Abstract
We identify here a novel class of loss-of-function alleles of uncoordinated locomotion (unc)-108, which encodes the Caenorhabditis elegans homologue of the mammalian small guanosine triphosphatase Rab2. Like the previously isolated dominant-negative mutants, unc-108 loss-of-function mutant animals are defective in locomotion. In addition, they display unique defects in the removal of apoptotic cells, revealing a previously uncharacterized function for Rab2. unc-108 acts in neurons and engulfing cells to control locomotion and cell corpse removal, respectively, indicating that unc-108 has distinct functions in different cell types. Using time-lapse microscopy, we find that unc-108 promotes the degradation of engulfed cell corpses. It is required for the efficient recruitment and fusion of lysosomes to phagosomes and the acidification of the phagosomal lumen. In engulfing cells, UNC-108 is enriched on the surface of phagosomes. We propose that UNC-108 acts on phagosomal surfaces to promote phagosome maturation and suggest that mammalian Rab2 may have a similar function in the degradation of apoptotic cells.
Introduction
During phagocytosis, a cell ingests objects >0.5 μm in diameter. The phagocytic removal of apoptotic and degenerating cells plays key roles in developmental processes such as organ sculpting, tissue remodeling, and axon pruning and actively prevents tissue injury, inflammation, and autoimmune responses (for reviews see Savill and Fadok, 2000; Fainzilber and Twiss, 2006).
During the development of Caenorhabditis elegans, somatic and germ cells undergo programmed cell death. Apoptotic cells can be visualized using Nomarski differential interference contrast (DIC) microscopy as highly refractile discs, referred to as “cell corpses” (Sulston and Horvitz, 1977). As in other metazoans, apoptotic cells in C. elegans are rapidly removed via phagocytosis (Ellis et al., 1991a). Somatic cell corpses are ingested by a variety of neighboring cells that include hypodermal cells, pharyngeal muscles, and intestinal cells (Sulston and Horvitz, 1977; Sulston et al., 1983; Zhou et al., 2001b). Apoptotic germ cells are specifically cleared by gonadal sheath cells, which wrap around the germ line syncytium (Gumienny et al., 1999). Once ingested, the cell corpse is sequestered within the phagosome, a compartment derived from the plasma membrane of the engulfing cell, where it is degraded (Robertson and Thomson, 1982).
Genetic screens for C. elegans mutants that contain persistent cell corpses have identified at least eight genes required for their removal: cell death abnormal (ced)–1, -2, -5, -6, -7, -10, and -12 (for review see Zhou et al., 2004) and dynamin 1 (dyn-1; Yu et al., 2006). Double mutant analyses suggest that they function in two parallel and partially redundant pathways to control the cell corpse removal: ced-2, -5, -10, and -12 in one pathway and ced-1, -6, -7, and dyn-1 in the other (Fig. 1 A; for review see Reddien and Horvitz, 2004; Yu et al., 2006).
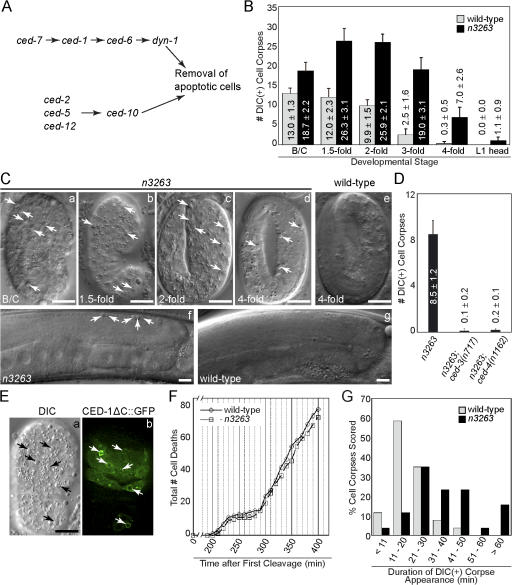
Figure 1.
n3263 mutants are defective in the removal of apoptotic cells. (A) Diagram of two parallel pathways that control the removal of apoptotic cells in C. elegans. (B) The number of cell corpses visible under DIC optics are reported as mean ± SD; n = 15. The bean/comma (B/C), 1.5-, 2-, 3-, and late 4-fold stages correspond to ∼380, 420, 460, 520, and 700 min after first cleavage (Yu et al., 2006). (C) DIC images of embryos (a–e) at the indicated stages. Anterior is on top. (f and g) DIC images of gonad arms in adult hermaphrodites 48 h after the L4 larval stage. Midbody is shown on the left and dorsal faces are on top. Cell corpses are marked with arrows. (D) Numbers of cell corpses in fourfold stage embryos are reported as mean ± SD (n = 15). (E) DIC (a) and fluorescence (b) images of an n3263 embryo. Arrows indicate cell corpses labeled with enriched CED-1ΔC::GFP from engulfing cells. (F) The total number of embryonic cell death events that occur 200–400 min after the first cleavage plotted against time. (G) Histogram summarizing the duration of the DIC(+) appearance of cell corpses in embryos. 30 cell corpses generated in embryos were scored. Bars, 10 μm.
In the first pathway, a CED-2/CrkII, CED-5/Dock180, and CED-12/ELMO1 protein complex activates CED-10/Rac1 GTPase to promote the reorganization of the actin cytoskeleton (Wu and Horvitz, 1998b; Reddien and Horvitz, 2000; Gumienny et al., 2001; Wu et al., 2001; Zhou et al., 2001a). In the second pathway, the CED-7/ABC transporter functions, at least in part, to facilitate the presentation of phosphatidylserine and perhaps other “eat me” signals on the surface of cell corpses to attract phagocytes (Wu and Horvitz, 1998a; Zhou et al., 2001b; Venegas and Zhou, 2007). The phagocytic receptor CED-1, which is expressed on the surface of engulfing cells, recognizes these eat me signals, clusters around cell corpses, and initiates engulfment (Zhou et al., 2001b). CED-6/GULP is a phosphotyrosine-binding domain–containing protein proposed to function as an adaptor for CED-1 (Liu and Hengartner, 1998; Zhou et al., 2001b; Su et al., 2002).
Several important events such as the reorganization of the cytoskeleton, the expansion and closure of the plasma membrane around a cell corpse, and the initiation of phagosome maturation must be coordinated in phagocytes to ensure the efficient clearance of cell corpses. Recently, we have shown that loss of function of dyn-1 (which encodes the large GTPase dynamin in C. elegans) results in severe defects in the engulfment and degradation of apoptotic cells (Yu et al., 2006). Under the regulation of CED-1 and -6, DYN-1 promotes the recruitment and fusion of intracellular vesicles to the surface of extending pseudopods and maturing phagosomes (Yu et al., 2006). The delivery of these vesicles is likely to provide membrane for cell surface extension and digestive enzymes for the degradation of apoptotic cells. An additional activity of CED-1 and -6 in actin polymerization mediated through CED-10 has also been suggested (Kinchen et al., 2005).
Besides dyn-1, not much is known about the genes that control intracellular vesicle trafficking during the removal of apoptotic cells. In this paper, we find a novel function for the gene unc-108 in the degradation of engulfed cell corpses.
Results
n3263 mutants are defective in the removal of both somatic and germ cell corpses
To identify additional genes required for the removal of apoptotic cells, we performed a genetic screen for late-stage embryos that contain multiple cell corpses visible using DIC microscopy (Ced phenotype; Zhou et al., 2001a; Yu et al., 2006). One of the recessive mutants is n3263.
During embryonic development, 113 somatic cells undergo apoptosis (Sulston et al., 1983). In wild-type embryos, many cell corpses are visible during midembryogenesis (from the bean to twofold stages), as a large number of cells die during this period (Fig. 1 B). However, by the late fourfold stage of embryogenesis (just before hatching), virtually all of these cell corpses are cleared because of their rapid engulfment and degradation (Fig. 1, B and C). n3263 mutants contain excess refractile discs that resemble cell corpses at both mid and late embryonic stages (Fig. 1, B and C).
Two lines of evidence indicate that these objects are cell corpses. First, loss-of-function mutations in the proapoptotic ced-3/caspase and ced-4/Apaf1 genes, which result in the blockage of developmental cell deaths (for review see Metzstein et al., 1998), prevent the appearance of cell corpse–like objects in n3263 mutants, which indicates that apoptosis is required for their generation (Fig. 1 D). Second, a CED-1ΔC::GFP reporter on the surface of engulfing cells (Zhou et al., 2001b) clusters around these objects in n3263 embryos, which suggests that they express the eat me signals and are thus likely to be apoptotic cells (Fig. 1 E).
To determine whether the extra cell corpses observed in n3263 mutant embryos result from excessive apoptosis, we recorded using DIC microscopy (see Materials and methods) each cell death that occurred 200–400 min after the first embryonic cell division. We found that the number as well as timing of cell deaths in n3263 embryos closely resembled that of the wild type (Fig. 1 F). These results indicate that the execution of programmed cell death remains largely unaffected. Thus, the excess cell corpses observed in n3263 embryos are unlikely to be a result of ectopic cell death.
Although n3263 mutants display more cell corpses during all embryonic stages compared with the wild type embryos, these cell corpses are eventually cleared so that by hatching almost none remain (Fig. 1, B and C). We recorded the duration for which individual cell corpses were visible under DIC optics (see Materials and methods). In wild-type embryos, the vast majority (90%, n = 30) of cell corpses disappeared within 30 min of their appearance and no cell corpse persisted for >50 min (Fig. 1 G). In contrast, 56.7% (n = 30) of cell corpses in n3263 mutants persisted for >30 min after their appearance (Fig. 1 G). Collectively, these observations suggest that the increased number of cell corpses observed in n3263 mutants is caused by a prolonged removal time.
About 300–500 germ cells undergo apoptosis in the adult hermaphrodite gonad. Dying germ cell nuclei in the distal section of the syncytial germline rapidly cellularlize and are engulfed by the gonadal sheath cell (Gumienny et al., 1999). Because of efficient removal, very few germ cell corpses are visible in the wild type at any given stage despite the continuous occurrence of apoptosis (Fig. 1 C and see Fig. 4 C). n3263 mutants display a more than threefold increase in germ cell corpses compared with the wild type at the same stage (Fig. 1 C and see Fig. 4 C), which indicates similar defects in their removal.
n3263 mutants are uncoordinated in locomotion
n3263 mutants display a recessive uncoordinated locomotion (Unc) phenotype (Fig. 2 D). These mutants are slow in forward locomotion. Although able to initiate backing upon a gentle tap to the head, they are unable to sustain this movement. This Unc phenotype cosegregated with the Ced phenotype during all the crosses performed (see Materials and methods), which indicates that these two defects may be caused by the same mutation. However, unlike the Ced phenotype, which can be rescued by the maternal wild-type gene product, the Unc phenotype is zygotic (see the following section; Fig. 2 E).
