Abstract
Monomeric human pancreatic RNase, devoid of any biological activity other than its RNA degrading ability, was engineered into a dimeric protein with a cytotoxic action on mouse and human tumor cells, but lacking any appreciable toxicity on mouse and human normal cells. This dimeric variant of human pancreas RNase selectively sensitizes to apoptotic death cells derived from a human thyroid tumor. Because of its selectivity for tumor cells, and because of its human origin, this protein represents a potentially very attractive, novel tool for anticancer therapy.
Among the RNases with bioactions other than their catalytic action (1) are seminal RNase from bull semen (BS-RNase) and Onconase from frog eggs, both endowed with a strong antitumor activity (2). Onconase is currently being tested in Phase III clinical trials (3). The cytotoxic action of BS-RNase is highly selective for malignant cells, and has been established in both in vitro and in vivo experiments (4–8). Some of the key steps of its mechanism of action have been elucidated (7, 9). The protein is concentrated on the extracellular matrix, is internalized by nonreceptor mediated endocytosis, and reaches the cytosol, where it selectively degrades ribosomal RNA and blocks protein synthesis, thus causing cell death. The cytosolic destination is reached, and cytotoxicity is exerted, only on malignant cells.
The antitumor activity of BS-RNase depends on its dimeric structure (5). BS-RNase is in fact the only dimeric RNase from the pancreatic-like RNase superfamily. Its identical subunits are linked by two adjacent intersubunit disulfides, involving cysteine-31 and -32 of each subunit, and are noncovalently associated through a small hydrophobic patch comprising Leu-28 and Met-29 of each subunit (10).
By protein engineering, RNase A, a classic monomeric protein, was transformed into a dimeric protein endowed with antitumor activity (11). This was achieved by the replacement of residues at positions 19, 28, 31, and 32 of RNase A chain with the corresponding residues of the BS-RNase subunit: i.e., a proline, a leucine, and two cysteines, respectively (11, 12). Thus, these residues were identified as the sufficient requirements for a monomeric RNase to be converted into a stable and biologically active dimer.
On the basis of these results, the project was conceived to transfer to a human RNase the key structural and functional determinants of BS-RNase antitumor action. Human pancreas RNase (HP-RNase) (13, 14), a monomeric RNase for which no special biological actions have been reported besides its RNA degrading ability, with >70% of its amino acid sequence identical to that of BS-RNase, was considered a logical candidate to reshape a human RNase into a BS-RNase-like dimeric RNase.
Here, we report the construction and characterization of a dimeric variant of HP-RNase, obtained by replacing four of its amino acid residues with the corresponding residues from BS-RNase sequence. This allowed the grafting of the intersubunit interface of BS-RNase on the human RNase, thus allowing its dimerization. The engineered dimeric protein, named HHP-RNase, was found to be enzymatically active and selectively cytotoxic for several malignant mouse and human cell lines.
MATERIALS AND METHODS
BS-RNase was purified from seminal vesicles as described (15, 16). Escherichia coli strain BL21 (DE3) pLysS and expression vector pET22b+ were provided by AMS Biotechnology (Milan); E. coli JM 101 strain was from Boehringer Mannheim; restriction enzymes were from Promega. Recombinant wild-type HP-RNase was a gift of A. Di Donato (University of Naples Federico II).
Site-Directed Mutagenesis.
The cDNA coding for HP-RNase (17), cloned in the BamHI/SalI cloning sites of a pUC118 vector, was mutated according to Kunkel (18) by using appropriate oligonucleotides synthesized by Beckman Analytical. In a first step, by using oligonucleotides 5′-CAGGTCGGAAGGGTATACTTTCTTAGATCTCGATTTT-3′ and 5′-TTAAGGTCGCAGTTGTGTAGCTGAGACTGAG-3′ as primers, a NdeI site was inserted immediately upstream of the Lys-1 codon, and an NdeI site, positioned within the coding region (nucleotides 34–39, starting from Lys-1 codon) was eliminated, without altering the amino acid sequence. In a second step, four residues were mutated as follows: Leu-28 for Gln, Cys-31 and Cys-32 for Arg, and Lys-34 for Asn, by using the oligonucleotide 5′-CGACCTTGGGTCATTTTACGGCAGCACATCATCAGGTTGCAGTACGTA-3′ as a primer. After each mutation step, the coding region within each recombinant plasmid, obtained from transformed JM 101 cells, was fully sequenced (19). The 407-bp NdeI/HindIII fragment, containing the entire coding sequence, then was cloned in the same sites of the expression vector pET22b+. The resulting plasmid was used to transform E. coli BL21 (DE3) pLysS competent cells.
