in the S17 system. Fig. 4 shows that although CD34+CD1a− cells generated CD4+CD45RA+CD123high pDC2, CD34+CD1a+ cells failed to develop into CD123high pDC2 upon coculture with S17 cells. In a parallel experiment, the same sorted CD34+CD1a+ cells developed into T cells in a fetal thymic organ culture system (data not shown), as was found previously 15 38. These results demonstrate that upon upregulation of CD1a, the cells lose their capacity to develop into CD123high pDC2. Note that coculture of CD34+CD1a− cells with S17 cells resulted in the appearance of CD123dimCD45RA+ cells, which were not observed in the cultures with CD34+ CD1a+ cells. The nature of these cells is presently under investigation.
Figure 4.
CD34+CD1a− but not CD34+CD1a+ cells are able to differentiate into CD123highCD45RA+ pDC2. CD34+CD1a− and CD1a+ thymocytes were purified and cocultured with S17 cells. After 4 d of culture, the cells were stained with CD123, CD4, and CD45RA.
Ectopic Expression of Id2 or Id3 in CD34+ Precursors Completely Inhibits Development of CD123high pDC2 but Not Development of Myeloid DC1.
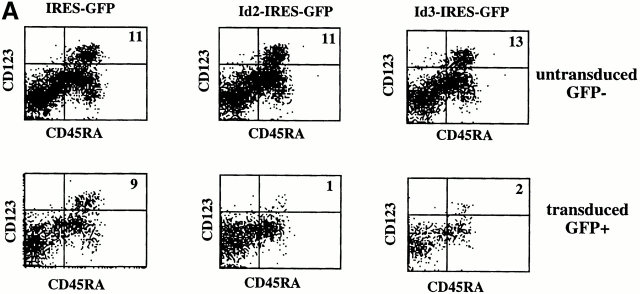
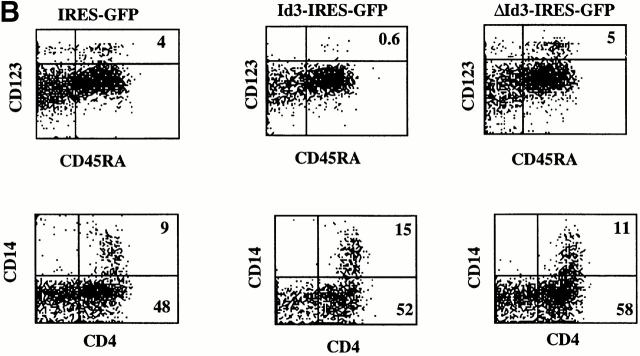
We have documented that enforced expression of Id3 into CD34+ precursors inhibited their capacity to develop into T and B cells without affecting their NK cell or CD14 myeloid precursor activity 13 14 15. These findings suggest a role for one or more Id protein(s) as switch factor(s) regulating T versus NK choices in the thymus and B versus NK choices in the bone marrow. Recently, it was reported that Id2−/− mice lack NK cells 16. In contrast, Id3−/− mice have normal NK cell development (Murre, C., personal communication). These observations strongly suggest that Id2 is the physiological factor determining lineage choices in the lymphoid compartment and predict that Id2 has an activity similar to that of Id3 in blocking T cell development. Indeed, ectopic expression of Id2 in CD34+CD1a− thymocytes that contain bipotential T/NK precursors 36 completely blocked T cell but not NK cell development in a fetal thymic organ culture (results not shown). To reveal a possible common developmental pathway of T cells, B cells, NK cells, and CD123high pDC2, we tested the effect of Id2 and Id3 overexpression on development of the latter cells. CD34+ CD38− fetal liver cells were purified and incubated overnight with SCF and IL-7 as indicated in Materials and Methods. The cells were then transduced with IRES-GFP, Id2-IRES-GFP, or Id3-IRES-GFP, and cocultured with S17. Fig. 5 A shows that compared with the GFP control, Id2 and Id3 strongly inhibited the generation of CD123highCD45RA+ cells. Not only the percentages of CD123highCD45RA+ cells were much lower, but also the absolute numbers of CD123highCD45RA+ cells were reduced >10-fold. To ensure that the inhibiting activity of the Id proteins is due to their capacity to sequester E proteins, we tested CD34+CD38− cell transduced with a mutated Id3 (ΔId3) that is unable to dimerize with bHLH factors 27. As expected, ΔId3 failed to inhibit pDC2 development (Fig. 5 B). As was also observed previously 14, generation of CD4+CD14+ cells in the S17 coculture was not inhibited by Id3 (Fig. 5 B), indicating that Id proteins do not inhibit generation of DC1 precursors (monocytes). Id3 slightly stimulated generation of CD4+CD14+ cells compared with the GFP only and ΔId3 in this experiment, but this effect was not observed in additional experiments. Also, Id2 did not affect generation of CD4+CD14+ cells in the cocultures of CD34+CD38− cells and S17 cells (results not shown).