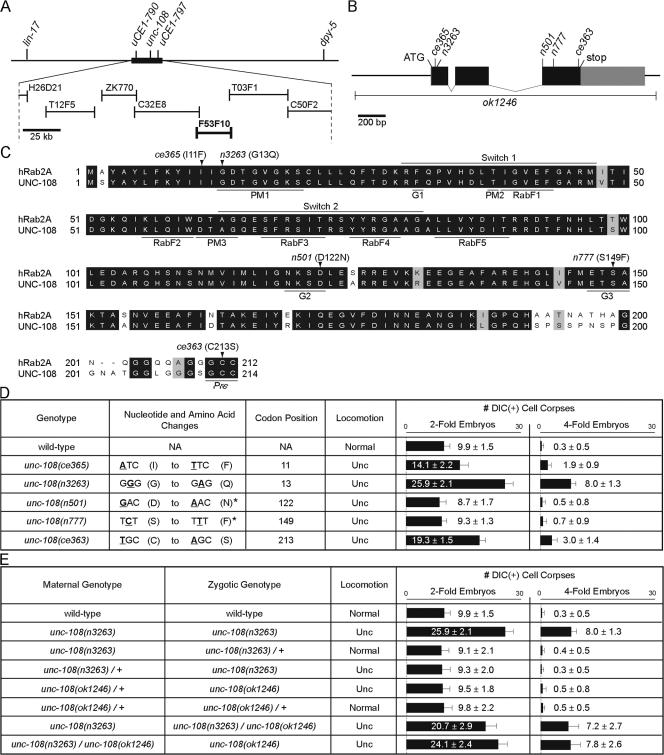
Figure 2.
unc-108 encodes a homologue of human Rab2 and controls locomotion and cell corpse removal. (A) Position of the unc-108 locus on chromosome I. Among the cosmid clones tested (lines with bars on both sides), F53F10 rescued the n3263 phenotypes. (B) Structure of unc-108. Exons (boxes) are connected by introns (lines). The coding region is shown in black and the untranslated region is in gray. Positions of mutations are indicated. (C) Alignment of human Rab2A and UNC-108. PM1–3, motifs that bind the phosphate groups of GTP and the Mg2+ cofactor; G1–G3, motifs that contact the guanine base; switch 1 and 2, domains predicted to undergo dramatic conformational change upon GTP hydrolysis; RabF, signature residues conserved among Rab family members (Pereira-Leal and Seabra, 2000, 2001). The positions and amino acid substitutions resulting from the five point mutations are indicated. Pre, prenylation site. (D and E) Molecular lesions and quantification of phenotypes in the indicated genetic backgrounds. The numbers of cell corpses are reported as mean ± SD (n = 15). (asterisks) Data from Simmer et al. (2003).
Cloning unc-108
We mapped the n3263 mutation to a 178-kb region on chromosome I and identified a cosmid clone, F53F10, that fully rescued the Ced and Unc phenotypes of n3263 (see Materials and methods; Figs. 2 A and 3 C). F53F10 contains unc-108 (F53F10.4), which encodes the C. elegans homologue of the small GTPase Rab2 (Fig. 2 B and C; Simmer et al., 2003). In n3263 animals, we identified a missense mutation in the unc-108 coding sequence that affects G13, a residue in the PM1 (phosphate-magnesium binding) motif that is absolutely conserved among Ras superfamily members (Fig. 2, B–D; Valencia et al., 1991). This mutation is predicted to severely inactivate Rab2 (see the following sections). We found that unc-108(+) cDNA under the control of ∼2.3 kb of its upstream regulatory sequence (Punc-108) completely rescued the Ced and Unc phenotypes of n3263 mutants (Fig. 3 C). Introduction of the G13Q mutation to unc-108 cDNA abolished its rescuing activity, demonstrating that this mutation causes the Ced and Unc phenotypes observed in n3263 animals (Fig. 3 C).
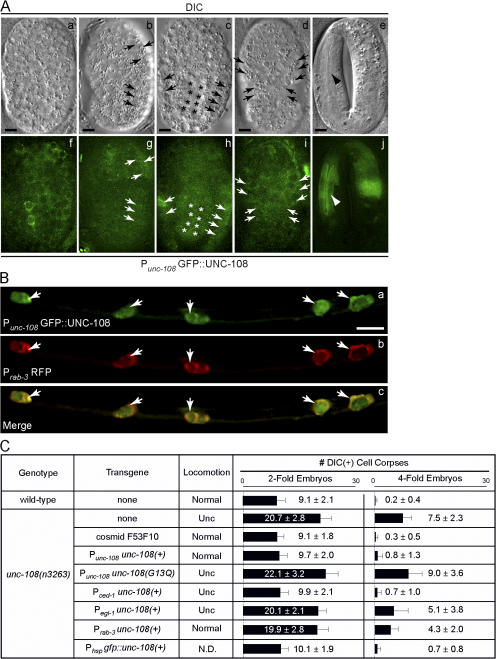
Figure 3.
unc-108 is strongly expressed and performs distinct functions in neurons and engulfing cells. (A) DIC (a–e) and corresponding GFP (f–j) images of wild-type embryos expressing Punc-108 gfp::unc-108(+). Anterior is shown on top. Cell types that express GFP include hypodermal (arrows) and intestinal (asterisks) cells and pharyngeal muscles (arrowhead). (B) Expression of the Punc-108 gfp::unc-108(+) and Prab-3 rfp reporters in neurons of the ventral cord (cell bodies are indicated with arrows) in an L2 larva. Bars, 5 μm. (C) Tissue-specific rescue of Unc and Ced defects of unc-108(n3263) mutants. Transgenes are introduced as extrachromosomal arrays. The number of DIC(+) cell corpses at two- and fourfold embryonic stages are reported as mean ± SD (n = 15). N.D., not determined.
Two distinct classes of unc-108 mutant alleles
We isolated two additional recessive alleles of unc-108 in a screen for mutations that improve the growth and reduce the hyperactive locomotion of a goa-1 (GαO, G protein O, α subunit) mutant (Williams et al., 2007). ce365 encodes an I11F substitution close to the N terminus, whereas ce363 results in a C213S mutation in the prenylation motif that may disrupt membrane localization of UNC-108 (Fig. 2, C and D). Both ce363 and ce365 mutants display Ced and Unc phenotypes (Figs. 2 D and S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1) and fail to complement unc-108(n3263) (not depicted). Furthermore, unc-108(RNAi) recapitulates the Ced and Unc defects of n3263, ce363, and ce365 (Fig. S1, B and C), which indicates that these phenotypes arise from the loss of unc-108 function.
ok1246, a deletion generated by the C. elegans Gene Knockout Consortium (http://www.wormbase.org), removes the entire coding region of unc-108 and most likely represents a null allele (Fig. 2 B). unc-108(ok1246) homozygotes descended from the unc-108(ok1246)/+ heterozygotes (unc-108 mok1246/+ zok1246; m, maternal genotype; z, zygotic genotype) display a recessive Unc phenotype characteristic of other loss-of-function mutants of unc-108. unc-108 mok1246/+ zok1246 animals cease development at L1 or L2 larval stages yet do not contain extra cell corpses at either two- or fourfold embryonic stages (Fig. 2 E). Similarly, n3263 homozygous embryos descended from n3263/+ mothers (unc-108 mn3263/+ zn3263) are non-Ced (Fig. 2 E).
We generated unc-108(n3263)/unc-108(ok1246) heterozygote progeny (unc-108 mn3263 zn3263/ok1246) and found they were viable and fertile yet exhibited Unc and Ced (Fig. 2 E). Furthermore, their ok1246 homozygous progeny (unc-108 mn3263/ok1246 zok1246) display Unc and Ced and arrest as L1- or L2-stage larvae (Fig. 2 E). These observations suggest that the loss of zygotic unc-108 is sufficient to cause the Unc phenotype but that loss of both maternal and zygotic unc-108 is required for the accumulation of cell corpses.
The severity of the Ced phenotype displayed by unc-108 mn3263/ok1246 zok1246 embryos is similar to that displayed by unc-108(n3263) homozygotes, which indicates that n3263 is a strong or possibly null allele with regard to the Ced phenotype. For this reason, we characterize unc-108(n3263) mutants in our subsequent experiments. We categorize n3263, ce363, ce365, and ok1246 as class I recessive Unc and Ced alleles. The total loss of zygotic unc-108 may also cause developmental arrest, as observed from unc-108 mok1246/+ zok1246. However, the possibility that this larval arrest is a result of a closely linked mutation has not been eliminated.
unc-108 mutants were originally isolated as dominant Unc mutants (Park and Horvitz, 1986). Two alleles, n501 and n777, bear mutations in conserved residues (D112N and S149F, respectively) in the G2 and G3 (guanine-binding) motifs, which form part of the pocket that holds the guanine base (Fig. 2 C; Valencia et al., 1991; Simmer et al., 2003). Results from structural modeling predict that the D112N and S149F mutations would result in the loss of hydrogen bonds that stabilize guanine nucleotide binding (see following section). Interestingly, although both display a Unc phenotype characteristic of all unc-108 mutants, neither n501 nor n777 has excess cell corpses in embryos (Fig. 2 D). We categorize these alleles as class II dominant-negative Unc non-Ced alleles.
Structural studies of point mutations in UNC-108
To predict the biochemical properties of the proteins encoded by dominant-negative class II mutants, we generated structural models of wild-type UNC-108 and the D122N and S149F mutant proteins. Although the structure of the GDP-bound human Rab2A has been studied (Eathiraj et al., 2005), the structure of GTP-bound conformation has not been solved. In order to model UNC-108 in the GTP-bound state, we used the structures for human Rab11A as templates (Pasqualato et al., 2004; Pasqualato and Cherfils, 2005). Rab11 belongs to the same subfamily as Rab2 (Pereira-Leal and Seabra, 2001) and is the closest homologue for which the GDP- and GTP-bound structures have been determined (Fig. S2, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1).