Protein Expression and Purification.
The recombinant protein was expressed by a single colony of freshly transformed cells, as described (20). The cell pellet from 1 liter of bacterial culture was resuspended in 40 ml of 0.1 M Tris·HCl, at pH 8, containing 5 mM Na2EDTA (sodium EDTA) and was incubated for 1.5 h at 4°C with 200 μg/ml lysozyme (Sigma). The inclusion bodies were isolated by centrifugation, followed by their resuspension by sonication in 20 ml of Tris buffer (see above), containing 2% Triton X-100 and 2 M urea. This procedure was repeated three times. The recombinant protein then was solubilized in 6 M guanidine-HCl in Tris buffer containing 0.1 M glutathione. After 2.5 h at room temperature, the sample was diluted with 20 volumes of 0.1 M Tris·acetate, at pH 8.4, containing 0.5 M l-arginine and was allowed to reoxidize at room temperature for 24 h in the presence of 1 mM oxidized glutathione. The protein then was concentrated by ultrafiltration to 30 ml, was dialyzed against 0.1 M Tris⋅HCl, at pH 8.4, containing 0.3 M NaCl, and was purified by gel-filtration chromatography on a HiLoad 26/60 Superdex 75 column (Amersham Pharmacia) equilibrated in the same buffer. More than 85% of the protein, named LCCK-HP-RNase, eluted as a monomer with mixed disulfides with glutathione moieties (12). Protein concentration was determined by amino acid analyses. The calculated extinction coefficient at 278 nm (1 mg/ml) was 0.57. The selective reduction of the mixed disulfides with a 10:1 molar excess of DTT, followed by dialysis to remove side products, allowed the spontaneous reoxidation of a dimeric protein. This was isolated by gel-filtration as described above and was purified further to homogeneity by RP-HPLC on a C4 column (Vydac, Hesperia, CA, 0.46 × 25 cm, 5-μm particles), equilibrated in 0.1% trifluoroacetic acid containing 15% acetonitrile. The protein was obtained by isocratic elution with 0.1% trifluoroacetic acid containing 24% acetonitrile at a flow rate of 1 ml/min. The removal of the Met−1 residue was performed by the use of Aeromonas proteolytica aminopeptidase (Sigma), in the conditions previously described (21), except for the presence in the incubation buffer of 0.3 M NaCl, necessary for the stability of the protein.
Cytotoxicity Assays.
Murine fibroblasts (BALB/c 3T3 cell line) and their counterpart obtained by transformation with SV40 (SVT2 cell line) were purchased from the American Type Culture Collection. The other cell lines were obtained as gifts or were prepared in the authors’ laboratories as described in the references. The cell lines used were from the following tumors, respectively: SJRH 30, rhabdosarcoma (22); UFK-NB-3, neuroblastoma (23); D-283, medulloblastoma (24); U-373, glioblastoma (25); Hep2, laryngeal carcinoma (26); PBF-M-1, melanoblastoma (8). TPC-1 (27) and NPA (28) cells were derived from two human papillary thyroid carcinomas. Both cell lines, as well as the diploid human fibroblast cell line [human diploid fibroblasts (HDF)] (29), were kindly provided by Alfredo Fusco (University of Catanzaro, Italy). SJRH 30, NPA, and TPC-1 cells were cultured in RPMI medium 1640; UFK-NB-3, D-283, and U-373 cells were cultured in Iscove’s Modified Dulbecco’s medium; the other cell lines were cultured in DMEM medium. Media were supplemented with 10% fetal calf serum (GIBCO/BRL), 50 units/ml penicillin, and 50 μg/ml streptomycin (Sigma). The assays on murine and human fibroblasts, and on thyroid human cell lines, were performed as described (6, 7). In brief, murine fibroblasts were plated in the presence or in the absence of the RNase under test, and growth curves and dose–response curves were determined by counting the cells that survived treatment at suitable time intervals. In the assays with human fibroblasts and thyroid tumor cells, the protein under test was added after 24 h of growth. Cell counts were determined in triplicate, and cells were routinely checked by the Trypan Blue exclusion test. In the assays with human tumor cell lines SRJH 30, UFK-NB-3, U 373, D-283, PBF-M-1, and Hep2, cells were seeded in 96-well plates (1 × 104/well in 100 μl of medium) and were incubated for 7 days at 37°C in the presence or absence of the RNase under test (0.4–200 μg/ml). Protein cytotoxicity was evaluated by the methyl tetrazolium bromide test (30). Dose–response curves were obtained by measuring in quadruplicate the percent of cell survival in the presence of the protein with respect to control cultures, grown in the absence of the protein.
