Abstract
Woronin bodies (WBs) are dense-core organelles that are found exclusively in filamentous fungi and that seal the septal pore in response to wounding. These organelles consist of a membrane-bound protein matrix comprised of the HEX protein and, although they form from peroxisomes, their biogenesis is poorly understood. In Neurospora crassa, we identify Woronin sorting complex (WSC), a PMP22/MPV17-related membrane protein with dual functions in WB biogenesis. WSC localizes to large peroxisome membranes where it self-assembles into detergent-resistant oligomers that envelop HEX assemblies, producing asymmetrical nascent WBs. In a reaction requiring WSC, these structures are delivered to the cell cortex, which permits partitioning of the nascent WB and WB inheritance. Our findings suggest that WSC and HEX collaborate and control distinct aspects of WB biogenesis and that cortical association depends on WSC, which in turn depends on HEX. This dependency helps order events across the organellar membrane, permitting the peroxisome to produce a second organelle with a distinct composition and intracellular distribution.
Introduction
Hyphae are the predominant mode of cellular organization in the fungi and, in most species, these filamentous cells are compartmentalized by perforate septa, which allow intercellular cooperation and communication. Woronin bodies (WBs) are peroxisome-related organelles that are unique to filamentous fungi, where they function to seal the septal pore in response to cellular wounding (Trinci and Collinge, 1974; Markham and Collinge, 1987). This organelle is centered on a fungus-specific protein, HEX, which uses peroxisome-targeting signal 1 (PTS1) for localization to the peroxisome matrix (Jedd and Chua, 2000; Managadze et al., 2007), where it self-assembles to produce a solid micrometer-scale protein assembly (Jedd and Chua, 2000; Yuan et al., 2003). In hex deletion mutants (Jedd and Chua, 2000; Tenney et al., 2000; Soundararajan et al., 2004; Maruyama et al., 2005) or hex mutants that disrupt self-assembly (Yuan et al., 2003), hyphae bleed protoplasm after cellular wounding and display a variety of secondary defects, including impaired invasive growth of the plant pathogen Magnaporthe grisea (Soundararajan et al., 2004). Thus, WBs provide an important adaptive function that supports the unique cellular architecture of the fungal hypha.
WBs are believed to form de novo through a process determined in part by apically biased hex gene expression (Tey et al., 2005). In apical hyphal compartments, HEX assemblies nucleate and grow and associate with the matrix face of the peroxisomal membrane to produce budding intermediates, which can be observed by electron microscopy (Brenner and Carrol, 1968; Wergin, 1973; Camp, 1977). These structures subsequently associate with the cell cortex in a manner that coincides with septation (Momany et al., 2002; Tey et al., 2005), and this presumably allows partitioning and inheritance of the newly formed WB organelle. The molecular mechanisms that determine this sequence of events are currently uncharacterized. In contrast, a large number of mutations affecting the biogenesis of yeast peroxisomes and mitochondria have been isolated and, interestingly, mutations that affect organellar inheritance frequently result in the accumulation of enlarged or morphologically abnormal organelles (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997; Fagarasanu et al., 2005; Cerveny et al., 2007), which suggests that where organelles proliferate by growth and division, morphogenesis and segregation can constitute an ordered pathway.
We used a forward genetic screen to isolate mutants defective in WB biogenesis and identified Woronin sorting complex (wsc), a PMP22/MPV17-related membrane protein that executes a dual function in WB morphogenesis and inheritance. In wsc mutants, HEX assemblies are formed but fail to associate with the peroxisomal membrane and instead move randomly in the matrix. These hybrid peroxisome-WB structures also fail to associate with the cell cortex and, as a result, accumulate to abnormally high levels in the apical compartment and are not segregated into subapical compartments. Here, we show that WSC assembles in the membrane to envelop HEX assemblies and produce asymmetrical nascent WBs. WSC also determines WB inheritance via cortical association. Cortical association depends critically on the level of membrane-associated WSC, which in turn depends on HEX. This dependency helps order morphogenesis and inheritance and permits the peroxisome to produce a derived organelle with a distinct function, composition, and subcellular distribution.
Results
A genetic screen identifies WB loss-of-function mutants
Mutants defective in WB biogenesis were identified by the visualization of the protoplasmic bleeding characteristic of WB loss of function. In brief, wild-type Neurospora crassa conidia were UV irradiated to 50% lethality, and ∼500,000 colonies were visually screened for the bleeding phenotype using a stereomicroscope. Mutants were backcrossed and assigned into 12 complementation groups using heterokaryon analysis. The majority of these mutants are devoid of HEX assemblies, and they accumulate a peroxisomal matrix marker (GFP-PTS1) in the cytosol and encode homologues of genes associated with various aspects of peroxisome assembly (Table I). We identified mutants in PEX3 and PEX16 that are involved in peroxisome membrane biogenesis, as well as peroxins associated with early (PEX14) and late (PEX1, 4, 6, 10, and 12) steps of the matrix protein import pathway. Multiple alleles of most of these genes were identified; however, 3 of 12 mutants were represented by only one allele and several essential peroxins were not recovered, which suggests that this screen has not yet been saturated. We also examined several deletions of predicted peroxins not recovered from our screen. HEX uses a PTS1 signal for matrix localization, and we found that the PTS1 receptor PEX5 is required for WB biogenesis, whereas PEX7 and PEX20, which are associated with the PTS2 matrix import pathway (Sichting et al., 2003), are not (Table I). In summary, results from the screen show that WB biogenesis requires most of the fungal peroxins and the PTS1, but not PTS2, matrix import pathways. One mutant retains the ability to produce peroxisomes and HEX assemblies (Table I) and presents a set of phenotypes that suggest a specific defect in WB biogenesis. We designated this mutant wsc.
Table I. Results from the mutant screen.
| Allelesa | Protein IDb | Genec | Woronind body function |
HEXd
assemblies |
PTS1d
import |
Function/domains |
|---|---|---|---|---|---|---|
| 5 | NCU08118 | pex1 | − | − | − | AAA-ATPase |
| 5 | NCU02070 | pex2 | − | − | − | Zinc binding, PMP |
| 2 | NCU06175 | pex3 | − | − | − | Membrane biogenesis, PMP |
| 1 | NCU02636 | pex4 | − | − | − | Ubiquitin-conjugating protein |
| 6 | NCU08373 | pex6 | − | − | − | AAA-ATPase |
| 6 | NCU00032 | pex8 | − | − | − | Matrix protein import, PMP |
| 2 | NCU03277 | pex10 | − | − | − | Zinc binding, PMP |
| 4 | NCU05245 | pex12 | − | − | − | Zinc binding, PMP |
| 3 | NCU03901 | pex14 | − | − | − | Docking receptor, PMP |
| 4 | NCU01850 | pex16 | − | − | − | Membrane biogenesis, PMP |
| 1 | NCU08332 | hex | − | − | + | WB core |
| 1 | NCU07842 | wsc | − | + | + | WB biogenesis |
| KO | NCU07662 | pex7 | + | + | + | PTS2 signal receptor |
| KO | NCU04062 | pex20 | + | + | + | PTS2 import |
| KO | NCU02960 | pex5 | − | − | − | PTS1 signal receptor |
| KO | NCU04301 | pex19 | − | − | − | PMP import receptor |
| KO | NCU02618 | pex13 | − | − | − | SH3 domain, PMP |
Number of alleles obtained is indicated. KO identifies mutants obtained from the Neurospora deletion project.
Neurospora gene ID (Colot et al., 2006).
Peroxin identities according to Sichting et al. (2003).
The presence (+) or absence (−) of the indicated function or structure. Analysis is described in Materials and methods.
wsc is required for WB biogenesis
To determine the structure of peroxisomes where HEX assemblies form, we examined the localization of peroxisome-targeted GFP (GFP-PTS1) and made time-lapse videos. In wild-type apical compartments, HEX assemblies are observed as distinct refractive structures (Jedd and Chua, 2000) found at the periphery of large peroxisomes with which they move in synchrony (Fig. 1 A and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1). In the wsc mutant, HEX assemblies are also observed in the matrix of large apical peroxisomes but here they move randomly within the peroxisome matrix (Fig. 1 A and Video 2). This suggests that WSC is required to recruit HEX assemblies to the matrix face of the peroxisome membrane, from which they can bud off to produce the WB organelle.
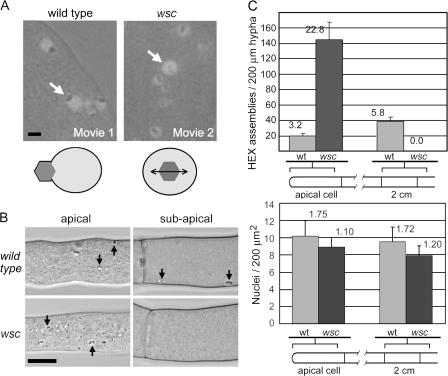
Figure 1.
wsc mutants are defective in WB maturation and inheritance. (A) HEX assemblies move in concert with peroxisomes in wild-type hyphae (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1) but move randomly in the matrix of wsc mutant peroxisomes (Video 2). The peroxisome matrix is revealed by peroxisome-targeted GFP, and HEX assemblies are observed as characteristic refractive structures by light microscopy. Arrows point to peroxisomes containing HEX assemblies. Bar, 1 μm. A schematic of results obtained from time-lapse microscopy is presented below. The double-headed arrow indicates random motion of the HEX assembly. (B) HEX assemblies accumulate in the apical cell of the wsc mutant and are not observed in subapical compartments. Representative apical and subapical compartments of wild-type and wsc mutant hyphae viewed by light microscopy are shown. Arrows point to HEX assembly. Bar, 10 μm. (C) The top shows a quantification of HEX assembly distribution in wild-type and mutant hyphae. The bottom shows the distribution of nuclei at similar positions in the hypha. Standard deviation is indicated with error bars.
Next, we examined the intracellular distribution of refractive HEX assemblies. Wild-type hyphae contain ∼20 HEX assemblies in the apical compartment and about twice as many are observed in a similar subapical interval found 2 cm behind the growth front. In contrast, in the wsc mutant, HEX assemblies are observed at approximately sevenfold higher levels in the apical compartment and are not observed in subapical compartments (Fig. 1, B and C), which suggests that the wsc mutant presents a second defect in the process of cortical association. The defect in segregation also accounts for the isolation of wsc from a loss-of-function screen, despite the mutant's ability to make peroxisomes and HEX assemblies (Table I). To begin to determine the specificity of the wsc mutant's effect on organellar segregation, we examined the spatial distribution of nuclei. In contrast to the abnormal distribution of HEX assemblies, nuclei show a similar distribution in wild-type and wsc mutant hyphae (Fig. 1 C, bottom), which suggests that WSC functions specifically in WB biogenesis.