The interactions that stabilize GTP binding in UNC-108 mirror those originally found in the human Ras (hRas) GTPase (Pai et al., 1989, 1990). Specifically, the C-terminal side chain of D122 (corresponding to hRas D119) is predicted to form hydrogen bonds with the endocyclic and exocyclic NH2 moieties of the guanine base (Fig. S2 C). Although S149 (hRas S145) does not form direct interactions with the guanine nucleotide, the hydroxyl group in its side chain may form a hydrogen bond with D122 (Fig. S2 C). This interaction stabilizes the G3 loop, particularly the position of A150 (hRas A146), whose main chain amine forms a hydrogen bond with the keto group of the guanine base (Pai et al., 1989, 1990). Although the D122N and S149F mutations are not predicted to dramatically alter the global structure of UNC-108, the positions of and the moieties present in the side chains of these mutations result in the loss of conserved hydrogen bonds that are required for binding guanine nucleotides (Fig. S2, D and E). Additionally, the aromatic ring of F149 might hinder the formation of a hydrogen bond between D122 and the endocyclic N of the guanine base.
The G13Q substitution encoded by unc-108(n3263) affects a residue that is absolutely conserved among members of the Ras superfamily of small GTPases. The amino acid backbone for position 13 (10 in hRas) is in a conformation that severely penalizes the replacement of glycine by any other amino acid (Pai et al., 1990). The replacement of G13 with a bulky residue (Q) in UNC-108 is thus predicted to result in a dramatic shift in the position of the PM1 loop, which is almost certain to disrupt the interaction with guanine nucleotides and may also affect the overall protein structure. In addition, G13 is part of the N-terminal β sheet implicated in interaction with downstream effectors of Rab2 (Tisdale, 2003; Tisdale et al., 2004). The G13Q mutation might also affect the association of UNC-108 with its effectors. Together, these effects are likely to result in a strong loss of UNC-108 activity.
unc-108 expression is detected in engulfing cell types and in neurons
We observed broad expression of a GFP::UNC-108 fusion protein under the control of Punc-108 in wild-type C. elegans. GFP signal was detected in pharyngeal, intestinal, and hypodermal cells that are known to function as phagocytes during embryogenesis (Fig. 3 A). In addition, we observed robust GFP expression in most if not all neurons, including head, ventral cord, and tail neurons in larvae and adults coexpressing Punc-108 gfp::unc-108(+) and a neural-specific Pr ab-3 rfp reporter (Fig. 3 B and not depicted; Choi et al., 2006).
unc-108 acts in engulfing cells for cell corpse removal and in neurons for normal locomotion
We examined whether the expression of unc-108(+) specifically in engulfing or dying cells could rescue the Ced phenotype of unc-108(n3263). The expression of unc-108(+) using engulfing cell-specific Pced-1 (Zhou et al., 2001b) rescues the Ced phenotype of unc-108(n3263) embryos, yet fails to rescue the Unc phenotype (Fig. 3 C). In contrast, the expression of unc-108(+) under the control of Pegl-1 in dying somatic cells (Conradt and Horvitz, 1999) does not significantly alleviate the Ced phenotype in unc-108(n3263) mutants (Fig. 3 C). These results indicate that the unc-108 activity in engulfing but not dying cells is sufficient for the removal of cell corpses.
However, although the expression of unc-108(+) under the control of neuron-specific Prab-3 does not rescue the Ced phenotype of unc-108(n3263) mutants, it completely rescues the Unc phenotype (Fig. 3 C), which implies that the abnormal locomotion results from the loss of unc-108 function in neurons. These findings suggest that unc-108 performs independent functions in engulfing cells and neurons. The expression of unc-108(+) in dying cells does not rescue the Unc phenotype (Fig. 3 C).
unc-108 is likely to function downstream of the engulfment ced genes
To determine if unc-108 functions in either of the two pathways acting in cell corpse removal (Fig. 1 A) or an independent pathway, we constructed double mutants between unc-108(n3263) and reference alleles of the seven engulfment ced genes. Double mutants of unc-108(n3263) and the null mutations ced-1(e1735), ced-5(n1812),or ced-7(n1996) (Wu and Horvitz, 1998a,b; Zhou et al., 2001b) display Ced phenotypes comparable in severity to that of ced-1(e1735), ced-5(n1812),or ced-7(n1996) single mutants (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1). Furthermore, unc-108(n3263) fails to enhance the Ced phenotype of either ced-1(e1735); ced-5(n1812) or ced-7(n1996); ced-5(n1812) double mutants (Fig. S3 A). These observations imply that unc-108 does not simply function exclusively in either one of the two pathways; otherwise, the Ced phenotype of at least one of the single mutants that function in parallel to unc-108 would be enhanced by the n3263 mutation. Furthermore, unc-108 does not appear to function in a third, independent pathway either, in which case the null mutant phenotypes in both of the known pathways should be enhanced by unc-108(n3263).
We observed an enhancement of the Ced phenotype of ced-2(n1994), ced-6(n2095), ced-10(n1993), and ced-12(n3261) single mutants by unc-108(n3263) (Fig. S3 A). However, ced-2(n1994), ced-6(n2095), ced-10(n1993), and ced-12(n3261) may represent partial loss-of-function alleles in corresponding genes (Liu and Hengartner, 1998; Reddien and Horvitz, 2000; Zhou et al., 2001a), which makes the interpretation of these results complicated. However, with the exception of unc-108(n3263); ced-6 (n2095), none of these double mutant combinations display a Ced phenotype significantly more severe than that of ced-1(e1735), ced-5(n1812),or ced-7(n1996) single null mutants, which is consistent with the conclusions drawn in the preceding paragraph.
Overexpression of ced-10 cDNA under the control of heat shock promoters has been found to bypass the requirement for its upstream regulators ced-2, ced-5, and ced-12 (Reddien and Horvitz, 2000; Gumienny et al., 2001; Wu et al., 2001; Zhou et al., 2001a). Furthermore, Phsp ced-10(+) overexpression can also alleviate the engulfment defects of ced-1, ced-6, and ced-7 mutants (Kinchen et al., 2005). We observed that ced-10(+) overexpression failed to reduce the number of cell corpses in unc-108(n3263) embryos even though a significant alleviation of the Ced phenotype was observed in ced-1(e1735) and ced-5 (n1812) embryos that overexpress Phsp ced-10(+) (Fig. S3 B). These observations indicate that unc-108 does not function upstream of ced-10. Rather, the results of double mutant analysis and ced-10(+) overexpression suggest, among other possibilities, that unc-108 may act downstream of both pathways.
The degradation of cell corpses is severely delayed in unc-108(n3263) mutants
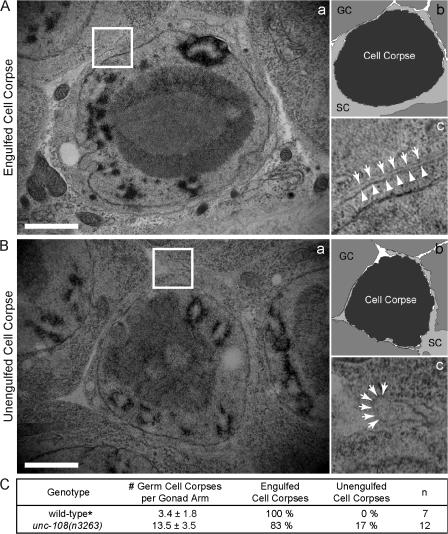
We performed transmission EM (TEM) on the hermaphrodite gonad to determine if cell corpses in unc-108(n3263) are engulfed (see Materials and methods). As in the wild type, apoptotic germ cells in unc-108(n3263) mutants cellularize and detach from the germline syncytium, appearing condensed and almost completely devoid of cytoplasm (Fig. 4, A and B). Phagosomal and plasma membranes are distinguishable around 10 of the 12 cell corpses that were scored, which indicates that these cell corpses have been internalized by gonadal sheath cells (Fig. 4 A). The two remaining cell corpses are not fully engulfed, although pseudopods from the sheath cells are seen extending around them (Fig. 4 B). In wild-type animals, all of the few germ cell corpses present are engulfed by sheath cells (Fig. 4 C; Yu et al., 2006). The presence of many additional germ cell corpses in unc-108(n3263) animals and the fact that most of them are internalized (Fig. 4 C) suggests that unc-108(n3263) mutants are primarily defective in the degradation of apoptotic cells.
Figure 4.
Most germ cell corpses in unc-108(n3263) mutants are engulfed but remain undegraded. TEM of gonadal tissues containing one engulfed (A) and one unengulfed (B) germ cell corpse in unc-108(n3263) adult hermaphrodites 48 h after the L4 larval stage. (b) A schematic diagram of panel a. GC, germ cell; SC, gonadal sheath cell. (c) 3× view of boxed region in panel a. Plasma membranes of sheath cells (arrows) and cell corpses (arrowheads) are marked. Bars, 1 μm. (C) Percentage of engulfed cell corpses as scored by TEM in hermaphrodites 48 h after the L4 stage. The numbers of germ cell corpses scored under DIC optics are reported as mean ± SD (n = 15). (asterisk) Data from Yu et al. (2006).
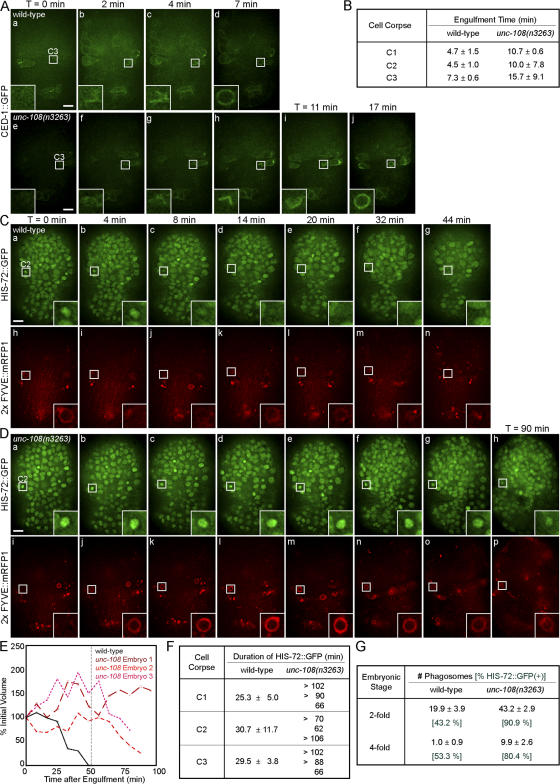
To characterize the specific defects caused by the unc-108(n3263) mutation in the removal of cell corpses, we used time-lapse imaging to monitor the engulfment and degradation of three particular cells (C1, C2, and C3) that are located close to the ventral surface of an embryo and die at approximately the same time (see Materials and methods and Fig. 7 A; Yu et al., 2006). Unless otherwise specified, the expression of fluorescent reporters was driven in engulfing cells by Pced-1.