For apoptotic analyses, 104 human malignant thyroid cells (NPA) or human normal fibroblasts (HDF) were plated in quadruplicate on different cover glasses inserted in separate wells of a 24-well plate; 24 h after plating, HHP-RNase (25 μg/ml) was added. After 72 h of treatment, the medium was removed, and the cells were fixed for 20 min in 3.7% formaldehyde and were washed and permeabilized for 5 min in 0.1% Triton. The cells then were washed with PBS and were stained for 30 min with Hoechst 33342 immunofluorescent reagent (Sigma) at a concentration of 0.5 μg/ml in PBS. The stained cells were observed under a Zeiss Axiophot microscope and were photographed.
Other Methods.
RNase activity on yeast RNA was assayed according to Kunitz (31). SDS/PAGE analyses were carried out as described (32). Protein sequence determinations were performed by using an Applied Biosystems sequencer, model 473A; amino acid analyses were done with a System Gold (Beckman) automated analyzer; and matrix-assisted laser desorption ionization mass spectra were done with a Voyager DE spectrometer (PerSeptive Biosystems, Framingham, MA).
RESULTS
Construction and Characterization of a Dimeric Mutant of HP-RNase.
The amino acid sequences of HP-RNase and BS-RNase are >70% identical, with 34 substitutions (Fig. 1). Four of these are located in a helical segment (helix-II, residues 24–34) that is connected to the N-terminal α-helix (residues 3–13) by a hinge loop (residues 16–22). In BS-RNase, the intersubunit interface is constituted by the helix-II segments of the two subunits (10). Hence, helix-II of HP-RNase was engineered to reproduce helix-II of BS-RNase by replacing the four different residues (Gln-28, Arg-31 and -32, and Asn-34) with those present at identical positions in BS-RNase. These included the two adjacent Cys residues for the two intersubunit disulfides and the Leu residue for a crucial hydrophobic interaction, both typical of the intersubunit interface of BS-RNase.
Figure 1.
Comparison of the amino acid sequence of human pancreas RNase (HP-RNase) with that of bovine seminal RNase (BS-RNase). For the latter, only the differences are indicated. Evidenced with a black background are the four BS-RNase residues grafted in the HP-RNase sequence to construct the dimeric mutant.
On the basis of these considerations, a synthetic cDNA (17), coding for HP-RNase and cloned in a pUC118 vector, was mutated as programmed and was expressed in E. coli as described in Materials and Methods. A molecular mass of ≈14 kDa was determined by SDS/PAGE for the recombinant protein. This was the major species in the bacterial cell extract prepared from cells transformed with the recombinant vector carrying the mutant HP-RNase cDNA (data not shown).
The mutant protein, sequestered in the inclusion bodies, was solubilized by a denaturing treatment and then was refolded in the presence of a glutathione/oxidized glutathione mixture. Gel-filtration chromatography, amino acid analyses, assays for the enzymatic activity, and SDS/PAGE analyses (see Fig. 2) indicated that the HP-RNase tetramutant (LCCK-HP-RNase) was a homogeneous, enzymatically active, monomeric derivative in which the two cysteines at positions 31 and 32 were linked through mixed disulfides with glutathione moieties.