The wsc mutant presents an additional phenotype. Initially, wsc colonies present wild-type growth and morphology. However, with age, they begin to present a poorly organized and irregular growth front (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1). We reasoned that this might be because of the stochastic occlusion of septal pores by ectopic HEX assemblies and a consequent disruption of the hyphal syncytium. Consistently, microscopic examination of wsc mutant hyphae revealed frequent intrahyphal hyphae (Fig. S1), which can occur if septal pores are sealed in undamaged hyphae. To determine the dependence of this phenotype on WB and peroxisome function, we created wsc-Δhex and wsc-pex14 double mutants (pex14 encodes a PMP essential for import of all matrix proteins; Niederhoff et al., 2005; Itoh and Fujiki, 2006). Both hex and pex14 mutants fully suppress the wsc growth phenotype (Fig. S1). These observations further support the idea that WSC functions exclusively in WB biogenesis.
wsc is a new member of the PMP22/MPV17 gene family
The wsc mutation was mapped by meiotic recombination and tightly linked to the sc mutant on the left arm of chromosome III (Fig. 2 A). To identify wsc, cosmids encompassing this region were transformed into the wsc mutant and two overlapping clones were found to complement the wsc phenotype. These shared four predicted genes, and one of these, NCU07842.2, was capable of independently complementing wsc. This gene encodes a predicted protein of 308 aa, which is suggested by hydropathy analysis to encode four transmembrane domains (Fig. 2 B). The wsc mutant was sequenced and found to contain a 7-bp duplication that induces a frame shift resulting in protein truncation at leucine 84. To confirm that the wsc mutant presents full loss-of-function phenotypes, we deleted NCU07842.2 and found that the deletion mutant presents phenotypes identical to those of wsc (unpublished data).
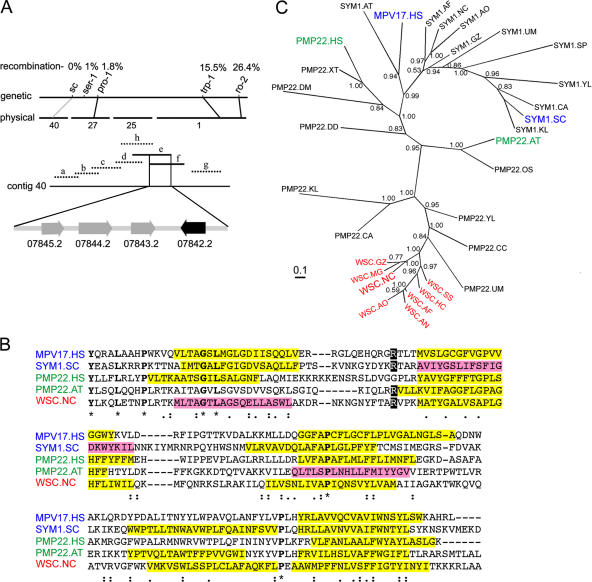
Figure 2.
Positional cloning, phylogenetic analysis, and alignment of WSC-related sequences. (A) The schematic shows a set of markers on the left arm of chromosome three. The recombination frequency between wsc and these markers is indicated at the top. Cosmids (a–h) encompassing contig 40 were assessed for their ability to complement the wsc mutant. Complementing cosmids (e and f) are drawn as solid lines and noncomplementing cosmids are drawn as dotted lines. Transformation with DNA fragments containing individual candidate genes identified NCU 07842.2 as wsc. (B) Alignment of WSC and selected homologous proteins. Regions predicted to encode membrane-spanning domains are highlighted in yellow (score > 500) and pink (score < 500). Identical (*), strongly conserved (:), and weakly conserved (.) residues are identified beneath the alignment. The arginine residue that is mutated in some individuals afflicted with MDDS is shaded with black. (C) Phylogenetic tree showing the relationship between WSC and homologous sequences. The tree was constructed using MrBayes3.1.2. The scale bar indicates 0.1 substitutions. Proteins that have been shown to reside in the peroxisome are shown in green and those known to target to mitochondria are shown in blue. WSC and close relatives in the Euascomycetes are labeled in red. AT, Arabidopsis thaliana; AF, Aspergillus fumigatus; AN, Aspergillus nidulans; AO, Aspergillus oryzae; CA, Candida albicans; CC, Coprinus cinereus; DD, Dictyostelium discoideum; DM, Drosophila melanogaster; GZ, Gibberella zea; HC, Histoplasma capsulatum; HS, Homo sapiens; KL, Kluyveromyces lactis; MG, Magnaporthe grisea; NC, Neurospora crassa; OS, Oryzae sativa; SC, Saccharomyces cerevisiae; SP, Schizosaccharomyces pombe; SS, Sclerotina sclerotinium; UM, Ustilago maydis; YL, Yarrowia lipolytica; XT, Xenopus tropicalis.
WSC is homologous to the PMP22/MPV17 family of proteins, and to better understand the relationship between these proteins, we constructed phylogenetic trees (Fig. 2 C). Close wsc homologues were identified in all sequenced Euascomycete genomes and, in contrast, some, but not all, other fungi contained more distantly related homologues. This fungal group is most closely related to mammalian and plant PMP22 proteins. PMP22 was originally identified as an abundant PMP (Fujiki et al., 1984) and its peroxisome targeting has been extensively studied (Diestelkotter and Just, 1993; Pause et al., 1997; Tugal et al., 1999; Brosius et al., 2002; Iida et al., 2003; Murphy et al., 2003); however, the function of PMP22 remains unknown. PMP22 and, to a lesser extent, WSC are also related to the conserved mitochondrial inner membrane protein MPV17, which has recently been associated with the human mitochondrial DNA depletion syndrome (MDDS; Spinazzola et al., 2006).
WSC envelops HEX assemblies to produce nascent Woronin bodies
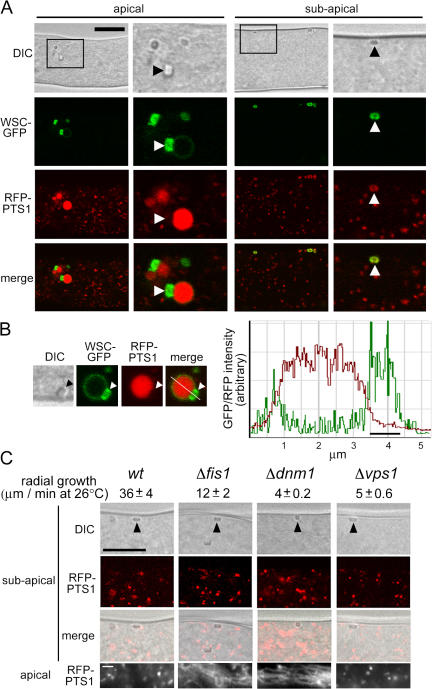
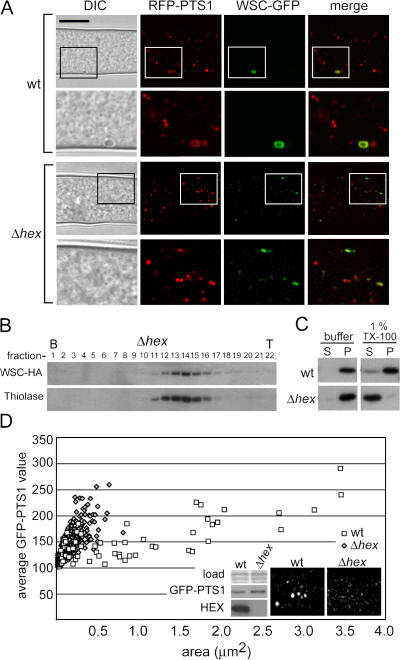
To examine the localization of WSC, we constructed HA-epitope and GFP-tagged versions of wsc. Both of these fully complement wsc loss-of-function phenotypes when expressed from the wsc promoter, which suggests that the fusion proteins preserve WSC function. To observe the peroxisome matrix and its relationship to HEX assemblies, we coexpressed WSC–GFP and peroxisome matrix–targeted RFP (RFP-PTS1). In apical compartments, two types of peroxisomes were observed: abundant small peroxisomes and infrequent large peroxisomes that usually contain HEX assemblies (Fig. 3). WSC–GFP is largely not detected in small peroxisomes but is detected in the membrane of large peroxisomes where it is enriched over budding HEX assemblies (Fig. 3 A), which suggests that these are nascent WBs. Line scans further show that WSC–GFP is found throughout the membrane of the nascent WB but is significantly enriched in the membrane that envelops the budding HEX assembly (Fig. 3 B). In subapical compartments, WSC–GFP is exclusively localized to cortex-associated WBs and these retain only traces of RFP-PTS1 (Fig. 3 A), which suggests that nascent WBs undergo membrane fission as they mature from apical into subapical compartments. This is consistent with time-lapse confocal microscopy showing membrane fission occurring after the initiation of cortical association (Tey et al., 2005).
Figure 3.
WSC is localized to nascent and mature WBs, and effect of deletion of dynamin-related proteins and FIS1. (A) WSC localizes over HEX assemblies in nascent WBs and surrounds HEX assemblies in mature WBs. WSC–GFP and RFP-PTS1 were coexpressed and localized in apical and subapical compartments by laser-scanning confocal microscopy. Bar, 10 μm. Magnified views of the region are indicated by a black square and shown at the right. Arrowheads point to refractive HEX assemblies. (B) WSC–GFP is concentrated over HEX assemblies in nascent WBs. Individual images of a single nascent WB are shown on the left and GFP and RFP fluorescence intensity are shown on the right across the indicated line. A black bar in the graph indicates the position of the HEX assembly, which can be seen in the DIC image. The line in the merge panel is 5.25 μm. (C) Effect of deletion of fis1, vps1, and dnm1 on WB assembly. DIC images show refractive HEX assemblies at the cell cortex (arrowheads) and RFP-PTS1 reveals the peroxisomal matrix. Bar, 10 μm. Bottom shows peroxisomes in the apical compartment, where Δdnm1 and Δfis1 present abnormal tubular peroxisomes. Bar, 1 μm. Radial growth for the indicated strains is shown in μm/min.