The engulfment of C1, C2, and C3 is monitored using CED-1::GFP, which clusters on the extending pseudopods (Zhou et al., 2001b; Yu et al., 2006). Although engulfment is initiated in unc-108(n3263) embryos, the speed of pseudopod extension appears to decrease and thus engulfment takes twice as long as in the wild type (Fig. 5, A and B; and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1). However, compared with the severe defects in phagosome maturation (see the following paragraphs), this prolonged engulfment only makes a minor contribution to the Ced phenotype of unc-108(n3263) mutants.
Figure 5.
unc-108(n3263) causes defects in the engulfment and degradation of apoptotic cells. Time-lapse images of embryos expressing Pced-1 ced-1::gfp (A) or coexpressing Phis-72 his-72::gfp and Pced-12× FYVE::mrfp1 (C and D). Insets are a 3× view of the boxed areas. The anterior is shown on top and the ventral side faces outward. (A) Engulfment of cell corpse C3 visualized by the clustering CED-1::GFP on extending pseudopods. Time (T) = 0 min, the time point immediately before budding pseudopods are detected. The enclosure of the CED-1::GFP circle marks the completion of engulfment. (B) Time required for the engulfment of C1, C2, and C3 in wild-type and unc-108(n3263) embryos given as mean ± SD (n = 3). (C and D) The degradation of C2 cell corpses. Nuclei of all cells are marked with HIS-72::GFP and phagosomal membranes are marked with 2× FYVE::mRFP1. T = 0 min, the completion of engulfment marked by recruitment of 2× FYVE::mRFP1. Bars, 5 μm. (E) Relative volumes of representative phagosomes containing C1 plotted against time. (F) Duration of apoptotic cell HIS-72::GFP in phagosomes. For wild-type samples, results are listed as mean ± SD (n = 3). Results for three individual cell corpses are reported for unc-108(n3263). (G) The number of phagosomes observed in twofold stage embryos reported as mean ± SD (n = 15). Numbers in green represent the percentage of phagosomes containing HIS-72::GFP(+) cell corpses.
To monitor cell corpse degradation, we developed reporters that label phagosomal membranes and the internalized cell corpses. Phosphatidylinositol-3-monophosphate (PI[3]P) is known to be enriched on the surface of early endosomes and phagosomes (Ellson et al., 2001; Vieira et al., 2001). By expressing the PI(3)P-specific reporter 2× FYVE::monomeric RFP 1 (mRFP1) in engulfing cells (Yu et al., 2006), we observed that PI(3)P was enriched on the surfaces of nascent phagosomes. Phagosomes are easily distinguishable from 2× FYVE-labeled endosomes because (a) they are much larger in size (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1), (b) the 2× FYVE–labeled phagosomes correspond to DIC(+) cell corpses (Fig. S4 C), and (c) nascent phagosomes are first labeled by clustered CED-1::GFP before 2× FYVE::mRFP1 (Fig. S4, A and B). 2× FYVE::mRFP1 remains associated with phagosomes until they become membrane remnants within an hour of the completion of engulfment (Fig. 5 C and Video 3). These results indicate that PI(3)P is a reliable marker for phagosomal surfaces. Furthermore, after the major wave of embryonic programmed cell deaths, which occurs 200–420 min after first cleavage (Sulston et al., 1983), very few cell corpses in engulfment ced mutants are labeled with 2× FYVE::GFP compared to the wild type (Fig. S4, C and D), which indicates that the detection of a 2× FYVE::GFP circle is a reliable marker for an engulfed cell corpse.
In unc-108(n3263) mutants, the appearance of the 2× FYVE::mRFP1 circles occurs with kinetics similar to the wild type (Figs. S4 and S5, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1), which suggests that the initial presentation of PI(3)P on phagosomes is unaffected. However, phagosomes in unc-108(n3263) retain much of their original volume for longer periods (Fig. 5, D and E; and Video 4). In some cases, these phagosomes were observed to increase in size during recording (Fig. 5 E).
We used a chromatin-associated HIS-72::GFP to monitor the degradation of phagosomal contents. HIS-72 encodes a variant of histone H3 that is broadly expressed in somatic cells (Ooi et al., 2006). In embryos coexpressing Phis-72 his-72::gfp and Pced-12× FYVE::mrfp1, we observed an increase in HIS-72::GFP signal in cell corpses, reflecting the condensation of nuclear DNA during apoptosis (Video 3). Shortly afterward, the condensed pyknotic nuclei labeled with HIS-72::GFP were seen within 2× FYVE::mRFP1(+) phagosomes (Fig. 5 C and Video 3). Within a period of 20–30 min, the HIS-72::GFP puncta decreased in size and subsequently disappeared, which indicates the degradation of nuclear chromosomes (Fig. 5, C and F; and Video 3). The decrease of GFP signal intensity might also be partially caused by the gradual acidification of the phagosomal lumen (see the following section). In contrast, the HIS-72::GFP signal in unc-108(n3263) remained within phagosomes for a minimum of 60 min and in most cases beyond ∼102 min (Fig. 5, D and F; and Video 4).
The failure of cell corpse degradation observed in unc-108(n3263) embryos is not limited to phagosomes containing C1, C2, and C3. We examined all phagosomes labeled by PI(3)P in two- and fourfold–stage embryos and observed that many more phagosomes persist in unc-108(n3263) compared with the wild type (Fig. S4 D). In addition, undigested chromatin was present in almost all phagosomes in unc-108(n3263) compared with only half in wild-type embryos (Fig. 5 G). Together, the persistence of phagosomes and the failure to degrade apoptotic cell chromatin suggests that unc-108(n3263) causes severe defects in phagosome maturation.
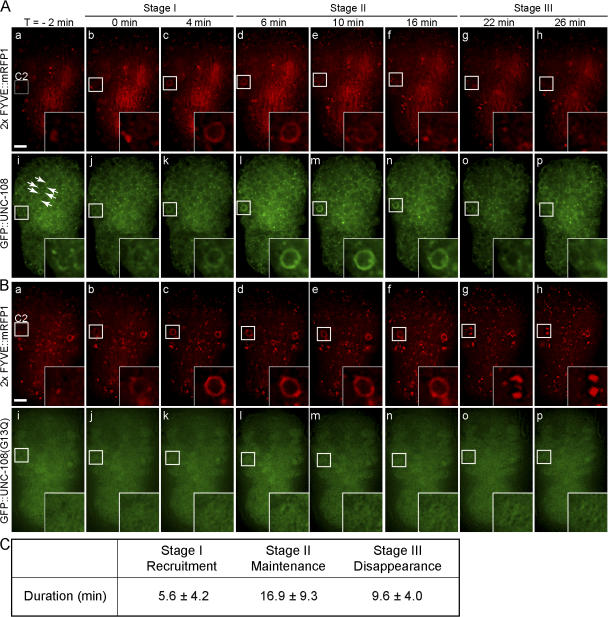
UNC-108 is transiently recruited to the surface of phagosomes
To test if UNC-108 acts directly on phagosomes to control the degradation of cell corpses, we analyzed the localization of a functional N-terminal GFP::UNC-108 fusion (Phsp gfp::unc-108(+); Fig. 3 C). GFP::UNC-108 is localized to small cytoplasmic puncta that resemble intracellular organelles (Fig. 6 A). In wild-type embryos, GFP::UNC-108 was recruited to the surface of nascent phagosomes labeled with 2× FYVE::mRFP1. Three distinct stages mark the dynamic localization of UNC-108 to phagosomes. During stage I, GFP::UNC-108(+) puncta are recruited to the surface of nascent phagosomes and gradually evolve into bright continuous circles (Fig. 6 A). These bright circles last for 10–20 min (stage II). During the subsequent period of∼10 min (stage III), GFP::UNC-108 gradually disappears from the phagosomal membrane (Fig. 6, A and C; and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1).
Figure 6.
UNC-108 is transiently enriched on phagosomal surfaces. Time-lapse images of wild-type embryos coexpressing Phsp gfp::unc-108 and Pced-12× FYVE ::mrfp1 (A) or Phsp gfp::unc-108(G13Q) and Pced-12x FYVE::mrfp1 (B). Insets are a 3× view of the boxed regions. The anterior is shown on top and the ventral side faces outward. Time (T) = 0 min, formation of a 2× FYVE::mRFP1 circle. (A) Transient enrichment of GFP::UNC-108 to phagosome membranes (i–p). GFP::UNC-108(+) puncta are indicated by arrows. (B) Diffuse cytoplasmic localization of GFP::UNC-108(G13Q) and the failure in recruitment to phagosomes (i–p). Bars, 5 μm. (C) Dynamic pattern of GFP::UNC-108 enrichment on phagosome surfaces in wild-type embryos. See text for the definition of the three stages. Results are reported as mean ± SD (n = 9).
We found that a GFP::UNC-108(G13Q) reporter (corresponding to the n3263 allele) appeared in a diffuse pattern in the cytoplasm and was not localized to the surface of phagosomes during cell corpse engulfment and degradation (Fig. 6 B and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1). This localization pattern is consistent with the behavior of inactive Ras superfamily G proteins (Zerial and McBride, 2001) and supports the genetic observations indicating that G13Q completely inactivates UNC-108.