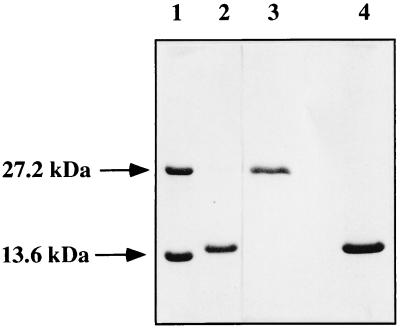
Figure 2.
SDS/PAGE electrophoresis on a 15% polyacrylamide gel of the monomeric and dimeric HP-RNase variants. Lanes: 1, dimeric BS-RNase and monomeric RNase A; 2, monomeric LCCK-HP-RNase; 3 and 4, dimeric HHP-RNase, analyzed under nonreducing (lane 3) and reducing (lane 4) conditions.
To obtain the dimeric form of the HP-RNase mutant, the mixed disulfides of the monomeric form were selectively reduced with DTT. The removal of the glutathione moieties allowed the protein to dimerize through the formation of intersubunit disulfide bridges between the exposed sulfydryl groups (20). The dimerized protein, indicated as HHP-RNase, was isolated by gel-filtration chromatography and was purified to homogeneity by RP-HPLC. The mass spectrometric analysis of the final product revealed that it was a homogeneous protein with a molecular mass of 28,716.7 Da, a value close to that expected for a dimeric HP-RNase (28,712 Da). The SDS/PAGE analysis under reducing conditions indicated that the protein migrated as a monomer, as expected for a dimer whose subunits are held together by intersubunit disulfides (Fig. 2). Its N-terminal sequence was that of wild-type HP-RNase, preceded by a methionine residue (Met−1), which was removed by treatment with an aminopeptidase (see Materials and Methods). When assayed on yeast RNA as a substrate, the specific activity of HHP-RNase was ≈20 Kunitz units/mg of protein, comparable to that of native wild-type HP-RNase.
The Selective Cytotoxic Action of HHP-RNase.
The cytotoxicity of HHP-RNase was tested on malignant SVT2 murine fibroblasts (BALB/c 3T3 cells transformed by SV40 virus). Growth curves were determined in the presence or in the absence of HHP-RNase, BS-RNase as a positive control, and HP-RNase as a negative control. Percent of cell survival was calculated from each curve obtained in the presence of the RNase being tested at 50 μg/ml, taking as 100% the cell survival in the absence of added RNases. Dimeric HHP-RNase was found to be a strong inhibitor of malignant fibroblast growth (see Fig. 3A). Although the effect was less pronounced than that of BS-RNase, it was highly selective for malignant cells, as the protein was found to be thoroughly inactive toward the parent nonmalignant line of 3T3 fibroblasts tested in the same experiment (Fig. 3A). Wild-type monomeric HP-RNase had no effect on the growth of any of the cell lines tested (data not shown).
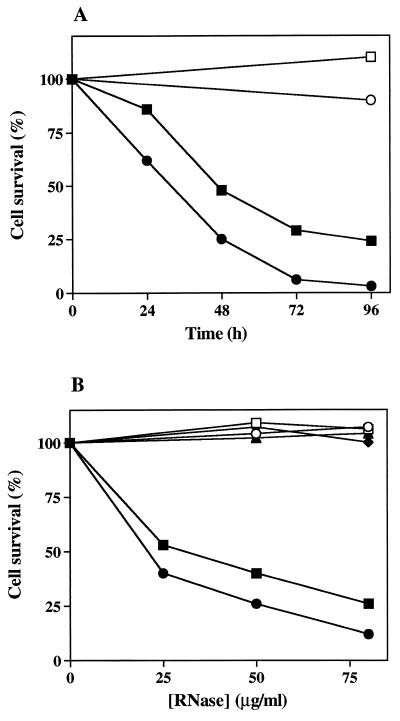
Figure 3.