To investigate the function of genes known to influence peroxisome division in other eukaryotes in this process, we deleted the N. crassa homologues of the dynamin-related proteins VPS1 (NCU04100.3) and DNM1 (NCU09808.3), which promote membrane constriction and fission in a variety of intracellular compartments including peroxisomes (Hoepfner et al., 2001; Koch et al., 2004; Kuravi et al., 2006), and the integral membrane protein FIS1 (NCU05313.3), which has been implicated in the division of mitochondria (Mozdy et al., 2000; Hoppins et al., 2007) and peroxisomes (Koch et al., 2005; Kuravi et al., 2006). To assess the ability of WBs to differentiate from peroxisomes, we constructed these deletions in an RFP-PTS1–expressing background. Deletion of vps1, dnm1, and fis1 resulted in significant defects in hyphal growth (Fig. 3 C). However, in all three cases, the deletion strains were able to segregate WBs containing quantities of RFP-PTS1, comparable to those seen in wild-type, into subapical compartments (Fig. 3 C). These can function, as assessed by their ability to seal septal pores in response to cellular wounding (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1). All three mutants presented phenotypes associated with peroxisome morphology and WB production. Peroxisomes were aberrantly tubular in apical compartments of Δfis1 and Δdnm1 mutants, and in all three mutants, nascent WBs were less apparent and hex assemblies in mature WBs were generally smaller than those observed in the wild-type control. This was especially true in the Δdnm1 mutant (Fig. 3 C). Collectively, these data suggest that N. crassa DNM1, VPS1, and FIS1 have general functions in peroxisome metabolism and influence but are not required for the differentiation of WBs from peroxisomes.
WB-associated WSC is detergent insoluble
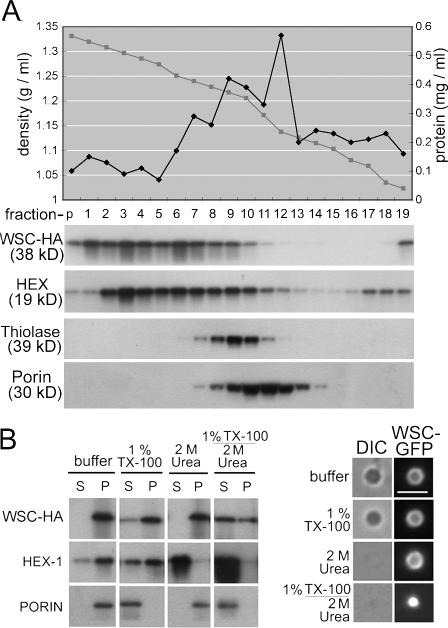
To further characterize the association of WSC with WBs, we used equilibrium density gradient centrifugation and fractionated organelles obtained from a WSC-HA–expressing strain. Consistent with its localization to WBs (Fig. 3), WSC-HA largely cofractionates with HEX into the densest region of the gradient, which contains only a minor fraction of total protein, and both WSC and HEX partly overlap with the peroxisomal matrix marker thiolase (Fig. 4 A). We next obtained an organellar fraction and separated this material into pellet and supernatant fractions after extraction with various chemical treatments (Fig. 4 B). Surprisingly, WSC is mostly resistant to extraction by 1% Triton X-100, whereas a control, the inner mitochondrial membrane protein porin, was solubilized by Triton X-100. Microscopic examination further shows that WSC remains associated with refractive WBs after detergent extraction (Fig. 4 B, right). HEX can be extracted from WBs by 2 M urea, but WSC–GFP remains in the pellet fraction and can be observed in an empty shell structure by fluorescence microscopy. Finally, under conditions that extract HEX and membranes, the WSC–GFP is partly solubilized; however, a significant amount remains in the pellet fraction and a compact punctate structure can be seen by fluorescence microscopy. Collectively, these data suggest that WSC associates with WBs in a detergent-resistant complex whose structural integrity may be partly independent of HEX.
Figure 4.
Biochemical characterization of WB-associated WSC. (A) HEX and WSC cofractionate. Organelles were separated on a 17–60% Nycodenz gradient and fractions were probed for various organellar markers. The graph shows the shape of the gradient and total protein distribution across the gradient. Anti-HEX and anti-HA reveal HEX and WSC and antithiolase provides a marker of the peroxisome matrix. The inner mitochondrial membrane protein porin reveals the distribution of mitochondria. (B) WSC is insoluble in Triton X-100. (left) 16 KgP was resuspended in buffer containing the indicated additions and then fractionated into pellet (P) and supernatant (S) fractions by centrifugation at 100,000 g for 45 min. Porin provides a control of an integral membrane protein. (right) An extract obtained from the WSC–GFP–expressing strain received the indicated treatments and was then examined by epifluorescence (WSC–GFP) and bright field (differential interference contrast) microscopy. Bar, 2 μm.
HEX determines a WSC-enriched peroxisomal subcompartment
Next, we examined the behavior of WSC in the absence of HEX. In contrast to its localization to the WB subcompartment in wild-type hyphae, in the absence of HEX, WSC–GFP localizes to abundant small peroxisomes that are mostly found in the cytosol (Fig. 5 A). Consistent with this localization, WSC-HA cofractionates with thiolase in equilibrium density centrifugation (Fig. 5 B). In addition, in the absence of HEX, WSC can be extracted from the membrane by detergent (Fig. 5 C). These observations suggest that in wild-type cells both the localization and detergent insolubility of WSC depend on HEX.
Figure 5.
HEX controls WSC localization and the development of large peroxisomes. (A) The localization of WSC–GFP was examined in the presence (wt) and absence (Δhex) of HEX. In wild-type hyphae, WSC is enriched in the WB membrane and is mostly excluded from abundant cytosolic peroxisomes. In the absence of hex, WSC is localized to small cytosolic peroxisomes. Bar, 10 μm. Magnified views of the region are indicated by a black square and shown below. (B) WSC cofractionates with thiolase in the absence of HEX. An organellar fraction was obtained from the hex deletion strain expressing WSC-HA and separated by density gradient centrifugation. The bottom (B) and top (T) of the gradient are indicated. (C) The detergent insolubility of WSC depends on HEX. An organellar fraction from wild-type and hex deletion strains was treated as indicated and fractionated into pellet (P) and supernatant (S) fractions by centrifugation at 100,000 g for 45 min. (D) Large apical peroxisomes are no longer observed in the hex deletion strain. GFP-PTS1–expressing wild-type and hex deletion strains were examined by laser-scanning confocal microscopy and peroxisome size was estimated using morphometric analysis as described in Materials and methods. Peroxisome size is plotted against mean GFP intensity (arbitrary units), which provides an estimate of GFP-PTS1 density in the peroxisome matrix. The inset panels show a representative image of the indicated strains as well as a Western blot showing that both strains express equal levels of GFP-PTS1.
To determine whether large peroxisomes resembling nascent WBs are still present in the hex deletion strain, we further examined the size distribution of apical peroxisomes, using GFP-PTS1–expressing strains and morphometric analysis. This analysis shows that large peroxisomes associated with WB production (Fig. 3) are absent from the hex deletion strain (Fig. 5 D). Also, the mean intensity of GFP-PTS1 shows a similar distribution in both strains, further suggesting that large peroxisomes do not accumulate matrix proteins to a higher concentration than small peroxisomes. Collectively, these data suggest that in the absence of HEX, WSC is trafficked by default to abundant small peroxisomes from which it is normally excluded. They further suggest that the matrix protein HEX is required for the differentiation of large WSC-enriched peroxisomes (nascent WBs).
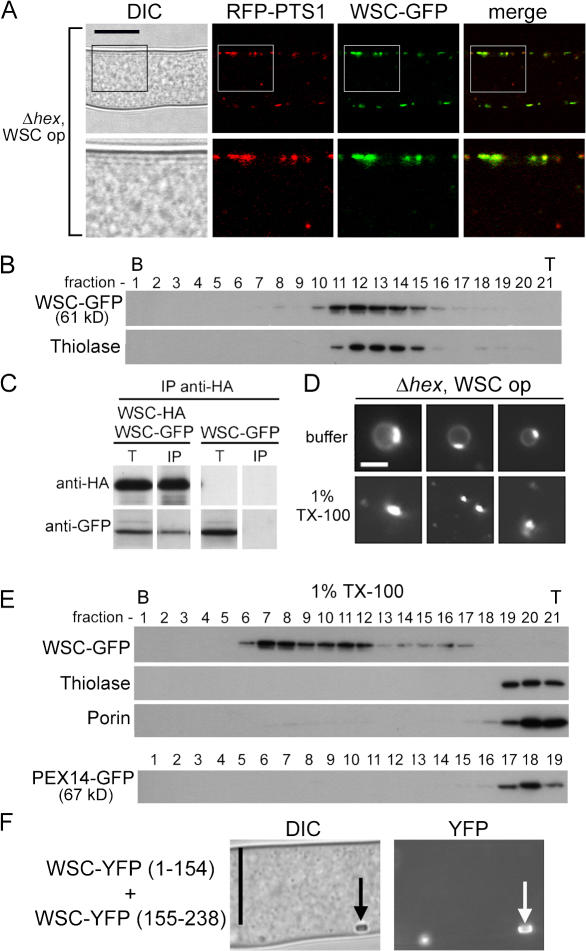
WSC overproduction promotes cortical association in the absence of HEX
In the hex deletion strain expressing WSC–GFP at endogenous levels, WSC is found at low but detectable levels in small peroxisomes and these are mostly localized to the cytosol (Fig. 5 A). WSC–GFP–containing peroxisomes were occasionally found at the cell cortex and these generally appeared enriched for WSC–GFP, which implies that WSC might be able to mediate cortex association in the absence of HEX. To examine the consequences of elevated levels of WSC, WSC–GFP was expressed from the hex promoter, which resulted in an approximately fivefold overproduction (unpublished data). In hex deletion hyphae (Fig. 6 A) as well as wild-type hyphae (not depicted), WSC–GFP overproduction results in relocalization of peroxisomes to the cell cortex of subapical compartments (Fig. 6 A), where they are immobilized and excluded from protoplasmic flow (Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1). Overproduced WSC–GFP also cofractionated with thiolase in equilibrium density centrifugation (Fig. 6 B), which suggests that its peroxisomal targeting is maintained under conditions of overproduction. These results suggest that WSC has an inherent ability to promote cortical association, which depends critically on its level in the membrane.