Figure 5.
Ectopic expression of Id2 or Id3 inhibits development of pDC2. Purified CD34+CD38− fetal liver cells were transduced with IRES-GFP, Id2-IRES-GFP, Id3-IRES-GFP (A), or Id3-IRES-GFP and ΔId3-IRES-GFP (B), and incubated with S17 cells for 4 d. After the culture, the cells were stained with antibodies against the indicated antigens. (A) Staining of untransduced GFP− and transduced GFP+ cells. (B) Staining of only the transduced GFP+ cells.
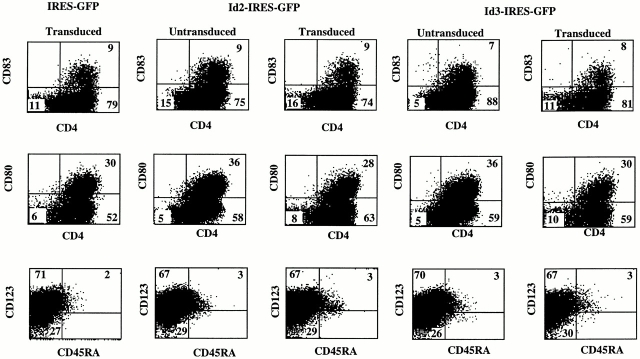
DCs can also be generated from CD34 precursor cells after culture in GM-CSF–containing media (39; for a review, see reference 40). These DCs are similar to monocyte-derived DCs (DC1 21) and most likely belong to the myeloid lineage. Fig. 6 shows that SCF, GM-CSF, and TNF-α–induced development of CD4+, CD83+, and CD80+ cells from CD34+CD38− fetal liver cells was not affected by Id2 or Id3 overexpression. Id2 and Id3 also did not affect the percentages of cells expressing CD1a, CD13, and CD40 (results not shown). Note that the in vitro–generated DC1 also contain cells with a high expression of CD123. However, the great majority of these cells did not express CD45RA. Importantly, generation of CD123+ cells in SCF, GM-CSF, and TNF-α was not inhibited by Id2 or Id3. Thus, the lack of CD123high pDC2 cells in the cultures of Id-transduced CD34+ cells with S17 is not the result of an inhibition of CD123 expression by Id proteins. These data clearly indicate a different mechanism of development of myeloid-derived DC1 and CD123high pDC2 and thus (mature) DC2. As expected, Id2 and Id3 also inhibited the generation of CD123high pDC2 from CD34+CD1a− thymocytes in the same way as that from CD34+CD38− fetal liver cells. However, the differentiation of CD34+CD1a− thymocytes into DC1 in medium containing SCF, GM-CSF, and TNF-α was not inhibited at all by Id2 or Id3 (results not shown).
Figure 6.
Ectopic expression of Id2 or Id3 does not affect differentiation of DC1. Purified CD34+CD38− fetal liver cells were transduced with IRES-GFP, Id2-IRES-GFP, and Id3-IRES-GFP, and cultured with SCF, GM-CSF, and TNF-α for 5 d. After the culture, the cells were stained with antibodies against the indicated antigens.