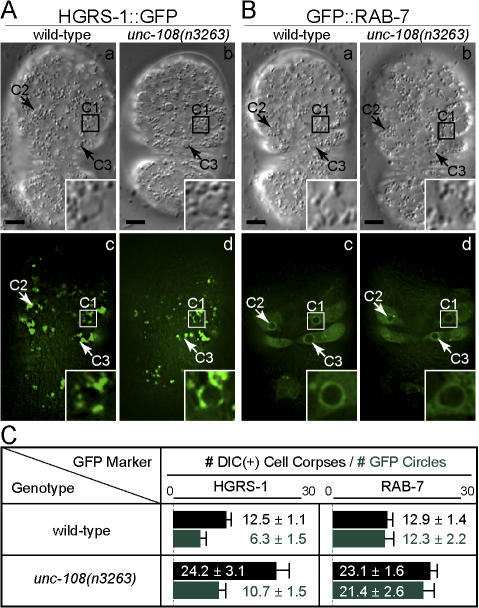
unc-108(n3263) causes severe defects in the delivery of lysosomes to phagosomes
To identify specific cellular processes regulated by UNC-108, we tested whether several events that are essential for phagosome maturation occur in unc-108(n3263) mutants. Components of the endocytic pathway are recruited to phagosomes to deliver membrane and protein cargo required for their maturation (for review see Vieira et al., 2002). Previously, using the marker hepatocyte growth factor–regulated tyrosine kinase substrate (HGRS-1; the C. elegans homologue of mammalian Hrs), we demonstrated that early endosomes are recruited to the surface of pesudopods and phagosomes during the removal of cell corpses (Yu et al., 2006). Here, using time-lapse recording, we observed that HGRS-1::GFP was recruited to phagocytic cups and nascent phagosomes in unc-108(n3263) embryos with kinetics indistinguishable from those observed in the wild type (Fig. 7 A and not depicted). In addition, a similar fraction of DIC(+) cell corpses are labeled with HGRS-1::GFP in wild-type and mutant embryos (Fig. 7 C), which suggests that the recruitment of early endosomes is not affected by the unc-108(n3263) mutation.
Figure 7.
The recruitment of HGRS-1 and RAB-7 is normal in unc-108(n3263) mutants. (A and B) DIC (a and b) and fluorescence (c and d) images of embryos expressing Pced-1 hgrs-1::gfp (A) or Pced-1 gfp::rab-7 (B). Insets are a 3× view of the boxed regions. The anterior is shown on top and the ventral side faces outward. Bars, 5 μm. (C) Number of DIC(+) cell corpses (black) and HGRS-1::GFP(+) and GFP::RAB-7(+) phagosomes (green) in twofold embryos. Results are reported as mean ± SD (n = 15).
We recently identified an essential role for the small GTPase RAB-7 in the removal of cell corpses in C. elegans. The GTP-bound form of RAB-7 is recruited to the surface of phagosomes shortly after the completion of engulfment and promotes the recruitment and fusion of lysosomal particles to phagosomes (unpublished data). Just as in the wild type, we found that virtually all phagosomes containing cell corpses in unc-108(n3263) embryos were labeled with GFP::RAB-7 (Fig. 7, B and C).
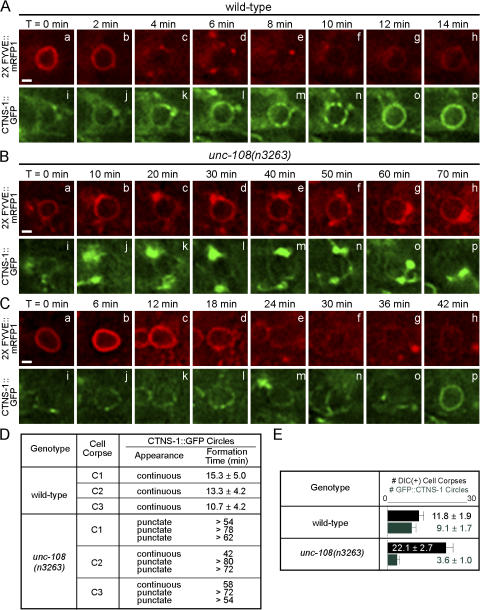
We then examined whether the recruitment of lysosomes to phagosomes was affected by unc-108(n3263) mutation. CTNS-1 is a C. elegans homologue of human lysosome-specific cystine transporter cystinosin (Town et al., 1998; Kalatzis et al., 2001) and was observed to colocalize on cytoplasmic puncta with a previously established lysosomal marker LMP-1 (unpublished data). In wild-type embryos, most DIC(+) cell corpses are surrounded by CTNS-1::GFP (unpublished data) circles, which indicates that lysosomes are recruited to maturing phagosomes (Fig. 8 E). We observed that CTNS-1::GFP was initially recruited to phagosomal surfaces as puncta. These puncta gradually progressed into a smooth circle, which suggests the subsequent fusion of lysosomes with the phagosomal membrane and the distribution of lysosomal membrane markers over phagosomal surfaces (Fig. 8 A and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1). In unc-108(n3263) mutants, we observed variable defects in the recruitment and fusion of CTNS-1::GFP(+) particles to phagosomes. In many cases, only a few CTNS-1::GFP(+) particles were observed to associate with phagosomal surfaces. Unlike in wild-type embryos, these puncta only covered a small portion of the phagosomal surface, which suggests that lysosomes are not efficiently recruited (Fig. 8, B and D; and Video 8). Quite often these particles did not evolve into continuous circles on phagosomal surfaces (Fig. 8, B and D; and Video 8), which suggests a failure of lysosomes to fuse with the phagosomal membrane (Fig. 8, B and D). In cases where lysosomes did fuse with phagosomes in unc-108(n3263), the progression of CTNS-1::GFP from phagosome-associating puncta to continuous circles took three to five times as long as in the wild type (Fig. 8, C and D; and Video 9). Consistent with these observations, a much smaller fraction of total phagosomes are covered with smooth CTNS-1::GFP(+) circles in unc-108(n3263) compared with the wild type at the twofold embryonic stage (12 vs. 77%; Fig. 8 E).
Figure 8.
unc-108(n3263) mutants are defective in the incorporation of lysosomes to phagosomes. (A–C) Time-lapse recording of phagosomes containing C2 in wild-type (A) and unc-108(n3263) (B and C) embryos coexpressing Pced-12× FYVE::mrfp1 and Pced-1 ctns-1::gfp. Bars, 1 μm. (D) Summary of the formation of CTNS-1::GFP circles on phagosomal surfaces over time. Formation time, the interval between the appearance of a 2× FYVE::mRFP1 circle and a CTNS-1::GFP circle on the same phagosome. Data for wild-type embryos are presented as mean ± SD (n = 3). Data for unc-108(n3263) embryos are individually listed, as the CTNS-1::GFP puncta on most phagosomes failed to convert to continuous circles within the recording period. (E) Number of DIC(+) cell corpses (black) and CTNS-1::GFP(+) phagosomes (green) in twofold embryos. Results are reported as mean ± SD (n = 15).
unc-108(n3263) mutants are defective in the acidification of phagosomal lumen
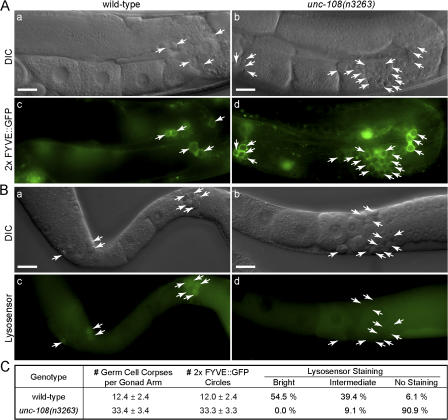
The acidification of the phagosomal lumen is an important step in the maturation of phagosomes. Low pH is required for the optimal activity of acid hydrolases and regulates key steps in membrane traffic (for review see Vieira et al., 2002). To measure the acidification of phagosome lumen, we stained animals using Lysosensor blue/yellow DND-160, a dual-emission wavelength dye that displays strong yellow fluorescence at a pH ≤ 5.0 (see Materials and methods).
To obtain a large number of cell corpses for analysis, we induced germ cell apoptosis in adult hermaphrodites by γ-ray irradiation (see Materials and methods). In wild-type animals, essentially all germ cell corpses are marked by 2× FYVE::GFP circles (Fig. 9, A and C), which indicates that germ cells undergoing DNA damaged–induced apoptosis are rapidly engulfed. Healthy germ cell nuclei display undetectable or faint Lysosensor staining, whereas 93.9% (n = 33) of phagosomes display intermediate to bright staining, reflecting their efficient acidification (Figs. 9, B and C). In unc-108(n3263) animals, only 9.1% (n = 33) of phagosomes are stained (Fig. 9), which suggests that unc-108 is required for the phagosome acidification.
Figure 9.
Acidification of phagosomal lumen is defective in unc-108(n3263) mutants. (A) DIC (a and b) and corresponding fluorescence (c and d) images showing 2× FYVE::GFP circles around germ cell corpses induced by γ-ray irradiation in adult hermaphrodites. The midbody is shown on the left and the dorsal side faces up. (B) DIC (a and b) and fluorescence (c and d) images of dissected gonads stained with Lysosensor DND-160 after γ-ray irradiation. Arrows indicate germ cell corpses or the corresponding phagosomes. Bars, 20 μm. (C) Quantification of the acidification defects in unc-108(n3263) mutants. The numbers of DIC(+) cell corpses and 2× FYVE::GFP phagosomes are reported as mean ± SD (n = 15). Bright, intermediate, and no Lysosensor staining correspond to a phagosome/germline signal intensity ratio of ≥2.0, >1.1 and < 2.0, and ≤1.1, respectively. For each genotype, 33 phagosomes were scored.
Discussion
Genetic studies reveal novel roles for UNC-108, the C. elegans Rab2
The biological functions of the Rab2 GTPase are much less characterized than several other Rab proteins. Although proteomic studies in Drosophila melanogaster and mammals have identified Rab2 as a component of phagosomes containing latex beads (Garin et al., 2001; Stuart et al., 2007), its role in phagosome maturation remains unknown. Mammalian Rab2 was previously implicated in protein sorting and recycling events during ER-to-Golgi transport (Chavrier et al., 1990; Tisdale et al., 1992; Tisdale and Jackson, 1998). In addition, a Rab2 effector complex was suggested to regulate vesicle traffic between Golgi cisternae (Short et al., 2001).
In this paper, we describe the isolation and characterization of the first loss-of-function alleles of unc-108, which encodes C. elegans Rab2. unc-108 mutations cause severe defects in the degradation of engulfed apoptotic cells. To our knowledge, this is the first finding of Rab2 function in phagosome maturation. In addition, we demonstrate that UNC-108 contributes to the efficient extension of pseudopods during the engulfment of cell corpses. By participating in both the engulfment and degradation of cell corpses, UNC-108 plays an important role in the completion of apoptosis, which is important for animal development and homeostasis.