Figure 6.
WSC overproduction promotes cortical association in the absence of HEX. (A) WSC can target peroxisomes to the cell cortex in the absence of hex. WSC–GFP was overproduced from the strong hex promoter (WSC op). Bar, 10 μm. Magnified views of the region are indicated by a black square and shown below. (B) Overproduced WSC–GFP cofractionates with thiolase. Fractionation was done as described for Fig. 4 A. The bottom (B) and top (T) of the gradient are indicated. (C) WSC is in oligomers. WSC-HA and WSC–GFP were coexpressed or WSC–GFP was expressed alone in the wsc deletion strain. Detergent extracts from these strains were precipitated with anti-HA, and Western blotting with anti-GFP reveals the degree of WSC–GFP coprecipitation. WSC–GFP alone provides a control for the specificity of anti-HA precipitation. (D) Peroxisomes from the hex deletion (Δhex) overproducing WSC–GFP contain detergent-insoluble WSC–GFP–enriched membrane domains. An organellar fraction from the indicated strain was examined by epifluorescence microscopy in the absence (buffer) and presence (1% TX-100) of detergent. Bar, 1 μm. (E) Detergent-insoluble WSC–GFP complexes sediment to densities higher than the peroxisomes in which they form (compare with B). The organellar fraction was treated with 1% Triton X-100 and separated as described in Fig. 6 B. Porin and thiolase provide controls for detergent extraction and, in a separate experiment, PEX14-GFP reveals the behavior of another PMP (bottom). The bottom and top of the gradient are indicated. (F) The WSC complex promotes BiFC. The indicated constructs were coexpressed in the wsc mutant background and the resulting strain was examined for YFP fluorescence. Arrows point to mature WBs. Bar, 10 μm.
WSC self-assembles into detergent-resistant oligomers
WB-associated WSC is detergent insoluble and, under normal conditions, the attainment of this state requires HEX (Fig. 5 C). However, it remains unclear to what extent the assembly and maintenance of the WSC complex requires WSC–HEX interactions, WSC–WSC interactions, or a combination of these. To begin to assess WSC–WSC interactions, we coexpressed WSC-HA and WSC–GFP from the native wsc promoter in the hex deletion strain and performed immunoprecipitation experiments. After anti-HA precipitation, significant amounts of WSC–GFP were coprecipitated (Fig. 6 C), which suggests that WSC has a tendency to form oligomers in the absence of HEX.
To further examine WSC oligomers, we obtained a crude organellar fraction from the hex deletion overproducing WSC and examined organelles by epifluorescence microscopy. Here, WSC–GFP–enriched domains could readily be observed as bright patches of fluorescence in the membrane of isolated peroxisomes. These structures varied in size and could be as many as several hundred nanometers in diameter (Fig. 6 D). In the presence of detergent, spherical peroxisomal profiles were lost but a heterogeneously sized population of WSC–GFP complexes persisted (Fig. 6 D). These complexes were further characterized by density gradient centrifugation where WSC complexes sediment to densities higher than the peroxisomes from which they are isolated (Fig. 6, compare B and E), whereas two controls, the peroxisome matrix marker thiolase and transmembrane protein porin, were solubilized and remained at the top of the gradient (Fig. 6 E). As an additional control we examined the PMP PEX14. In contrast to WSC, PEX14 remains at the top of the density gradient after detergent extraction, which suggests that the behavior of WSC is not a general feature of PMPs.
We also examined the possible association of WSC complexes with detergent-insoluble lipid rafts (Bagnat et al., 2000) and found that isolated WSC complexes do not float in density gradients that isolate lipid rafts (unpublished data), which suggests that WSC complexes are detergent insoluble primarily because of persistent protein–protein interactions and not because of association with lipid rafts.
To further examine the packing of WSC, we used bimolecular fluorescence complementation (BiFC; Hu and Kerppola, 2003). Expression of either half of YFP fused to WSC (WSC–YFP1-154 or WSC–YFP155-238) alone results in complementation of the wsc mutant phenotypes but fails to produce YFP fluorescence (unpublished data). In contrast, coexpression of WSC–YFP1-154 and WSC–YFP155-238 results in bright WB-associated YFP fluorescence (Fig. 6 F), which suggests that WB-associated WSC is assembled into a tightly packed complex that allows fluorescence complementation.
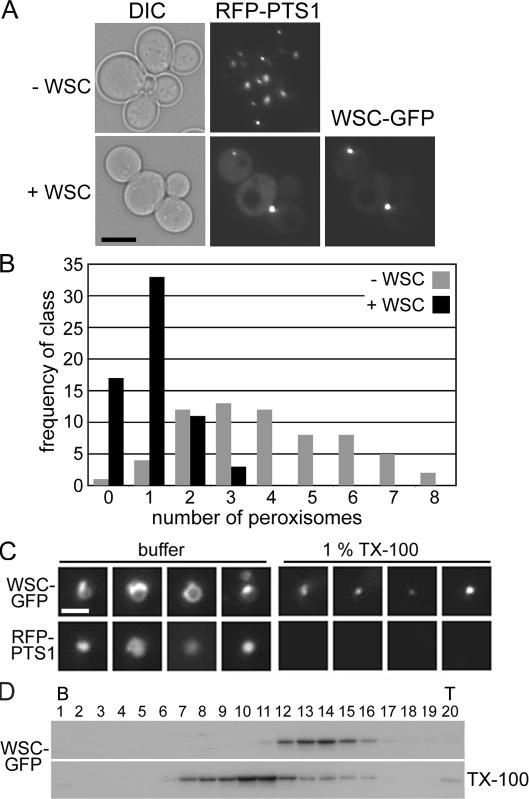
Next, we investigated the assembly of WSC in yeast. Here, WSC–GFP is localized to punctate structures that colocalize with RFP-PTS1 (Fig. 7 A), which suggests that WSC is targeted to the yeast peroxisome. However, WSC-expressing cells contain significantly fewer peroxisomes compared with empty vector controls (Fig. 7 B) and also tend to accumulate the RFP-PTS1 marker in the cytosol (Fig. 7 A). The peroxisome-inhibitory effect of WSC did not allow us to use the yeast system to investigate the physical association of WSC and HEX (see next section). However, WSC-expressing strains did allow a further examination of requirements for WSC complex assembly. In extracts prepared from yeast, WSC is also found in detergent-insoluble complexes (Fig. 7, C and D), which have a similar behavior in density gradients to those isolated from N. crassa (compare Figs. 6 and Figs.7). This further suggests that WSC can self-assemble not only in the absence of HEX but also in the absence of other WB-associated functions.
Figure 7.
WSC forms detergent-resistant complexes in the yeast peroxisome and also inhibits peroxisome biogenesis. (A) Yeast were transformed with an RFP-PTS1–expressing plasmid to reveal peroxisomes and then transformed with an empty plasmid (−WSC) or a WSC–GFP–expressing plasmid (+WSC). Bar, 5 μm. (B) The effect of WSC on peroxisome numbers was quantified by counting punctate signal from RFP-PTS1 in the indicated strains. 64 cells were counted for each strain. (C) WSC–GFP forms detergent-insoluble complexes in yeast. Examination of peroxisomes by fluorescence microscopy in the absence (buffer) and presence (1% TX-100) of detergent was conducted as described in Fig. 6 D. Bar, 1 μm. Panels presented in this figure were enlarged using the scaling function in Photoshop software. (D) Fractionation by density gradient centrifugation of extracts prepared from WSC–GFP–expressing yeast was done as described in Fig. 6 E in the absence (top) and presence (TX-100) of detergent. The bottom (B) and top (T) of the gradient are indicated.
MDDS-causing mutations also abolish WSC function and complex assembly
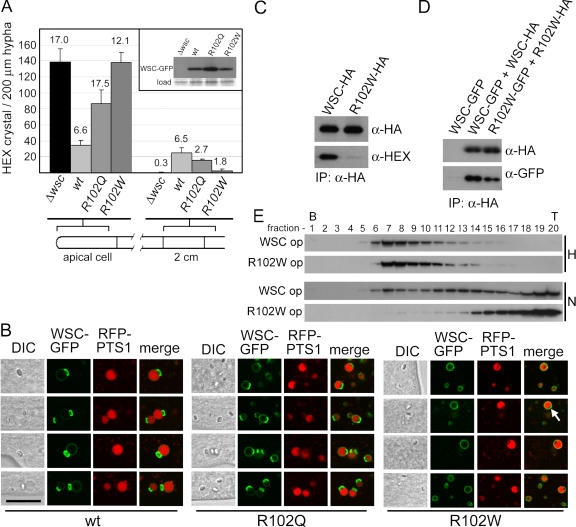
The wsc homologue MPV17 is mutated in certain forms of infantile hepatocerebral MDDS, and two of three identified mutations, R51Q and R51W (Spinazzola et al., 2006), occur in a residue that is largely conserved in wsc/MPV17/PMP22 genes (Fig. 2 B). We introduced these into the analogous position in genomic wsc and targeted wild-type and mutant versions to the his-3 locus (Aramayo and Metzenberg, 1996) to ensure equal levels of gene expression.
When these mutations are introduced into the yeast MPV17 orthologue SYM1, R51Q produces a partially functional protein and R51W destroys Sym1p function (Spinazzola et al., 2006). These mutations debilitate WSC function in a similar manner. wscR102Q produces a partial defect in WB inheritance, whereas wscR102W displays a near total defect (Fig. 8 A). Both mutants are targeted to the membrane of HEX assembly–containing peroxisomes and in keeping with their degree of loss of function, wscR102Q shows a slight defect in association with HEX assemblies, whereas wscR102W is almost totally defective in the production of asymmetrical nascent WBs and is found uniformly localized in the membrane (Fig. 8 B).
Figure 8.