Discussion
In previous studies we have documented that ectopic expression of Id3 but not of ΔId3 inhibited development of primitive hematopoietic precursors into T and B cells 13 14. In contrast, NK cell development was stimulated by Id3 13. The recent observation that Id2−/− mice lack NK cells 16, whereas NK cells are normal in Id3−/− mice (41; Murre, C., personal communication), strongly suggests that Id2 is the relevant switch factor for T/NK development in vivo. Consistent with this notion, we found that ectopic expression of Id2 inhibits development of T and B cells but not NK cells (results not shown), similarly as found previously for Id3 13. These data strongly suggest that Id2 positively regulates the development of NK cells and at the same time shuts off the capacity of precursor cells to develop into T and B cells. To test the effects of ectopic expression of Id2 and Id3 on the development of pDC2, we employed our observation that the murine stromal cell line S17 induces development of these cells from CD34+ cells. The mechanism by which S17 stimulates pDC2 development is unknown but it is possible that this involves cell–cell contact and a soluble factor. Blom et al. in this issue demonstrated that Flt-3L induces CD34+CD45RA− cells to differentiate into pDC2 42. As murine Flt-3L interacts with human Flt-3 43, this cytokine may be involved in S17-mediated induction of pDC2 development. Using this assay, we demonstrate that Id2 and Id3 but not ΔId3 strongly blocked the development of primitive CD34+CD38− fetal liver cells and CD34+ CD1a− thymic precursors into CD123high pDC2. In contrast, Id3 and Id2 had no effect on S17-induced development of CD34+CD38− cells into CD4+CD14+ pDC1. Moreover, neither Id2 nor Id3 inhibited SCF/GM-CSF/TNF-α–induced DC1 development of CD34+CD38− fetal liver cells and thymic CD34+CD1a− cells, respectively. The differential effect of Id2 and Id3 on the development of CD123high pDC2 compared with that on SCF/GM-CSF/TNF-α–induced DC1 development indicates that distinct mechanisms regulate differentiation of these two DC lineages and strongly suggests distinct developmental origins.
Cell transfer studies in the mouse support a model in which T cells and thymic DCs are intrathymically generated from a common precursor. As the thymic precursors are unable to develop into myeloid cells, thymic DCs are considered to be lymphoid rather than myeloid related (18; for a review, see reference 35). The observation that CD123high pDC2 develop from CD34+CD1a− thymic precursors could therefore be consistent with the notion that these cells are of lymphoid origin. However, it should be noted that M-CSF-R+CD34+ cells have been found in the human thymus 44. Upon culture with M-CSF and GM-CSF, those cells can develop into DCs via a CD14+ intermediate 44, indicating that the human thymus does contain precursor cells with myeloid DC potential. The observations that CD34+CD1a− thymocytes can develop into DCs in SCF, GM-CSF, and TNF-α, and that this is not blocked by Id2 or Id3 (results not shown), are consistent with this notion. The presence of myeloid DC precursors in the human thymus indicates that the argument that a certain DC type is of lymphoid origin because their precursors are present in the thymus is not valid, at least for the human system. However, several characteristics of thymic CD123high pDC2 support the notion of a lymphoid origin of these cells. These include the presence of CD2, CD5, and CD7, which are expressed on T and NK cells but in general not on myeloid cells, and the lack of expression of typical myeloid cell–associated markers CD13 and CD33 19. Moreover, both the CD123high pDC2 generated from CD34+ cells (Fig. 2) as well as ex vivo–isolated pDC2 19 express pTα transcripts. The fact that these transcripts are expressed in pre-T cells 45, uncommitted T/NK precursors 46, and pDC2 19 strongly suggests a common developmental origin of T cells and pDC2. Therefore, we believe the hypothesis that CD123high pDC2 are of lymphoid origin to be a valid one. However, we realize that final proof requires clonal analyses that demonstrate the development of single precursors into CD123high pDC2 and other lymphocyte lineages but not into myeloid cells. As CD123high pDC2 cells expand very poorly in S17 cocultures, culture conditions that induce extensive expansion of CD123high pDC2 should be established before clonal analyses of the DC precursor activity of putative CLP populations can be performed.