Both class I (recessive Ced and Unc) and class II (dominant Unc and non-Ced) unc-108 alleles display severe defects in locomotion. unc-108 is strongly expressed in all neurons and restricted expression of unc-108(+) cDNA in these cells specifically rescued the Unc phenotype of class I mutants. These observations, together with the previously described role for Rab2 in regulating protein sorting during vesicular transport, suggest that unc-108 may function in neurons during synaptic or dense-core vesicle biogenesis, neurotransmitter release, or receptor trafficking. It is possible that the neuronal and engulfment functions of unc-108 are related on the molecular level (see the following section).
Our genetic analyses indicate that n3263, a class I allele, causes a severe or total loss of UNC-108 activity, particularly with regards to the removal of apoptotic cells. Consistent with this conclusion, the G13Q mutation may result in overall protein structure changes. Two other class I alleles (ce363 and ce365) also appear to result in the loss of unc-108 activity. In addition, both the Ced and Unc phenotypes were reproduced by RNAi knockdown of unc-108. These results argue against the possibility that class I mutations affect a process not under the control of unc-108. Thus, locomotion and phagosome maturation are two biological events that rely on the function of wild-type unc-108.
Structural models suggest that the two class II mutations (D122N and S149F) may partially affect the binding of the guanine nucleotide and the activation of this G protein. These mutant proteins may sequester guanine nucleotide exchange factors from wild-type UNC-108 in a heterozygous strain, eliciting a dominant-negative effect similar to that observed in GTP binding–deficient Rab2 mutants (Tisdale et al., 1992). The D122N and S149F mutant proteins may possess relatively low levels of UNC-108 activity, which is sufficient for the removal of cell corpses but not for supporting the neuronal regulation of locomotion.
Molecular functions of Rab2 in the removal of apoptotic cells
Previous work has demonstrated that lipids and proteins are delivered to phagosomes to drive phagosome maturation (for review see Vieira et al., 2002). Given the documented role for Rab2 in regulating protein sorting and recycling (Tisdale and Jackson, 1998), it is possible that the export of digestive enzymes and other factors required for phagosome maturation might be impaired in unc-108(n3263) mutants. However, we have not yet identified obvious defects in exocytosis in engulfing cells. For example, the transmembrane receptor CED-1 is normally localized to the cell surface and able to cluster around neighboring cell corpses in unc-108(n3263) embryos (Fig. 1 E), suggesting that its transport from ER to the plasma membrane is relatively normal.
Phagosome maturation involves the sequential recruitment of different signaling molecules, the incorporation of intracellular organelles, and the gradual acidification of the phagosomal lumen (for review see Vieira et al., 2002). We examined these events in unc-108(n3263) mutants and observed that the initial presentation of PI(3)P, the incorporation of early endosomes, and the recruitment of RAB-7 are normal. Instead, we identified defects in the recruitment and fusion of lysosomes with phagosomes as well as a failure in phagosome acidification. The transient recruitment of UNC-108 to the surface of phagosomes suggests that it may play a direct role in driving the incorporation of lysosomes to phagosomes. The effect of the G13Q mutation, which disrupts both the phagosomal localization and the activity UNC-108, is consistent with this hypothesis.
Rab GTPases are best characterized by their functions in driving the fusion of cellular membranes, primarily through organizing a protein complex that facilitates the tethering of two surfaces (Cai et al., 2007). For instance, yeast Rab7 specifically promotes the homotypic vacuole fusions using this mechanism (Cai et al., 2007). In addition, mammalian Rab7 was implicated in the recruitment of lysosomal particles to phagosomal surfaces through its effector Rab-interacting lysosomal protein (Harrison et al., 2003). Perhaps UNC-108 employs mechanisms similar to those used by Rab7 to promote the recruitment and fusion of lysosomes to phagosomes. Because both C. elegans UNC-108 and RAB-7 are recruited to phagosomal surfaces to promote the degradation of apoptotic cells (unpublished data), these Rab GTPases might perform related functions during phagosome maturation. Membrane fusion is required for most if not all trafficking events, including ER–Golgi trafficking, exocytosis, vesicle trafficking along the endocytic pathway, and phagosome maturation. If Rab2 acts as a tethering factor, it may also act in neurons to facilitate the generation or secretion of synaptic or dense-core vesicles.
It remains unclear how UNC-108 regulates phagosome acidification. V-type ATPases, proton pumps essential for phagosome acidification, are abundant on lysosomal membranes (Beyenbach and Wieczorek, 2006). Thus, the incorporation of lysosomes into phagosomes, a process that requires UNC-108 activity, might provide a source of V-type ATPases on phagosomal membranes. Alternatively, UNC-108 may regulate V-type ATPase activity directly through unknown mechanisms.
The strict regulation of UNC-108 localization may be applied through the switch between the GDP-bound, inactive form and the GTP-bound, active form. UNC-108 is recruited to phagosomal surfaces after the presentation of PI(3)P, which suggests the presence of an upstream signaling pathway. C. elegans DYN-1 is known to transiently reside on extending pseudopods and nascent phagosomes to control the incorporation of intracellular vesicles to phagocytic membranes (Yu et al., 2006). UNC-108 may be one of the downstream effectors regulated by DYN-1.
Our study of two specific physiological consequences of the loss of UNC-108 activity suggests the requirement of this protein in multiple cellular events. Based on the high-level homology between UNC-108 and human Rab2, the phagosomal localization of both proteins, and the knowledge that many aspects of apoptosis are regulated by conserved mechanisms, we propose that Rab2 performs evolutionarily conserved functions. Mammalian Rab2 may play important roles in the removal of apoptotic cells and perhaps even other phagocytic targets such as invading pathogens and degenerating axons. In mammals, Rab2 may thus take part in important immune responses such as self-tolerance and host cell defense.
Materials and methods
Mutations and strains
C. elegans was grown as described previously by Brenner (1974). N2 was used as the wild type and CB4856 was used as the single nucleotide polymorphism–mapping strain. Mutations and integrated arrays were prepared as described in Riddle et al. (1997) except when noted as follows: LG I: bli-3(e767), lin-17(n677), unc-108(n3263, ce363, and ce365 (this study); unc-108(ok1246) (C. elegans Gene Knockout Consortium); sem-4(n1378), dpy-5(e61), ced-1(e1735), ced-12(n3261) (Zhou et al., 2001a), and ok46 (myo-2::gfp, pes-10::gfp, and gut::gfp; a gift from M. Nonet, Washington University, St. Louis, MO); LG II: rrf-3(pk1426) (Simmer et al., 2002); LG III: ced-4(n1162), ced-6(n2095), and ced-7(n1996); LG IV: ced-2(n1994), ced-3(n717), ced-5(n1812), and ced-10(n1993); LG V: unc-76(e911) and zuIs178 (Phis-72 his-72::gfp; a gift of J. Priess, Fred Hutchinson Cancer Center, Seattle, WA; and R. Waterston, University of Washington, Seattle, WA; Ooi et al., 2006); LG X: lin-15(n765). Transgenes were introduced using standard methods (Jin, 1999). pWH17 (Pegl-13 gfp; Hanna-Rose and Han, 1999), pL15EK (lin-15(+); Clark et al., 1994), and pUNC76 (unc-76(+); Bloom and Horvitz, 1997) were used as coinjection markers. Transgenic animals are identified by GFP expression or the rescue of lin-15(n765) or unc-76(e911).
Molecular cloning of unc-108
We assigned n3263 to chromosome I by linkage to the markers sem-4(n1378) and ok46. n3263 was originally isolated in a strain containing the sem-4(n1378) mutation, which causes an egg laying defect (Egl phenotype). n3263 sem-4(n1378) hermaphrodites were crossed with wild-type males and 159 of 200 homozygous sem-4(n1378) F2 progeny contained Ced F3 embryos. n3263 hermaphrodites were also crossed to ok46 males. From this cross, 4 out of 100 F3 embryos were both Ced and GFP+ (i.e.,at least heterozygous for the dominant marker ok46). A bli-3(e767) n3263 sem-4(n1378) strain was crossed to wild-type males. From this cross, 24 of 25 Egl non-Bli F2 recombinants produced Ced progeny and 1 of 25 Bli non-Egl F2 recombinants gave rise to non-Ced progeny, which indicates that n3263 is flanked by these two markers. We narrowed the interval where n3263 is located using single nucleotide polymorphism (SNP) markers (Wicks et al., 2001). In brief, we crossed CB4856 males to bli-3(e767) n3263 sem-4(n1378) and lin-17(n677) n3263 dpy-5(e61) hermaphrodites, cloned F2 recombinants that were either Bli non-Egl, non-Bli Egl, Lin non-Dpy, or non-Lin Dpy, and then scored for the presence of selected SNP markers and the n3263 Ced and Unc phenotypes in the F3 generation. The left and right boundaries of the n3263 locus were SNPs uCE1-790 and uCE1-797 (Fig. 2A). Cosmids T28F2, D1037, H26D21, T12F5, ZK770, C32E8, F53F10, T03F1, C50F2, and F21A9 were injected either singly or in groups of three (at 10 ng/μl per cosmid) and transgenic animals were scored for the rescue of the Ced and Unc phenotypes. The molecular lesion in n3263 mutants was identified by sequencing the unc-108 locus.
Microscopy
Specimens were mounted on agarose pads and imaging was performed at 20–25°C. For DIC microscopy, a microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with a digital camera (AxioCam MRm; Carl Zeiss, Inc.) and imaging software (AxioVision; Carl Zeiss, Inc.) was used. A 100× Plan Neofluar objective (NA 1.30) was used with Immersol 518N oil (Carl Zeiss, Inc.). Cell corpses were counted at the indicated stages. The description of embryonic stages and time-lapse recording protocol were performed as described previously (Yu et al., 2006). To record the duration of cell corpses, embryos ∼350 min after the first cleavage were recorded for 80 min at 3-min intervals. 30 cell corpses per genotype were analyzed. To count the number of cell death events, we recorded embryos from 160–460 min after the first embryonic division at 3-min intervals. For each time point, 40 z sections at 0.5-μm intervals were taken. For fluorescent imaging, a microscope system (DeltaVision; Applied Precision) with an inverted microscope (IX70; Olympus) and a digital camera (CoolSNAP; Photometrics) was used. A 100× UPlan Apo objective (NA 1.35) was used with immersion oil (n = 1.514, DeltaVision; Applied Precision). Serial z sections in 0.5-μm intervals spanning entire embryos were captured. Time-lapse imaging of fluorescent markers was performed as described previously (Yu et al., 2006). Images were deconvolved using softWoRx software (Applied Precision). Fluorescent images were analyzed using ImageJ. The radius (r) of each phagosome was determined using the measuring tool and the volume was approximated by the formula 4/3 πr3 (volume of a sphere).