MDDS-causing point mutations abolish WSC function and complex assembly. (A) The wsc deletion mutant was complemented with the indicated version of wsc–gfp and the distribution of HEX assemblies was quantified in apical and subapical compartments. Standard deviation is shown. These mutants do not dramatically alter steady-state levels of protein as revealed by a Western blot of extracts from the indicated strains (inset). (B) Capping is abolished by the R102W mutation. The indicated versions of WSC–GFP were localized using laser-scanning confocal microscopy. RFP-PTS1 reveals the peroxisome matrix. Arrow points to weak capping occasionally observed in the R102W mutant. Bar, 5 μm. (C) Evidence that WSC and HEX physically interact and effect of the R102W mutation on this interaction. Detergent extracts were prepared from strains expressing WSC-HA or WSC-R102W-HA from the native wsc promoter and anti-HA antibodies were used to precipitate WSC. Anti-HEX antibodies reveal the degree of HEX coprecipitation. (D) The R102W mutation affects WSC–WSC interaction. Extracts obtained from strains expressing the indicated versions of WSC were examined for WSC coprecipitation as described in Fig. 6 C. (E) Aberrant WSC–WSC complexes in the R102W mutant. Extracts were prepared from the indicated strains, treated with detergent, and separated by density gradient centrifugation, as in Fig. 6 E, using buffer H or buffer N.
WSC is required to recruit HEX assemblies to the matrix face of the peroxisomal membrane (Fig. 1), where it envelops them in a WSC-enriched membrane domain (Fig. 3). To obtain evidence for the physical association between WSC and HEX and to further dissect the defect engendered by the R102W mutation, we immunoprecipitated WSC-HA from detergent-treated extracts and examined the coprecipitation of HEX. Wild-type WSC is coprecipitated with HEX, and the R102W mutation produces a clear defect in this association (Fig. 8 C), which suggests that defects in the production of nascent WBs are, at least in part, caused by a defect in the formation of a WSC–HEX complex. Next, we examined the effect of the R102W mutation on the WSC–WSC interaction and found that the R102W mutation produces a moderate but reproducible defect in WSC coprecipitation when compared with wild-type WSC (Fig. 8 D). To further assess self-assembly, we compared the behavior of wild-type and mutant WSC complexes in density gradients. After detergent extraction, the R102W mutant displays a sedimentation pattern similar to WSC; however, when we used a buffer that favors disassembly of the WSC complex, the R102W mutant showed a greater tendency to disassemble and was significantly shifted to less dense regions of the gradient compared with wild-type WSC (Fig. 8 E). Collectively, these data show that MDDS-defined mutations abolish WSC function and interfere with WSC–HEX and WSC–WSC complex assembly.
Discussion
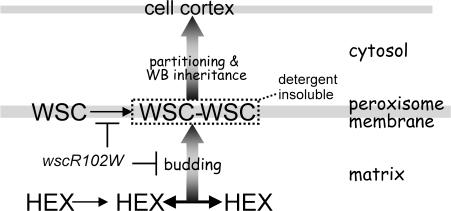
Peroxisomes proliferate through the division of preexistent peroxisomes (Titorenko and Rachubinski, 2001; Schrader and Fahimi, 2006; Motley and Hettema, 2007) and a growing body of evidence suggests that they can also form de novo from ER-derived precursors (Hoepfner et al., 2005; Tam et al., 2005; Kim et al., 2006; Titorenko and Mullen, 2006; Motley and Hettema, 2007). Mature peroxisomes are largely homogenous in a given cell type, and where they display specialized functions, e.g., in different types of plant peroxisomes, the distinction appears to be mediated by tissue-specific gene expression (Olsen et al., 1993; Nishimura et al., 1996). In this paper, we show that fungal peroxisomes engender WBs through a process that couples the self-assembly of HEX in the matrix with the assembly of WSC in the membrane (Fig. 9). This ultimately results in the production of two peroxisomal compartments, which are distinct in function and subcellular localization. HEX and WSC appear to control distinct steps in WB biogenesis. In the early stages, the development of large peroxisomes containing WSC depends on HEX, whereas in the later steps, the production of asymmetrical nascent WBs and their delivery to the cell cortex depend on WSC. WSC can form membrane-associated oligomers in the absence of HEX (Fig. 6 D); however, these are smaller than those formed in the presence of HEX (Fig. 3) and do not take the form of budding profiles. Thus, under normal conditions, WSC self-assembly appears to be patterned by HEX oligomers.
Figure 9.
Model of WB biogenesis. HEX self-assembles (double-headed arrow) in the peroxisome matrix and is recruited to the matrix face of the peroxisome membrane by interaction with WSC. A combination of WSC–WSC and HEX–WSC interactions concentrate WSC in the membrane and promote the production of asymmetrical nascent WBs. WSC complexes can then trigger cortical association, which allows partitioning of the nascent WB and allows segregation of the newly formed organelle into subapical compartments.
In the absence of HEX, peroxisomes containing low levels of WSC mostly fail to associate with the cortex (Fig. 5 A). In contrast, WSC overproduction results in the relocalization of WSC-enriched peroxisomes to the cortex (Fig. 6 A and Video 3). This suggests that cortical association requires a threshold level of membrane-associated WSC and, further, implies that under normal conditions, the interaction between HEX assemblies and WSC orders WB biogenesis by making the process of cortical association dependent on the assembly of HEX (Fig. 9).
Several findings demonstrate the functional and physical association of HEX assemblies and WSC. First, in wild-type hyphae, HEX assemblies are found at the peroxisome periphery in budding intermediates (Video 1). In contrast, in the absence of WSC, these fail to associate with the peroxisome membrane and move randomly in the matrix (Video 2). Second, WSC is localized to HEX assembly–containing peroxisomes and this localization depends on HEX (Figs. 3 and Figs.5). Third, HEX can be found in a complex with WSC (Figs. 4 and 8 C). Fourth, The MDDS-derived mutants result in loss-of-function phenotypes (Fig. 8 A), which are correlated with defects in the formation of asymmetrical nascent WBs (Fig. 8 B) and interaction between WSC and HEX (Fig. 8 C). The finding that MDDS-defined mutations also interfere with WSC function with a similar allelic specificity to their effects on Sym1p further suggests that members of this gene family conserve a similar mode of function.
In yeast, WSC also assembles into peroxisome-associated detergent-insoluble complexes, but here, it inhibits peroxisome proliferation and matrix protein import (Fig. 7). This effect might be achieved through a form of membrane occlusion where tightly woven WSC complexes restrict the activity or access of other PMPs. In principle, membrane occlusion by WSC complexes may also provide a mechanism to exclude peroxisomal functions from the WB membrane. This is consistent with the recent finding that two components of the matrix import pathway, PEX13 and PEX14, are largely excluded from a WB-enriched fraction (Managadze et al., 2007).
The WSC homologue Pmp22 has been characterized in plant (Tugal et al., 1999; Murphy et al., 2003) and animal systems (Diestelkotter and Just, 1993; Pause et al., 1997; Brosius et al., 2002; Iida et al., 2003). Like other PMPs, Pmp22 is targeted to the membrane posttranslationally (Fujiki et al., 1984; Diestelkotter and Just, 1993) through the PMP chaperone/import receptor Pex19p (Brosius et al., 2002; Shibata et al., 2004). PMPs are targeted to different peroxisome-related compartments. Some are targeted to very early ER-associated compartments, whereas others are targeted to late peroxisomal compartments and this requires additional layers of regulation, which are poorly understood (Heiland and Erdmann, 2005; Titorenko and Mullen, 2006). Pmp22 does not appear to define a subset of plant (Murphy et al., 2003) or mammalian (Brosius et al., 2002; Iida et al., 2003) peroxisomes. In contrast, WSC is preferentially localized to HEX-containing peroxisomes (Figs. 3 and 5 A). Moreover, the deletion of hex results in the localization of WSC to abundant small peroxisomes, from which it is normally excluded (Fig. 5 A), and the loss of large peroxisomes. This implies that the matrix protein HEX controls both peroxisome size and the membrane targeting of WSC.
How might this control be exerted? We speculate that a stochastic process, e.g., the intraperoxisomal nucleation of HEX oligomers, favors the subsequent growth of these peroxisomes, possibly through a feedback mechanism, to produce nascent WBs. In the fungus Yarrowia lipolytica, the density of the matrix protein acyl–CoA oxidase controls peroxisome division (Guo et al., 2003) and this provides another example of a matrix protein controlling peroxisomal fate. Peroxisomal matrix proteins are often assembled in the cytosol and imported into peroxisomes as oligomers (Leon et al., 2006). It will be interesting to investigate whether HEX is also imported in an oligomeric form. In this case, association with nascent WSC could provide one mechanism for HEX to control WSC membrane targeting.
WBs in Δdnm1, Δvps1, and Δfis1 mutants are functional and do not contain excessive quantities of RFP-PTS1 (Fig. 3 C), which suggests that they are differentiating and segregating from peroxisomes in a relatively normal manner. Δdnm1, Δvps1, and Δfis1 mutants have effects on WB biogenesis; however, it remains unclear whether these reflect a primary function or a secondary consequence of impaired hyphal growth or effects on upstream events in peroxisome biogenesis. Surprisingly, none of these mutants contained obviously reduced peroxisome numbers or abnormally enlarged spherical peroxisomes like those observed in yeast (Hoepfner et al., 2001; Kuravi et al., 2006). Instead, the Δdnm1 and Δfis1 mutants develop networks of tubular peroxisomes similar to those observed after knockdown of their mammalian counterparts, DLP1 (Koch et al., 2004) and hFis1 (Koch et al., 2005). This suggests that N. crassa peroxisomes may bear more similarity to their mammalian counterparts than to their yeast counterparts.
HEX assemblies in nascent WBs can be almost fully surrounded by the WSC complex and this results in constriction of the membrane connecting the two halves of the nascent structure (Fig. 8 B, wt). Δvps1 and Δdnm1 mutants each impair hyphal growth, which suggests that they cannot functionally substitute for one another. However, it remains possible that they can both promote fission of nascent WBs with equal efficacy. Alternatively, or in addition, the nascent structure may be partitioned through a mechanism where cortical association counteracts cytoskeleton-mediated forces that allow the organelle to be pulled apart. This is consistent with time-lapse video microscopy that shows the fission of the nascent WB occurring after the onset of cortical association (see Video 1 of Tey et al., [2005], at http://www.molbiolcell.org/cgi/content/full/E04-10-0937/DC2). Identification of the molecules that link the peroxisomes of filamentous fungi to the cytoskeleton should help resolve the mechanism of partitioning.