Assuming that CD123high pDC2 are of lymphoid origin, a model presents itself in which Id proteins control lymphoid development by allowing NK cell development and limiting the options of lymphoid precursors to develop into T and B cells and CD123high pDC2. It is likely that the relevant Id protein is Id2, as Id2-deficient mice 16, but not Id3-deficient mice (Murre, C., personal communication), lack NK cells. The phenotype of Id2−/− mice strongly suggests that certain bHLH factors (the E proteins, HEB, E2A, and E2-2 or other yet to be defined factors) inhibit the development of NK cells. Expression of Id1, Id3 14, and Id2 (results not shown) is relatively high in uncommitted CD34+CD38− fetal liver cell precursors compared with more differentiated CD34+CD38+ precursors. Therefore, it is conceivable that in microenvironments, where Id2 is not downregulated in lymphoid precursors, NK cells are generated from these precursor cells because bHLH factors that block NK development are sequestered. These may be the same bHLH factors that positively control T cell, B cell, and CD123high pDC2 development.
Id protein expression is downregulated during B cell development 47, releasing E2A proteins that control, either directly or indirectly, the transcription of PAX-5 48. PAX-5 in turn consolidates B cell development by shutting off the options of precursor cells to develop into other lineages 8. A similar mechanism may operate in the thymus for T cells and CD123high pDC2. We have recently shown that Id3 overexpression in early precursor cells inhibits T cell development at the T/NK branchpoint 13 and now show that Id2 has the same activity. Ectopic expression of Id3 in committed T cell precursors still blocks development of TCR-α/β1 cells 15, indicating that continued activities of bHLH factors during early stages of T cell development are required to complete this process. Interestingly, Id3-overexpressing pre-T cells acquire the capacity to develop into cells with the phenotype and functional activities of NK cells but with TCR gene rearrangements 15. Thus, pre-T cells may be able to develop into another cell type when crucial bHLH factors are sequestered by overexpression of Id3. This situation is similar to that with PAX-5 and B cell development, as “pre-B” cells lacking PAX-5 have the capacity to develop into various other lineages, including T and NK cells 8 9. It is tempting to speculate that sequestering bHLH factors inhibits the activity of a T cell equivalent of PAX-5, releasing a block on alternative developmental options of pre-T cells.
Several bHLH factors have now been implicated in T cell development. These include the bHLH factors E12, E47, HEB 10 11, and the class VI HLH factor HES-1 12. Elucidation of the identity of class I bHLH factors critical for the development of CD123high pDC2 will come from analyses for the presence of the murine equivalent of these cells in mouse strains with E protein deficiencies.
Acknowledgments
We thank C. Murre and R. de Groot for providing us with constructs and Ken Dorshkind for giving us the S17 cell line
This work was supported by Netherlands Organization for Science grant 805-17-531 to H. Spits and by grants (HD29341 and HD37597) from the National Institutes of Health to C. Uittenbogaart.
Footnotes
C.H. Uittenbogaart's present address is the Dept. of Pediatrics, and the Department of Microbiology and Immunology, UCLA School of Medicine, Los Angeles, CA 90095-1747.
Abbreviations used in this paper: CLP, common lymphoid precursor; bHLH, basic helix-loop-helix; DC, dendritic cell; GFP, green fluorescent protein; HPRT, hypoxanthine ribosyltransferase; Id, inhibitor of DNA binding; ΔId3, mutated Id3; IRES; internal ribosomal entry site; pDC, pre-DC; pTα, pre-TCR-α; SCF, stem cell factor.