TEM
Preparation of specimens for TEM by high-pressure fixation has been described previously (Shaham, 2006). unc-108(n3263) animals were collected 48 h after the mid-L4 stage, mounted in specimen hats using Escherichia coli OP50, and high-pressure frozen (HPM 010; Bal-tec). Samples were freeze-substituted into 2% osmium tetroxide in acetone for 3 d at −90°C. Fixed specimen were rinsed in 100% acetone several times and embedded in Embed 812 resin (Electron Microscopy Sciences). Serial sections of 50 nm in thickness were collected onto Formvar-coated slot grids (Fullham), poststained for 1 h in a 2% aqueous solution of uranyl acetate, and counterstained in Reynold's lead citrate. Sections were observed in a TEM (H-7500; Hitachi) equipped with a charge-coupled device camera (Sapera; Gatan).
Lysosensor staining
Hermaphrodites 24 h past the L4 larval stage were exposed to 180 Gy of γ radiation (Gammacell 1000 137Cs source; Atomic Energy of Canada Limited). 4 h after irradiation, animals were mounted on a microscope slide with M9 buffer supplemented with 5 mM Lysosensor yellow/blue DND-160 (Invitrogen). The anterior gonad arms were dissected out by cutting open animals at the second pharyngeal bulb using a 21-gauge needle. Specimens were examined using the Axioplan microscope described in the Microscopy section. Excitation and emission wavelengths were 360 and 535 nm, respectively. Fluorescence signal intensity was measured using ImageJ. We observed that healthy germ cells displayed a fluorescence signal 10% brighter than the germline syncytium. No or faint staining corresponds to a ratio of germ cell corpse to syncytium signal intensity of ≤1.1, intermediate staining corresponds to a ratio >1.1 and <2, and bright staining corresponds to a ratio ≥2.
Plasmid construction
The 2.3-kb promoter of unc-108 (Punc-108) was PCR amplified from cosmid F53F10 (A. Coulson, Sanger Institute, Cambridge, UK) using primers PM149 and PM150 and cloned into the HindIII–PstI sites of pPD49.26 to generate pAOLO125. unc-108(+) cDNA was amplified using primers PM146 or PM148 and PM147 and cloned into pCRII-TOPO to generate pAOLO112 and pAOLO113. To generate pAOLO135 (Punc-108 unc-108(+)), the SmaI–KpnI fragment from pAOLO112 was cloned into the corresponding sites of pAOLO125. The BamHI–KpnI fragment from pAOLO135 was cloned into the same sites in pAOLO89 (Pced-1 in pPD49.26) to generate pAOLO132 (Pced-1 unc-108(+)). The SmaI–SpeI fragment from pAOLO135 was cloned into pBC99 (Conradt and Horvitz, 1999) to generate pAOLO134 (Pegl-1 unc-108(+)). Prab-3 was PCR amplified from rab-3::rfp (Choi et al., 2006) using primers PM183 and PM184. To generate pAOLO160 (Prab-3 unc-108(+)), the 5′ end of the Prab-3 PCR product blunted at 5′ and digested with PstI at 3′ was cloned into the HindIII (blunt)–PstI sites of pAOLO135. The G13Q mutation was generated in pAOLO112 by site-directed mutagenesis (QuikChange; Stratagene) using primers PM167 and PM168 to generate pAOLO143. The SmaI–KpnI fragment from pAOLO143 was cloned into the same sites in pAOLO125 to generate pAOLO173. To generate N-terminal GFP fusions, unc-108(+) (ApaI–KpnI fragment from pAOLO113) and unc-108(n3263) (PCR amplified from pAOLO143 using primers PM148 and PM147) were first fused to a gfp cDNA without a stop codon, and the gfp::unc-108 cDNA cassettes (SmaI–KpnI fragments) were cloned into the corresponding sites in pAOLO125 to generate pAOLO136 (Punc-108 gfp::unc-108(+)) and into those of pPD41.78 and pPD41.83 (heat-shock promoters Phsp-16.2 and Phsp-16.41, respectively; Stringham et al., 1992) to generate pAOLO121 and pAOLO122 (Phsp gfp::unc-108(+)) and pAOLO210 and pAOLO211 (Phsp gfp::unc-108(n3263)). pIH1 (Pced-12× FYVE::mrfp1) was generated by substituting the mrfp1 cDNA (Campbell et al., 2002) for gfp in Pced-12× FYVE::gfp (Yu et al., 2006). pAOLO230 (unc-108 RNAi construct) was generated by cloning the HindIII–XhoI fragment from pAOLO112 into the corresponding sites of vector pPD129.36. Primer sequences are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1). Information regarding pPD vectors (gifts from A. Fire, Stanford University, Stanford, CA) is available at http://www.addgene.org/Andrew_Fire.
RNAi treatment
RNAi treatment was performed by feeding as described previously (Fraser et al., 2000). In brief, L4 larvae were treated to control (vector pPD129.36) or unc-108 (I-1J21 [Fraser et al., 2000] or pAOLO230) RNAi. The numbers of germ cell corpses was scored in the P0 generation 48 h after the transfer to RNAi plates. The locomotion phenotype was scored in the F1 generation.
Heat shock expression
Extrachromosomal arrays containing pAOLO121 and pAOLO122 (Phsp gfp::unc-108(+)) or pAOLO210, pAOLO211 (Phsp gfp::unc-108(n3263)), and pIH1 (Pced-12× FYVE::mrfp1) were introduced to animals of the appropriate genetic background. Transgenic adults were incubated at 33°C, allowed to lay eggs for 1 h, and then removed. Embryos were incubated at 25°C for 2 h before the start of time-lapse recording. To overexpress ced-10(+), extrachromosomal arrays containing pPR45, pPR46 (Phsp ced-10(+); Reddien and Horvitz, 2000), and pWH17 (Pegl-13 gfp) were introduced to animals of the appropriate genetic background. Transgenic adults were incubated at 33°C, allowed to lay eggs for 1 h, and then removed. Embryos were then incubated at 25°C until they are scored at either two- or fourfold stages for the number of cell corpses.
Structural modeling
Structural models for UNC-108 were generated using the protein structure homology modeling platform SWISS-MODEL (Schwede et al., 2003). Crystal structures for GDP-bound human Rab2A (PDB ID 1Z0A) and GDP- (PDB ID 10IV) and GTPγS-bound human Rab11A (PDB ID 10IW) were used as templates. The predicted structural models were then aligned to GDP-bound Rab2 using the Protein3Dfit algorithm (Lessel and Schomburg, 1994) and the resulting structures were viewed using PyMOL (DeLano Scientific). Root mean square deviation values of the corresponding α-C atoms of the structural predictions were calculated using Protein3Dfit.
Online supplemental material
Fig. S1 shows the phenotypes caused by ce363, ce365, and unc-108(RNAi). Fig. S2 shows structural modeling of UNC-108. Fig. S3 shows epistatic analysis between unc-108 and the engulfment of ced genes. Fig. S4 shows characterization of the 2× FYVE reporter as a phagosome marker. Fig. S5 shows the kinetics of 2× FYVE::mRFP1 recruitment to phagosomes in unc-108(n3263). Table S1 lists primer sequences. Videos are fluorescence time-lapse displays. Videos 1 and 2 show the engulfment of cell corpse C3 in wild-type (Video 1) and unc-108(n3263) (Video 2). Videos 3 and 4 show degradation of cell corpse C2 in wild-type (Video 3) and unc-108(n3263) (Video 4). Video 5 shows enrichment of UNC-108 on the phagosome containing cell corpse C2. Video 6 shows mislocalization of UNC-108(G13Q). Video 7 shows recruitment of lysosomes to the phagosome containing C2 in the wild type. Videos 8 and 9 show defects in lysosome recruitment in unc-108(n3263). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708130/DC1.
Supplemental Material
Acknowledgments
We thank H.R. Horvitz for his support during isolation of n3263; S. Williams and N. Charlie for isolating ce363 and ce365; D.H. Hall and K. Nguyen for help with TEM; A. Coulson, A. Fire, M. Nonet, J. Priess, R. Waterston, the C. elegans Gene Knockout Consortium, and the Caenorhabditis Genetics Center for reagents; X. He for the DeltaVision microscope; A. Biter for assistance with structural modeling; H.J. Bellen and T. Shin for helpful comments; and S. Yamamoto, M. Prabhakaran, and I. Hansen for technical assistance.
This work was supported by grants from the National Institutes of Health (GM067848), the Rita Allen Foundation, and the March of Dimes Foundation to Z. Zhou and grants from the National Institutes of Health (GM080765) and the Oklahoma Center for the Advancement of Science (HR06-078) to K.G. Miller.
Abbreviations used in this paper: Ced, cell death abnormal; DIC, differential interference contrast; DYN-1, dynamin 1; HGRS-1, hepatocyte growth factor–regulated tyrosine kinase substrate; mRFP1, monomeric RFP 1; PI(3)P, phosphatidylinositol-3-monophosphate; TEM, transmission EM; Unc, uncoordinated locomotion.