Yeast peroxisomal segregation is controlled by a “tug-of-war” mechanism, where cortical anchoring counteracts actomyosin-dependent transport into daughter cells (Fagarasanu et al., 2006), and mutants defective in cortical association accumulate reduced numbers of enlarged peroxisomes (Fagarasanu et al., 2005). Similarly, yeast mitochondrial segregation depends on a balance between retention at the mother cell cortex (Yang et al., 1999; Cerveny et al., 2007) and actomyosin-dependent transport into daughter cells (Itoh et al., 2002; Boldogh et al., 2003). Mutations that disrupt this balance result in defects in segregation and the accumulation of enlarged and morphologically abnormal mitochondria (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997; Cerveny et al., 2007).
The proteins controlling the segregation of yeast peroxisomes, mitochondria, and fungal WBs are unrelated at the sequence level and probably independently evolved. However, these systems all use cortical association to control organellar segregation and tend to couple morphogenesis and segregation, suggesting that they have converged on a common basic strategy. Unlike yeast peroxisomes and mitochondria, WB biogenesis from peroxisomes is an asymmetrical process that results in physically and functionally distinct compartments. The dual function of WSC determines this asymmetry by binding and enveloping HEX assemblies and promoting WB segregation through cortical association. Continued studies in diverse systems should provide further insights into the basic mechanisms of organellar biogenesis as well as the innovations that allow each organelle to fulfill its unique biological imperatives.
Materials and methods
Culture conditions, fungal strains, and identification of mutants
Vogel's N synthetic medium was used for growth in solid and liquid medium (Davis and de Serres, 1970), and N. crassa conidia were transformed by electroporation (Turner et al., 1997) or, in the case of cosmids, by chemical transformation of spheroplasts (Vollmer and Yanofsky, 1986). Crosses were performed as previously described (Davis and de Serres, 1970). For the induction of colonial growth, conidia were grown in plating medium as previously described (Davis and de Serres, 1970). Gene deletions were constructed through the direct transformation of deletion fragments obtained from fusion PCR using sets of oligos (Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1) that fuse 5′ and 3′ homology regions to the hygromycin resistance cassette. dnm1 and vps1 mutants do not produce spores and are infertile. These mutants were maintained in heterokaryons and obtained in the haploid state by plating and PCR screening of resultant colonies. A list of strains used in this study can be found in Table S1.
Mutants were obtained by UV irradiation of wild-type (FGSC #465) N. crassa conidia to ∼50% lethality. Mutated conidia were plated in a top-agar medium that induces tip lysis and colonial growth (Davis and de Serres, 1970), and after 3 d, colonies were screened on a stereomicroscope for excessive bleeding of protoplasm. To aid in the detection of mutants, the dye phyloxin B was added to the medium to a concentration of 7.5 μg/ml. Mutants selected in this manner were backcrossed using standard methods, and heterokaryons were used to identify complementation groups (Davis and de Serres, 1970). Mutants in pex genes were identified either by complementation using genomic fragments of candidate pex genes or through noncomplementation of deletion strains (Colot et al., 2006) obtained from the Fungal Genetics Stock Center. The linkage tester strain alcoy (Perkins, 1991) was used for mapping the wsc mutant and, after preliminary assignments to the chromosome were made, various regional markers were selected and used for fine mapping. To identify the wsc gene, we transformed cosmid clones (Kelkar et al., 2001) encompassing the closely linked sc marker and identified H122 C5 and H004 G10 clones as wsc-complementing fragments. PCR was used to amplify genomic fragments encompassing wsc candidates, and NCU07842.2 was found to fully complement wsc mutant phenotypes.
Mutant characterization
To obtain the results shown in Table I, representatives from each complementation group were examined for the presence of HEX assemblies using light microscopy, and transformation with the GFP-PTS1 marker (Tey et al., 2005) was used to visualize peroxisomes. WB function was assessed as previously described (Jedd and Chua, 2000).
To count HEX assemblies, the indicated strains were grown on Vogel's solid medium with appropriate supplements, and blocks of agar were excised and transferred to a microscope slide. The largest hyphae were selected and all HEX assemblies in a single field were counted. At a magnification of 1,000, the field of view has a diameter of 200 μm, and this was used to standardize the length of the hypha that were counted. To examine the apical compartment, the hyphal tip was placed at the edge of the field of view. Subapical compartments were examined ∼2 cm behind the growth front. To count nuclei, mutant and wild-type strains expressing a nucleus-targeted GFP (Freitag et al., 2004) were examined. In this case, apical and subapical compartments were photographed and nuclei in a region corresponding to 200 μm2 were counted. The data shown are the mean of at least ten measurements.
Biochemical methods
Mycelium was grown in Vogel's liquid medium with the appropriate supplements and harvested by filtration through Miracloth (EMD). The mycelium was quickly washed with ice-cold water, and excess liquid was removed by sandwiching the mycelium between several layers of paper towels. The pancake of mycelium was then frozen and ground to a powder in liquid nitrogen using a mortar and pestle. To prepare extracts, this frozen powder was resuspended in buffer containing 2 mM PMSF and a protease inhibitor cocktail (Roche). This lysate was applied to a 50-μm cell strainer and centrifuged at 1,000 g for 2 min to remove hyphae and obtain a crude cellular extract. The pellet and supernatant from this step were mixed and centrifuged at 100 g for 2 min to remove cellular debris. For density gradient centrifugation, the extract was prepared in buffer H (20 mM Hepes, pH 6.8, 200 mM Sorbitol, 50 mM potassium acetate, and 1 mM MgCl2) or buffer N (10 mM Tris-Hcl, pH 7.4, 150 mM NaCl, and 1 mM MgCl2) and centrifuged at 16,000 g for 45 min to obtain an organellar pellet (16 KgP), and this was resuspended in fresh buffer and applied to a preformed 17–60% Nycodenz gradient (Gentaur Inc.) prepared in the same buffer and centrifuged in a swinging bucket rotor (SW41; Beckman Coulter) at 27,000 rpm for 7 h at 4°C. For detergent extraction, Triton X-100 was added to the resuspended organellar pellet to a final concentration of 1% (vol/vol) and incubated on ice for 30 min before separation by density gradient centrifugation. Fractions were collected from the bottom of the tube using a peristaltic pump. For extraction experiments shown in Figs. 4 and Figs.5, the 16-Kg pellet was resuspended in buffer N and adjusted to the indicated conditions by the addition of 4× concentrated buffers. Extractions were performed for 30 min on ice and then centrifuged at 100,000 g for 45 min to obtain pellet and supernatant fractions. These were resuspended to equal volumes and protein distribution was determined by ECL Western blotting (Millipore). Primary antibodies were obtained from the following sources: antithiolase, R. Rachubinski (University of Alberta Edmonton, Alberta, Canada); anti-porin, F. Nargang (University of Alberta Edmonton); anti-GFP, Clontech Laboratories, Inc.; anti-HEX, Jedd and Chua (2000); and peroxidase-coupled anti-HA, Roche. Secondary antibodies were obtained from the following sources: HRP-coupled anti–Rabbit IgG (H + L) for use with anti-HEX, anti-Porin, and anti-GFP antibodies, Jackson ImmunoResearch Laboratories; and HRP-coupled anti–guinea pig IgG for use with antithiolase, Dako.
Immunoprecipitation
For WSC–WSC and WSC–HEX coimmunoprecipitation, a volume of frozen powder equal to ∼0.25 ml was resuspended in 0.5 ml of buffer N containing 1% Triton X-100. The crude extract was centrifuged at 800 g for 5 min to remove cellular debris. The supernatant was recovered and BSA was added to a final concentration of 0.1 mg/ml. A packed volume of 15 μl of agarose-coupled anti-HA antibodies (Roche) was added and binding was performed for 1 h at 4°C. Beads were washed four times in 0.5 ml of buffer N containing 1% Triton X-100 and the precipitate was released by boiling in SDS-PAGE loading buffer. Precipitates were resolved on SDS-PAGE and Western blotting was used to reveal the indicated proteins.
Construction of plasmids
Plasmid construction is described in this section and a list of plasmids used in this study can be found in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1). Sequences of oligonucleotides used for PCR and mutagenesis can be found in Table S3. Restriction sites and mutants were introduced using the QuikChange site-directed mutagenesis kit (Stratagene).
A modified hygromycin B resistance-encoding plasmid.
To construct a pBluescript II SK + (BS + SK; Stratagene) –based cloning vector containing the hygromycin resistance cassette and useful NotI and PacI restriction sites, a PacI site was first introduced into BS + SK, using primers BS + SK.PacI.f and BS + SK.PacI.r. The hygromycin cassette was then amplified from pCSN43 (Staben et al., 1989), using primers HYGr.NotI and HYGr.PacI, and cloned into the modified BS + SK using NotI and PacI restriction sites. The PacI site was then mutated, using primers PacI.kill.1 and PacI.kill.2, and a new PacI site was reintroduced into the vicinity of the NotI site using primers PacI.create.1 and PacI.create.2, resulting in GJP #961.
Genomic wsc.
To construct a plasmid carrying a genomic fragment of wsc, wsc was amplified from genomic DNA using primers 7842.f-2 and 7842.r-2 and this fragment was cloned into the TOPOII vector, producing GJP #1001. The wsc genomic fragment was then subcloned to GJP #961 using NotI and PacI, and this produced the plasmid GJP #1010.
wsc deletion.
A wsc deletion cassette was constructed using fusion PCR. The 3′ and 5′ frank of wsc were amplified using GJP #1001 as a template and the primers 7842.P-1, 7842.P-2 and 7842.P-3, and 7842.P-4, respectively. The hygromycin B resistance cassette was amplified from the plasmid GJP #961 using primers 7842.P-5 and 7842.P-6. These three fragments were purified and fused using primers 7842.P-1 and 7842.P-4. The fused product was cloned into the TOPOII vector (Invitrogen), producing GJP #1009. This vector was transformed into a mus51 deletion strain (FGSC #9179) by electroporation, and homologous integrants were identified by PCR. Deletion strains were then backcrossed to remove the mus51 mutation.
GFP and epitope tagging of wsc.