References
- Galy A., Travis M., Cen D., Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Murre C., Bain G., van Dijk M.A., Engel I., Furnari B.A., Massari M.E., Matthews J.R., Quong M.W., Rivera R.R., Stuiver M.H. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Massari M.E., Murre C. Helix-loop-helix proteinsregulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H. The protein Ida negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bain G., Maandag E.C., Izon D.J., Amsen D., Kruisbeek A.M., Weintraub B.C., Krop I., Schlissel L., Feeney A.J., van Roon M., Murre C. E2A proteins are required for proper B cell development and initiation of Ig gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Soriano P., Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Heavey B., Rolink A.G., Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- Rolink A.G., Nutt S.L., Melchers F., Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5- deficient B-cell progenitors. Nature. 1999;401:603–606. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- Bain G., Engel I., Robanus Maandag E.C., te Riele H.P.J., Voland J.R., Sharp L.L., Chun J., Huey B., Pinkel D., Havran W.L., Murre C. E2A deficiency leads to abnormalities in αβ and γδ T cell development, perturbations in V(D)J rearrangement and to the rapid development of T-cell lymphomas. Mol. Cell. Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Cheng P., Weintraub H. B lymphocyte development is regulated by the combined dosage of three basic-helix-loop-helix genes, E2A, E2-2 and HEB. Mol. Cell. Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Hattori M., Nakamura E., Nakanishi S., Minato N., Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk M.H.M., Blom B., Nolan G., Stegmann A.P.A., Bakker A.Q., Weijer K., Res P.C.M., Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleco A.C., Stegmann A.P., Heemskerk M.H., Couwenberg F., Bakker A.Q., Weijer K., Spits H. Genetic modification of human B-cell developmentB-cell development is inhibited by the dominant negative helix loop helix factor Id3. Blood. 1999;94:2637–2646. [PubMed] [Google Scholar]
- Blom B., Heemskerk M.H., Verschuren M.C., van Dongen J.J., Stegmann A.P., Bakker A.Q., Couwenberg F., Res P.C., Spits H. Disruption of alpha beta but not of gamma delta T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2793–2802. doi: 10.1093/emboj/18.10.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y., Gyotuku J.-I., Ogawa M., Nishikawa G.-I., Katsura Y., Gachelin G., Nakauchi H. Characterization of c-kit positive intrathymic stem cells that are restricted to lymphoid differentiation. J. Exp. Med. 1993;178:1283–1292. doi: 10.1084/jem.178.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardavin C., Wu L., Shortman K. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 1993;362:761–763. doi: 10.1038/362761a0. [DOI] [PubMed] [Google Scholar]
- Res P.C., Couwenberg F., Vyth-Dreese F.A., Spits H. Expression of pTalpha mRNA in a committed dendritic cell precursor in the human thymus. Blood. 1999;94:2647–2657. [PubMed] [Google Scholar]
- Grouard G., Rissoan M.C., Filgueira L., Durand I., Banchereau J., Liu Y.J. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissoan M.C., Soumelis V., Kadowaki N., Grouard G., Briere F., de Waal Malefyt R., Liu Y.J. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–1186. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- Sorg R.V., Kogler G., Wernet P. Identification of cord blood dendritic cells as an immature CD11c− population. Blood. 1999;93:2302–2307. [PubMed] [Google Scholar]
- Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., Liu Y.J. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- Cella M., Jarrossay D., Facchetti F., Alebardi O., Nakajima H., Lanzavecchia A., Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Bocarsly P., Feldman M., Mendelsohn M., Curl S., Lopez C. Human mononuclear cells which produce interferon-alpha during NK(HSV-FS) assays are HLA-DR positive cells distinct from cytolytic natural killer effectors. J. Leukoc. Biol. 1988;43:323–334. doi: 10.1002/jlb.43.4.323. [DOI] [PubMed] [Google Scholar]
- Sandberg K., Gobl A.E., Funa K., Alm G.V. Characterization of the blood mononuclear leucocytes producing alpha interferon after stimulation with herpes simplex virus in vitro, by means of combined immunohistochemical staining and in situ RNA-RNA hybridization. Scand. J. Immunol. 1989;29:651–658. doi: 10.1111/j.1365-3083.1989.tb01169.x. [DOI] [PubMed] [Google Scholar]