References
- Beyenbach, K.W., and H. Wieczorek. 2006. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209:577–589. [DOI] [PubMed] [Google Scholar]
- Bloom, L., and H.R. Horvitz. 1997. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc. Natl. Acad. Sci. USA. 94:3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, H., K. Reinisch, and S. Ferro-Novick. 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 12:671–682. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier, P., R.G. Parton, H.P. Hauri, K. Simons, and M. Zerial. 1990. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 62:317–329. [DOI] [PubMed] [Google Scholar]
- Choi, J., K.L. Richards, H.N. Cinar, and A.P. Newman. 2006. N-ethylmaleimide sensitive factor is required for fusion of the C. elegans uterine anchor cell. Dev. Biol. 297:87–102. [DOI] [PubMed] [Google Scholar]
- Clark, S.G., X. Lu, and H.R. Horvitz. 1994. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 137:987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt, B., and H.R. Horvitz. 1999. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 98:317–327. [DOI] [PubMed] [Google Scholar]
- Eathiraj, S., X. Pan, C. Ritacco, and D.G. Lambright. 2005. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 436:415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, R.E., D.M. Jacobson, and H.R. Horvitz. 1991. a. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics. 129:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson, C.D., K.E. Anderson, G. Morgan, E.R. Chilvers, P. Lipp, L.R. Stephens, and P.T. Hawkins. 2001. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr. Biol. 11:1631–1635. [DOI] [PubMed] [Google Scholar]
- Fainzilber, M., and J.L. Twiss. 2006. Tracking in the WldS—the hunting of the SIRT and the luring of the Draper. Neuron. 50:819–821. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., R.S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408:325–330. [DOI] [PubMed] [Google Scholar]
- Garin, J., R. Diez, S. Kieffer, J.F. Dermine, S. Duclos, E. Gagnon, R. Sadoul, C. Rondeau, and M. Desjardins. 2001. The phagosome proteome: insight into phagosome functions. J. Cell Biol. 152:165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny, T.L., E. Lambie, E. Hartwieg, H.R. Horvitz, and M.O. Hengartner. 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 126:1011–1022. [DOI] [PubMed] [Google Scholar]
- Gumienny, T.L., E. Brugnera, A.C. Tosello-Trampont, J.M. Kinchen, L.B. Haney, K. Nishiwaki, S.F. Walk, M.E. Nemergut, I.G. Macara, R. Francis, et al. 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 107:27–41. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose, W., and M. Han. 1999. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans.Development. 126:169–179. [DOI] [PubMed] [Google Scholar]
- Harrison, R.E., C. Bucci, O.V. Vieira, T.A. Schroer, and S. Grinstein. 2003. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol. Cell. Biol. 23:6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y. 1999. Transformation. In C. elegans, A Practical Approach. I.A. Hope, editor. Oxford University Press, Oxford. 69–96.
- Kalatzis, V., S. Cherqui, C. Antignac, and B. Gasnier. 2001. Cystinosin, the protein defective in cystinosis, is a H+-driven lysosomal cystine transporter. EMBO J. 20:5940–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen, J.M., J. Cabello, D. Klingele, K. Wong, R. Feichtinger, H. Schnabel, R. Schnabel, and M.O. Hengartner. 2005. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 434:93–99. [DOI] [PubMed] [Google Scholar]
- Lessel, U., and D. Schomburg. 1994. Similarities between protein 3-D structures. Protein Eng. 7:1175–1187. [DOI] [PubMed] [Google Scholar]
- Liu, Q.A., and M.O. Hengartner. 1998. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell. 93:961–972. [DOI] [PubMed] [Google Scholar]
- Metzstein, M.M., G.M. Stanfield, and H.R. Horvitz. 1998. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 14:410–416. [DOI] [PubMed] [Google Scholar]
- Ooi, S.L., J.R. Priess, and S. Henikoff. 2006. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet. 2:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai, E.F., W. Kabsch, U. Krengel, K.C. Holmes, J. John, and A. Wittinghofer. 1989. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 341:209–214. [DOI] [PubMed] [Google Scholar]
- Pai, E.F., U. Krengel, G.A. Petsko, R.S. Goody, W. Kabsch, and A. Wittinghofer. 1990. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9:2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E.C., and H.R. Horvitz. 1986. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics. 113:821–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato, S., and J. Cherfils. 2005. Crystallographic evidence for substrate-assisted GTP hydrolysis by a small GTP binding protein. Structure. 13:533–540. [DOI] [PubMed] [Google Scholar]
- Pasqualato, S., F. Senic-Matuglia, L. Renault, B. Goud, J. Salamero, and J. Cherfils. 2004. The structural GDP/GTP cycle of Rab11 reveals a novel interface involved in the dynamics of recycling endosomes. J. Biol. Chem. 279:11480–11488. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J.B., and M.C. Seabra. 2000. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301:1077–1087. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J.B., and M.C. Seabra. 2001. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313:889–901. [DOI] [PubMed] [Google Scholar]
- Reddien, P.W., and H.R. Horvitz. 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2:131–136. [DOI] [PubMed] [Google Scholar]
- Reddien, P.W., and H.R. Horvitz. 2004. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 20:193–221. [DOI] [PubMed] [Google Scholar]
- Riddle, D.L., T. Blumenthal, B.J. Meyer, and J.R. Priess, editors. 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1222 pp. [PubMed]
- Robertson, A.M.G., and J.N. Thomson. 1982. Morphology of programmed cell death in the ventral nerve cord of Caenorhabditis elegans larvae J. Embryol. Exp. Morphol. 67:89–100. [Google Scholar]
- Savill, J., and V. Fadok. 2000. Corpse clearance defines the meaning of cell death. Nature. 407:784–788. [DOI] [PubMed] [Google Scholar]
- Schwede, T., J. Kopp, N. Guex, and M.C. Peitsch. 2003. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31:3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham, S., editor. 2006. Methods in cell biology. In WormBook. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.49.1, http://www.wormbook.org.
- Short, B., C. Preisinger, R. Korner, R. Kopajtich, O. Byron, and F.A. Barr. 2001. A GRASP55-Rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155:877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., M. Tijsterman, S. Parrish, S.P. Koushika, M.L. Nonet, A. Fire, J. Ahringer, and R.H. Plasterk. 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12:1317–1319. [DOI] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A.M. van der Linden, E. Kuijk, P.V. van den Berghe, R.S. Kamath, A.G. Fraser, J. Ahringer, and R.H. Plasterk. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1:E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringham, E.G., D.K. Dixon, D. Jones, and E.P. Candido. 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell. 3:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, L.M., J. Boulais, G.M. Charriere, E.J. Hennessy, S. Brunet, I. Jutras, G. Goyette, C. Rondeau, S. Letarte, H. Huang, et al. 2007. A systems biology analysis of the Drosophila phagosome. Nature. 445:95–101. [DOI] [PubMed] [Google Scholar]
- Su, H.P., K. Nakada-Tsukui, A.C. Tosello-Trampont, Y. Li, G. Bu, P.M. Henson, and K.S. Ravichandran. 2002. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J. Biol. Chem. 277:11772–11779. [DOI] [PubMed] [Google Scholar]
- Sulston, J.E., and H.R. Horvitz. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56:110–156. [DOI] [PubMed] [Google Scholar]
- Sulston, J.E., E. Schierenberg, J.G. White, and N. Thomson. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119. [DOI] [PubMed] [Google Scholar]
- Tisdale, E.J. 2003. Rab2 interacts directly with atypical protein kinase C (aPKC) ι/λ and inhibits aPKCι/λ-dependent glyceraldehyde-3-phosphate dehydrogenase phosphorylation. J. Biol. Chem. 278:52524–52530. [DOI] [PubMed] [Google Scholar]
- Tisdale, E.J., and M.R. Jackson. 1998. Rab2 protein enhances coatomer recruitment to pre-Golgi intermediates. J. Biol. Chem. 273:17269–17277. [DOI] [PubMed] [Google Scholar]
- Tisdale, E.J., J.R. Bourne, R. Khosravi-Far, C.J. Der, and W.E. Balch. 1992. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale, E.J., C. Kelly, and C.R. Artalejo. 2004. Glyceraldehyde-3-phosphate dehydrogenase interacts with Rab2 and plays an essential role in endoplasmic reticulum to Golgi transport exclusive of its glycolytic activity. J. Biol. Chem. 279:54046–54052. [DOI] [PubMed] [Google Scholar]
- Town, M., G. Jean, S. Cherqui, M. Attard, L. Forestier, S.A. Whitmore, D.F. Callen, O. Gribouval, M. Broyer, G.P. Bates, et al. 1998. A novel gene encoding an integral membrane protein is mutated in nephropathic cystinosis. Nat. Genet. 18:319–324. [DOI] [PubMed] [Google Scholar]
- Valencia, A., P. Chardin, A. Wittinghofer, and C. Sander. 1991. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 30:4637–4648. [DOI] [PubMed] [Google Scholar]
- Venegas, V., and Z. Zhou. 2007. Two alternative mechanisms that regulate the presentation of apoptotic-cell engulfment signal in Caenorhabditis elegans. Mol. Biol. Cell. 18:3180–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, O.V., R.J. Botelho, L. Rameh, S.M. Brachmann, T. Matsuo, H.W. Davidson, A. Schreiber, J.M. Backer, L.C. Cantley, and S. Grinstein. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, O.V., R.J. Botelho, and S. Grinstein. 2002. Phagosome maturation: aging gracefully. Biochem. J. 366:689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S.R., R.T. Yeh, W.R. Gish, R.H. Waterston, and R.H. Plasterk. 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28:160–164. [DOI] [PubMed] [Google Scholar]
- Williams, S.L., S. Lutz, N.K. Charlie, C. Vettel, M. Ailion, C. Coco, J.J. Tesmer, E.M. Jorgensen, T. Wieland, and K.G. Miller. 2007. Trio's Rho-specific GEF domain is the missing Gαq effector in C. elegans. Genes Dev. 21:2731–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y.C., and H.R. Horvitz. 1998. a. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 93:951–960. [DOI] [PubMed] [Google Scholar]
- Wu, Y.C., and H.R. Horvitz. 1998. b. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 392:442–443. [DOI] [PubMed] [Google Scholar]
- Wu, Y.C., M.C. Tsai, L.C. Cheng, C.J. Chou, and N.Y. Weng. 2001. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell. 1:491–502. [DOI] [PubMed] [Google Scholar]
- Yu, X., S. Odera, C.H. Chuang, N. Lu, and Z. Zhou. 2006. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell. 10:743–757. [DOI] [PubMed] [Google Scholar]
- Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., E. Caron, E. Hartwieg, A. Hall, and H.R. Horvitz. 2001. a. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev. Cell. 1:477–489. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., E. Hartwieg, and H.R. Horvitz. 2001. b. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 104:43–56. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., P.M. Mangahas, and X. Yu. 2004. The genetics of hiding the corpse: engulfment and degradation of apoptotic cells in C. elegans and D. melanogaster. Curr. Top. Dev. Biol. 63:91–143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.