GFP and 3×HA epitope tag were amplified from plasmid pEGFP-N1 (Clontech Laboratories, Inc.) and pREP41 (Craven et al., 1998), using primers eGFP-3′-XbaI, eGFP-3′-SphI and HA.3′-XbaI, and HA.3′-SphI, respectively. These two fragments were cloned into the TOPOII vector and sequenced. To introduce these two fragments into the genomic version of wsc, site-directed mutagenesis was used to create XbaI and SphI restriction sites at the 3′ end of the wsc structural gene, using primers 7842–3′-Mut-1, 7842–3′-Mut-2 and 7842–3′-Mut-3, and 7842–3′-Mut-4, respectively. This produced GJP #1050. GFP and 3×HA were subcloned into GJP #1050 using XbaI and SphI to create in-frame fusions, resulting in GJP #1081 and GJP #1064, respectively.
Targeting wsc derivatives to the his-3 locus.
To construct both GFP and epitope-tagged wsc genomic fragments for his-3 targeting, wsc–GFP and wsc-3×HA were amplified from GJP #1081 and GJP #1064, using primers 7842.f-2 and 7842.r-3. These two fragments were cloned into pBM60 (Margolin et al., 1997) using NotI and SpeI restriction sites, producing GJP #1245 and GJP #1353.
Plasmids for wsc overexpression.
A wsc–gfp fragment was amplified with primers wsc5′-NotI and wsc3′-PacI, using GJP #1245 as a template. This fragment was cloned directly into the TOPOII vector, sequenced, and subcloned into the hex promoter containing expression vector GJP #1406, using NotI and PacI. This produced GJP #1500. The mutations R102Q and R102W were introduced into epitope-tagged (GJP #1353) and GFP-fused (GJP #1245) versions of wsc using site-directed mutagenesis with primers wsc-Mut7-f, wsc-Mut7-r (R102Q) and wsc-Mut8-f, and wsc-Mut8-r (R102W). This produced the plasmids GJP #1322 and 1372, containing the wscR102Q, and GJP #1324 and 1361, containing the wscR102W mutation.
Constructs for WSC–WSC immunoprecipitation.
The R102W mutation was introduced into epitope-tagged (GJP #1064) and GFP-fused (GJP #1081) versions of wsc, resulting in GJP #1561 and 1564. For WSC-R102W-GFP overexpression, the R102W mutation was introduced into the hex promoter–controlled version of wsc (GJP #1500), producing plasmid GJP #1821.
BiFC.
To express half of YFP fused to WSC, YFP1-154 and YFP155-238 were amplified from plasmid pEYFP-N1 (Clontech Laboratories, Inc.) with primers EYFP-I-3′-XbaI, EYFP-I-3′-SphI and EYFP-II-3′-XbaI, and EYFP-II-3′-SphI. After sequencing, these two partial YFP fragments were cloned into GJP #1050 using XbaI and SphI to create in-frame fusions, resulting in GJP #1302 and 1304.
Targeting RFP to the peroxisome matrix.
To construct a version of RFP that serves as marker of peroxisomal matrix, the RFP fragment was amplified from pRSET–B mCherry (Shaner et al., 2004) using primers NotI.pts1.ch.5′ and PacI.pts1.ch.3′, which introduce NotI and PacI restriction sites and append the PTS1 tripeptide SRL to the C terminus of RFP. The expression plasmid GJP #613 contains a genomic fragment of the hex gene where an intron has been introduced into the 3′ UTR (Tey et al., 2005). The RFP-PTS1 fragment was subsequently cloned into this vector to produce GJP #1406.
PEX14-GFP fusion protein.
A pex14 fragment was amplified from genomic DNA, using primers Pex14-SpeI and Pex14-PacI. After sequencing, this fragment was cloned into plasmid pMF272 with SpeI and PacI to create an in-frame fusion, resulting in plasmid GJP #1530.
Plasmids for expression in yeast.
To express WSC–GFP in yeast, a wsc–gfp cDNA was amplified by RT-PCR using total mRNA from GSF #629 using primers wsc-gfp-5′-SpeI and wsc-gfp-3′-XhoI. To express RFP-PTS1 in yeast, an rfp-pts1 fragment was amplified from GJP #1406 using PCR and primers mCherry-5′-SpeI and mCherry-3′-XhoI. These two fragments were first cloned into the TOPOII vector for sequencing and then subcloned into plasmid p416GPD and p413GPD to produce GJP #1662 and 1712, respectively.
Microscopy
All imaging was done at room temperature, ∼26°C. For confocal microscopy, a microscope (Meta Upright; Carl Zeiss, Inc.) fitted with a 100×/1.4 NA oil immersion objective lens (Plan-apochromat) was used. Differential interference contrast and fluorescence images were simultaneously obtained. Enhanced GFP was imaged using 488-nm excitation and a 500–530-nm band-pass filter, and RFP was imaged using 543-nm excitation and a 565–615-nm band-pass filter. The pinhole diameter was set at 1 airy unit, and for simultaneous imaging of enhanced GFP and RFP the single-track function was used. To quantify peroxisome size and mean levels of GFP-PTS1 shown in Fig. 5 D, we used Metamorph software (MDS Analytical Technologies) and integrated morphometry analysis. The micrographs shown in Figs. 1 (A and B), 4 B, 6 (D and F), and 7 (A and C) were taken using an epifluorescence microscope (BX51; Olympus) equipped with a 100×/1.4 NA oil immersion objective (UplanSapo) and a digital camera (Coolsnap ES; Photometrics) controlled by Metamorph software. The videos were acquired using the stream acquisition function. The figures were created using Photoshop 7.0 software (Adobe) and, in some cases, where the brightness of the image needed adjustment, we used the image-adjustments-curves function.
Sequence analysis
WSC-related sequences were obtained from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST) and the Fungal Genome Project at the Broad Institute (www.broad.mit.edu/annotation/fgi). These were aligned using ClustalX (Thompson et al., 1997) and the alignment was subjected to Bayesian-based tree construction using MrBayes3.1.2 (Huelsenbeck and Ronquist, 2001). After the burn-in phase, every 100th generation for 40,000 generations was considered. The 50% majority tree is shown with percent posterior probabilities shown at each node. For the prediction of transmembrane domains highlighted in Fig. 2 B, sequences were submitted to the TMpred server (http://www.ch.embnet.org/software/TMPRED_form.html). Predicted transmembrane domains scoring >500 are highlighted in yellow and those scoring <500 are highlighted in pink. The sequences that were used to create Fig. 2 C are available under the following accession nos.: PMP22.AT, CAB77915; PMP22.CA, XP_717821; PMP22.CC, EAU92685.1; PMP22.DD, XP_635840; PMP22.DM, AAF47541; PMP22.HS, XP_001126107; PMP22.KL, CAH00328; PMP22.OS, XP_483851; PMP22.UM, XP_756258; PMP22.XT, NP_001006885; PMP22.YL, XP_505475; SYM1.AF, XP_748225; SYM1.AO, BAE66121; SYM1.AT, AAN46791; SYM1.CA, XP_711828; SYM1GZ, XP_380906; SYM1.HS, NP_002428; SYM1.KL, XP_456102; SYM1.NC, XP_963854; SYM1.SC, NP_013352; SYM1.SP, CAA93564; SYM1.UM, XP_759354; SYM1.YL, XP_502197; WSC.AF, XP_756042; WSC.AN, XP_680527; WSC.AO, BAE61217; WSC.GZ, XP_384295; WSC.HC, EAS28198; WSC.MG, XP_359485; and WSC.NC, XP_963103.
Online supplemental material
Fig. S1 shows that wsc growth defects are suppressed by hex and pex14 mutation. Fig. S2 shows that Δvps1, Δdnm1, and Δfis1 produce functional WBs as assessed by the response to cellular wounding. Video 1 shows time-lapse microscopy showing that HEX assemblies are attached to the membrane of wild-type peroxisomes. Video 2 shows time-lapse microscopy showing that HEX assemblies move randomly in the matrix of wsc mutant peroxisomes. Video 3 shows time-lapse microscopy showing that WSC-enriched peroxisomes are attached to the cell cortex and excluded from protoplasmic flow when WSC is overproduced in the hex deletion mutant. Table S1 shows the N. crassa strains used in this study. Table S2 lists plasmids used in this study. Table S3 lists oligonucleotides used for PCR and site-directed mutagenesis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200705049/DC1.
Supplemental Material
Acknowledgments
We thank Rick Rachubinski and Frank Nargang for generously providing antibodies and the Neurospora genome project knockout group for providing some of the deletion mutants used in this study. We also thank Steve Cohen, Fred Berger, and Davis Ng for comments on the manuscript and Temasek Life Science Laboratory attachment students Ling-wei Cheong, Wei Liang Ng, and Qi Chun Toh for help with initial stages of mutant screening.
The authors declare that they have no competing financial interest.
Abbreviations used in this paper: BiFC, bimolecular fluorescence complementation; MDDS, mitochondrial DNA depletion syndrome; PTS1, peroxisome-targeting signal 1; WB, Woronin body; WSC, Woronin sorting complex.