- Loveys D.A., Streiff M.B., Kato G.J. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acid Res. 1996;24:2813–2820. doi: 10.1093/nar/24.14.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella T.M., Nolan G.P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Hanenberg H., Hashino K., Konishi H., Hock R.A., Kato I., Williams D.A. Optimization of fibronectin-assisted retroviral gene transfer into human CD34+ hematopoietic cells. Hum. Gene Ther. 1997;8:2193–2206. doi: 10.1089/hum.1997.8.18-2193. [DOI] [PubMed] [Google Scholar]
- Yssel H., De Vries J.E., Koken M., van Blitterswijk W., Spits H. Serum-free medium for the generation and the propagation of functional human cytotoxic and helper T cell clones. J. Immunol. Methods. 1984;72:219–227. doi: 10.1016/0022-1759(84)90450-2. [DOI] [PubMed] [Google Scholar]
- Res P., Martínez-Cáceres E., Jaleco A.C., Noteboom E., Weijer K., Spits H. CD34+CD38dim cells in the human thymus can differentiate into T, natural killer and dendritic cells but are distinct from stem cells. Blood. 1996;87:5196–5206. [PubMed] [Google Scholar]
- Rawlings D.J., Quan S.G., Kato R.M., Witte O.N. Long-term culture system for selective growth of human B-cell progenitors. Proc. Natl. Acad. Sci. USA. 1995;92:1570–1574. doi: 10.1073/pnas.92.5.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings D.J., Quan S., Hao Q.L., Thiemann F.T., Smogorzewska M., Witte O.N., Crooks G.M. Differentiation of human CD34+CD38− cord blood stem cells into B cell progenitors in vitro. Exp. Hematol. 1997;25:66–72. [PubMed] [Google Scholar]
- Olweus J., BitMansour A., Warnke R., Thompson P.A., Carballido J., Picker L.J., Lund-Johansen F. Dendritic cell ontogenya human dendritic cell lineage of myeloid origin. Proc. Natl. Acad. Sci. USA. 1997;94:12551–12556. doi: 10.1073/pnas.94.23.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortman K., Vremec D., Corcoran L.M., Georgopoulos K., Lucas K., Wu L. The linkage between T-cell and dendritic cell development in the mouse thymus. Immunol. Rev. 1998;165:39–46. doi: 10.1111/j.1600-065x.1998.tb01228.x. [DOI] [PubMed] [Google Scholar]
- Sánchez M.-J., Muench M.O., Roncarolo M.G., Lanier L., Phillips J.H. Identification of a common T/NK cell progenitor in human fetal thymus. J. Exp. Med. 1994;180:569–576. doi: 10.1084/jem.180.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez C., Trigueros C., Fernández E., Toribio M.L. The development of T and non-T cell lineages from CD34+ human thymic precursors can be traced by the differential expression of CD44. J. Exp. Med. 1995;181:475–483. doi: 10.1084/jem.181.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom B., Verschuren M.C., Heemskerk M.H., Bakker A.Q., van Gastel-Mol E.J., Wolvers-Tettero I.L., van Dongen J.J., Spits H. TCR gene rearrangements and expression of the pre-T cell receptor complex during human T-cell differentiation. Blood. 1999;93:3033–3043. [PubMed] [Google Scholar]
- Caux C., Dezutter Dambuyant C., Schmitt D., Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Rivera R.R., Johns C.P., Quan J., Johnson R.S., Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- Blom B., Ho S., Antonenka S., Liu Y.-J. Generation of interferon α–producing pre-dendritic cell (pre-DC)2 from human CD34+ hematopoietic stem cells. J. Exp. Med. 2000;192:1785–1795. doi: 10.1084/jem.192.12.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum C., Culpepper J., Campbell D., McClanahan T., Zurawski S., Bazan J.F., Kastelein R., Hudak S., Wagner J., Mattson J. Ligand for FLT3/FLK2 receptor tyrosine kinase regulates growth of haematopoietic stem cells and is encoded by variant RNAs. Nature. 1994;368:643–648. doi: 10.1038/368643a0. [DOI] [PubMed] [Google Scholar]
- Dalloul A.H., Patry C., Salamero J., Canque B., Grassi F., Schmitt C. Functional and phenotypic analysis of thymic CD34+CD1a− progenitor-derived dendritic cellspredominance of CD1a+ differentiation pathway. J. Immunol. 1999;162:5821–5828. [PubMed] [Google Scholar]
- von Boehmer H., Fehling H.J. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- Carlyle J.R., Zuniga-Pflucker J.C. Requirement for the thymus in alphabeta T lymphocyte lineage commitment. Immunity. 1998;9:187–197. doi: 10.1016/s1074-7613(00)80601-9. [DOI] [PubMed] [Google Scholar]
- Allman D., Li J., Hardy R.R. Commitment to the B lymphoid lineage occurs before DH-JH recombination. J. Exp. Med. 1999;189:735–740. doi: 10.1084/jem.189.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G., Robanus Maandag E., te Riele H., Feeney A.J., Sheehy A., Schlissel M., Shinton S.A., Hardy R.R., Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]