Effects of dimeric HHP-RNase on murine fibroblasts. Malignant SVT2 fibroblasts (closed symbols) or normal 3T3 fibroblasts (open symbols) were grown in the absence or in the presence of HHP-RNase (squares), BS-RNase (circles), monomeric LCCK-HP-RNase (triangles), or wild-type HP-RNase (rhombs). Cell survival values (means of triplicates) are expressed as percent of the corresponding values obtained in control cultures grown in the absence of protein. (A) Cell survival in the presence or in the absence of 50 μg/ml RNase. (B) Dose–response curves after 48 h of incubation.
These results were confirmed by dose–response curves, shown in Fig. 3B. HHP-RNase, as well as BS-RNase, was strongly and selectively cytotoxic for malignant fibroblasts, incubated 48 h with increasing amounts of protein, whereas wild-type HP-RNase and monomeric LCCK-HP-RNase were ineffective. The IC50 value calculated for HHP-RNase, and defined as the protein concentration corresponding to 50% of cell growth inhibition, was 30 μg/ml, comparable to that measured for BS-RNase (21 μg/ml).
The N-terminal Lys-1, as in the native protein, was essential for the antitumor action of HHP-RNase, as the recombinant protein was found to be inactive when tested before treatment with aminopeptidase to remove Met−1 (data not shown). Also, the dimeric structure was found to be essential for the cytotoxic activity of the RNase, as monomeric LCCK-HP-RNase, even after removal of its Met−1 residue, did not affect cell survival (Fig. 3B).
An investigation on the nature of the adverse effects of HHP-RNase on malignant cell cultures was carried out with the sensitive 3T3-SVT2 cell line. By the Trypan Blue exclusion test, it was found that the effect of HHP-RNase on malignant cells was cytotoxic rather than cytostatic. Only ≈30% of the originally plated cells were recovered as viable cells after 72 h of culture in the presence of 20 μg/ml of HHP-RNase. The number of viable cells decreased to 10% of the plated cells when the concentration of the RNase was increased to 100 μg/ml.
The effects of HHP-RNase also were tested on a series of human tumor cells, derived from tumors of different origin, by determining dose–response curves in which the effect of HHP-RNase (0.4–200 μg/ml) was measured on cell survival. The results, expressed as IC50 values, are summarized in Table 1 and compared with the values obtained with seminal RNase tested under the same conditions on the same cell lines.
Table 1.
Cytotoxic effect of dimeric HHP-RNase on human tumor cells
| Tumor | Cell line | IC50, μg/ml
|
|
|---|---|---|---|
| HHP-RNase | BS-RNase | ||
| Rhabdomyosarcoma | SJRH 30 | 2.0 | 2.3 |
| Neuroblastoma | UFK-NB-3 | 2.6 | 0.4 |
| Medulloblastoma | D-283 | 17.9 | 14.3 |
| Glioblastoma | U 373 | 33.6 | 7.1 |
| Melanoblastoma | PBF-M-1 | 38.9 | 54.8 |
| Laryngeal carcinoma | Hep2 | 85.7 | 35.7 |
The IC50 values represent the protein concentrations that produce 50% of the total cytotoxic effect, measured for each cell line in a dose–response curve with a range of protein concentration of 0.4–200 μg/ml.
The results obtained indicate that the stably dimeric derivative of human RNase is highly cytotoxic for all of the tumor cells tested, although with varying degrees of cytotoxicity. Comparison with the antitumor activity of BS-RNase revealed that dimeric HHP-RNase is as effective as BS-RNase on certain cell lines (from rhabdomyosarcoma and medulloblastoma), even more active on melanoblastoma cells, and less active on others (from neuroblastoma, glioblastoma, and laryngeal carcinoma). RNase A and monomeric wild-type HP-RNase, tested in the same experiments, had no effects on any of the tumor cell lines tested (data not shown).