References
- Aramayo, R., and R.L. Metzenberg. 1996. Gene replacements at the his-3 locus of Neurospora crassa Fungal Genetics Newsletter. 43:9–13. [Google Scholar]
- Bagnat, M., S. Keranen, A. Shevchenko, A. Shevchenko, and K. Simins. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 97:3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, K.H., L.F. Sogo, and M.P. Yaffe. 1997. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 136:545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I.R., D.W. Nowakowski, H.C. Yang, H. Chung, S. Karmon, P. Royes, and L.A. Pon. 2003. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 14:4618–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, D.M., and G.C. Carrol. 1968. Fine-structural correlates of growth in hyphae of Ascodesmis sphaerospora. J. Bacteriol. 95:658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius, U., T. Dehmel, and J. Gartner. 2002. Two different targeting signals direct human peroxisomal membrane protein 22 to peroxisomes. J. Biol. Chem. 277:774–784. [DOI] [PubMed] [Google Scholar]
- Burgess, S.M., M. Delannoy, and R.E. Jensen. 1994. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 126:1375–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp, R.R. 1977. Association of microbodies, Woronin bodies, and septa in intercellular hyphae of Cymadothea trifolii Can. J. Bot. 55:1856–1859. [Google Scholar]
- Cerveny, K.L., S.L. Studer, R.E. Jensen, and H. Sesaki. 2007. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev. Cell. 12:363–375. [DOI] [PubMed] [Google Scholar]
- Colot, H.V., G. Park, G.E. Turner, C. Ringelberg, C.M. Crew, L. Litvinkova, R.L. Weiss, K.A. Borkovich, and J.C. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA. 103:10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven, R.A., D.J. Griffiths, K.S. Sheldrick, R.E. Randall, I.M. Hagan, and A.M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 221:59–68. [DOI] [PubMed] [Google Scholar]
- Davis, R.H., and F.J. de Serres. 1970. Genetic and microbiological research techniques for Neurospora crassa Methods Enzymol. 17:79–143. [Google Scholar]
- Diestelkotter, P., and W.W. Just. 1993. In vitro insertion of the 22-kD peroxisomal membrane protein into isolated rat liver peroxisomes. J. Cell Biol. 123:1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu, A., M. Fagarasanu, G.A. Eitzen, J.D. Aitchison, and R.A. Rachubinski. 2006. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell. 10:587–600. [DOI] [PubMed] [Google Scholar]
- Fagarasanu, M., A. Fagarasanu, Y.Y. Tam, J.D. Aitchison, and R.A. Rachubinski. 2005. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 169:765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag, M., P.C. Hickey, N.B. Raju, E.U. Selker, and N.D. Read. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897–910. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., R.A. Rachubinski, and P.B. Lazarow. 1984. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc. Natl. Acad. Sci. USA. 81:7127–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, T., Y.Y. Kit, J.M. Nicaud, M.T. Le Dall, S.K. Sears, H. Vali, H. Chan, R.A. Rachubinski, and V.I. Titorenko. 2003. Peroxisome division in the yeast Yarrowia lipolytica is regulated by a signal from inside the peroxisome. J. Cell Biol. 162:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiland, I., and R. Erdmann. 2005. Biogenesis of peroxisomes. Topogenesis of the peroxisomal membrane and matrix proteins. FEBS J. 272:2362–2372. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., M. van den Berg, P. Philippsen, H.F. Tabak, and E.H. Hettema. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner, D., D. Schildknegt, I. Braakman, P. Philippsen, and H.F. Tabak. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 122:85–95. [DOI] [PubMed] [Google Scholar]
- Hoppins, S., L. Lackner, and J. Nunnari. 2007. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76:751–780. [DOI] [PubMed] [Google Scholar]
- Hu, C.D., and T.K. Kerppola. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J.P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755. [DOI] [PubMed] [Google Scholar]
- Iida, R., T. Yasuda, E. Tsubota, H. Takatsuka, M. Masuyama, T. Matsuki, and K. Kishi. 2003. M-LP, Mpv17-like protein, has a peroxisomal membrane targeting signal comprising a transmembrane domain and a positively charged loop and up-regulates expression of the manganese superoxide dismutase gene. J. Biol. Chem. 278:6301–6306. [DOI] [PubMed] [Google Scholar]
- Itoh, R., and Y. Fujiki. 2006. Functional domains and dynamic assembly of the peroxin Pex14p, the entry site of matrix proteins. J. Biol. Chem. 281:10196–10205. [DOI] [PubMed] [Google Scholar]
- Itoh, T., A. Watabe, E.A. Toh, and Y. Matsui. 2002. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:7744–7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd, G., and N.H. Chua. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2:226–231. [DOI] [PubMed] [Google Scholar]
- Kelkar, H.S., J. Griffith, M.E. Case, S.F. Covert, R.D. Hall, C.H. Keith, J.S. Oliver, M.J. Orbach, M.S. Sachs, J.R. Wagner, et al. 2001. The Neurospora crassa genome: cosmid libraries sorted by chromosome. Genetics. 157:979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, P.K., R.T. Mullen, U. Schumann, and J. Lippincott-Schwartz. 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A., G. Schneider, G.H. Luers, and M. Schrader. 2004. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117:3995–4006. [DOI] [PubMed] [Google Scholar]
- Koch, A., Y. Yoon, N.A. Bonekamp, M.A. McNiven, and M. Schrader. 2005. The role of Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 16:5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi, K., S. Nagotu, A.M. Krikken, K. Sjollema, M. Deckers, R. Erdmann, M. Veenhius, and I. Van der Klei. 2006. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119:3994–4001. [DOI] [PubMed] [Google Scholar]
- Leon, S., J.M. Goodman, and S. Subramani. 2006. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim. Biophys. Acta. 1763:1552–1564. [DOI] [PubMed] [Google Scholar]
- Managadze, D., C. Wurtz, M. Sichting, G. Niehaus, M. Veenhuis, and H. Rottensteiner. 2007. The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic. 8:687–701. [DOI] [PubMed] [Google Scholar]
- Margolin, B.S., M. Freitag, and E.U. Selker. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genetics Newsletter. 44:34–36. [Google Scholar]
- Markham, P., and A.J. Collinge. 1987. Woronin bodies of filamentous fungi. FEMS Microbiol. Lett. 46:1–11. [Google Scholar]
- Maruyama, J., P.R. Juvvadi, K. Ishi, and K. Kitamoto. 2005. Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 331:1081–1088. [DOI] [PubMed] [Google Scholar]
- Momany, M., E.A. Richardson, C. Van Sickle, and G. Jedd. 2002. Mapping Woronin body position in Aspergillus nidulans Mycologia. 94:260–266. [PubMed] [Google Scholar]
- Motley, A.M., and E.H. Hettema. 2007. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy, A.D., J.M. McCaffery, and J.M. Shaw. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, M.A., B.A. Phillipson, A. Baker, and R.T. Mullen. 2003. Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol. 133:813–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhoff, K., N.M. Meindl-Beinker, D. Kerssen, U. Perband, A. Schafer, W. Schliebs, and W.H. Kunau. 2005. Yeast Pex14p possesses two functionally distinct Pex5p and one Pex7p binding sites. J. Biol. Chem. 280:35571–35578. [DOI] [PubMed] [Google Scholar]
- Nishimura, M., M. Hayashi, A. Kato, K. Yamaguchi, and S. Mano. 1996. Functional transformation of microbodies in higher plant cells. Cell Struct. Funct. 21:387–393. [DOI] [PubMed] [Google Scholar]
- Olsen, L.J., W.F. Ettinger, B. Damsz, K. Matsudaira, M.A. Webb, and J.J. Harada. 1993. Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell. 5:941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause, B., P. Diestelkotter, H. Heid, and W.W. Just. 1997. Cytosolic factors mediate protein insertion into the peroxisomal membrane. FEBS Lett. 414:95–98. [DOI] [PubMed] [Google Scholar]
- Perkins, D.D. 1991. Neurospora alcoy linkage tester stocks with group VII marked, and their use for mapping translocation. Fungal Genetics Newsletter. 38:83. [Google Scholar]
- Schrader, M., and H.D. Fahimi. 2006. Growth and division of peroxisomes. Int. Rev. Cytol. 255:237–290. [DOI] [PubMed] [Google Scholar]
- Shaner, N.C., R.E. Campbell, P.A. Steinbach, B.N. Giepmans, A.E. Palmer, and R.Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572. [DOI] [PubMed] [Google Scholar]
- Shibata, H., Y. Kashiwayama, T. Imanaka, and H. Kato. 2004. Domain architecture and activity of human Pex19p, a chaperone-like protein for intracellular trafficking of peroxisomal membrane proteins. J. Biol. Chem. 279:38486–38494. [DOI] [PubMed] [Google Scholar]
- Sichting, M., A. Schell-Steven, H. Prokisch, R. Erdmann, and H. Rottensteiner. 2003. Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol. Biol. Cell. 14:810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, L.F., and M.P. Yaffe. 1994. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 126:1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan, S., G. Jedd, X. Li, M. Ramos-Pamplona, N.H. Chua, and N.I. Naqvi. 2004. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell. 16:1564–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzola, A., C. Viscomi, E. Fernandez-Vizarra, F. Carrara, P. D'Adamo, S. Calvo, R.M. Marsano, C. Donnini, H. Weiher, P. Strisciuglio, et al. 2006. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 38:570–575. [DOI] [PubMed] [Google Scholar]
- Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, J. Kinsey, and E. Selker. 1989. Use of a bacterial Hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genetics Newsletter. 36:79–81. [Google Scholar]
- Tam, Y.Y., A. Fagarasanu, M. Fagarasanu, and R.A. Rachubinski. 2005. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 280:34933–34939. [DOI] [PubMed] [Google Scholar]
- Tenney, K., I. Hunt, J. Sweigard, J.I. Pounder, C. McClain, E.J. Bowman, and B.J. Bowman. 2000. hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31:205–217. [DOI] [PubMed] [Google Scholar]
- Tey, W.K., A.J. North, J.L. Reyes, Y.F. Lu, and G. Jedd. 2005. Polarized gene expression determines Woronin body formation at the leading edge of the fungal colony. Mol. Biol. Cell. 16:2651–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin, and D.G. Higgins. 1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko, V.I., and R.A. Rachubinski. 2001. The life cycle of the peroxisome. Nat. Rev. Mol. Cell Biol. 2:357–368. [DOI] [PubMed] [Google Scholar]
- Titorenko, V.I., and R.T. Mullen. 2006. Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinci, A.P.J., and A.J. Collinge. 1974. Occlusion of septal pores of damaged hyphae of Neurospora crassa by hexagonal crystals. Protoplasma. 80:57–67. [DOI] [PubMed] [Google Scholar]
- Tugal, H.B., M. Pool, and A. Baker. 1999. Arabidopsis 22-kilodalton peroxisomal membrane protein. Nucleotide sequence analysis and biochemical characterization. Plant Physiol. 120:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, G.E., T.J. Jimenez, S.-K. Chae, R.A. Baasiri, and K.A. Borkovich. 1997. Utilization of the Aspergillus nidulans pyrG gene as a selectable marker for transformation and electroporation of Neurospora crassaFungal Genet. Newsl. 44:57–59. [Google Scholar]
- Vollmer, S.J., and C. Yanofsky. 1986. Efficient cloning of genes of Neurospora crassa. Proc. Natl. Acad. Sci. USA. 83:4869–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wergin, W.P. 1973. Development of Woronin bodies from microbodies in Fusarium oxysporum f.sp lycopersici. Protoplasma. 76:249–260. [DOI] [PubMed] [Google Scholar]
- Yang, H.C., A. Palazzo, T.C. Swayne, and L.A. Pon. 1999. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr. Biol. 9:1111–1114. [DOI] [PubMed] [Google Scholar]
- Yuan, P., G. Jedd, D. Kumaran, S. Swaminathan, H. Shio, D. Hewitt, N.H. Chua, and K. Swaminathan. 2003. A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 10:264–270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.