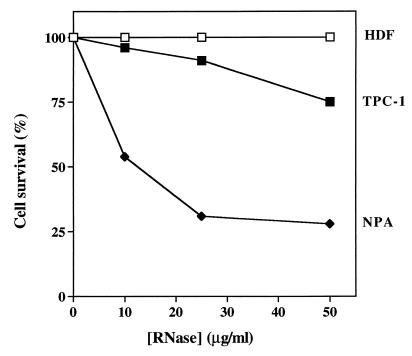
The selectivity for malignant cells of the cytotoxic action of HHP-RNase was tested also for human cells by using normal fibroblasts and thyroid tumor cells. The protein had no effect on normal human diploid fibroblasts (see Fig. 4) but had an evident cytotoxic effect on cells derived from thyroid papillary carcinomas. Moreover, there was a clear correlation between the degree of malignancy of a cell line and its sensitivity to the cytotoxic effect of HHP-RNase. TPC-1 and NPA cells, both of which are derived from human papillary thyroid carcinomas, behave differently, in that the TPC-1 cell line displays a rather benign phenotype compared with the NPA cell line (33). We found that TPC-1 cells were only mildly affected by HHP-RNase whereas NPA cells were killed even by low concentrations of the protein.
Figure 4.
Effects of dimeric HHP-RNase on normal HDFs (open squares), human papillary thyroid carcinoma cells TPC-1 (closed squares), and NPA (closed rhombs). Cell survival values, expressed as in Fig. 3, were calculated after 72 h of treatment.
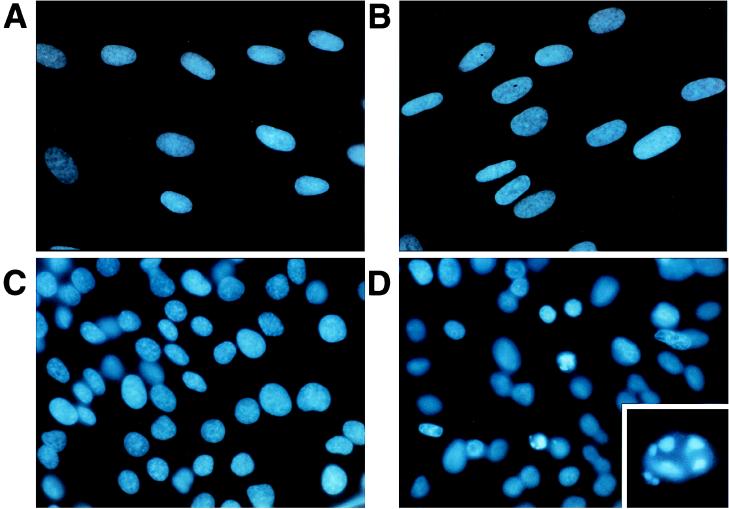
To inspect the nature of cell death induced by HHP-RNase on tumor cells, both a malignant and a normal cell line were treated with the protein and were analyzed for apoptosis. No differences were appreciated for HDFs between untreated or treated (for 72 h with 25 μg/ml of HHP-RNase) cells, thus confirming that the enzyme has no detectable effect on normal cells (see Fig. 5 A and B). On the contrary, a clear induction of apoptosis by HHP-RNase was appreciable in treated thyroid tumor NPA cells, with nuclei condensation and fragmentation (Fig. 5D) whereas no signs of apoptosis were detectable in untreated cells (Fig. 5C). In the inset of Fig. 5D, a magnification of apoptotic bodies is shown. A quantitative estimate of the extent of apoptosis gave values ranging from 20 to 27% of apoptotic cells after 72 h of culture in the presence of 25 μg/ml of HHP-RNase. A similar degree of apoptotic cell death was observed for NPA cells treated with HHP-RNase under the same conditions when different techniques such as DNA laddering or flow cytometric analyses were used (data not shown).
Figure 5.
Tumor cells sensitized to apoptosis by HHP-RNase. HDFs and human thyroid tumor (NPA) cells were grown, were exposed for 72 h to HHP-RNase (25 μg/ml), and were fixed and stained with Hoechst immunofluorescent reagent. (A) Untreated HDF cells. (B) HDF cells exposed to HHP-RNase. (C) Untreated NPA cells. (D) NPA cells exposed to HHP-RNase. The inset shows a fragmented and condensed apoptotic nucleus from an NPA treated cell. (A–D, ×360; D Inset, ×2,400.)
DISCUSSION
The results reported here indicate that the selective cytotoxicity of BS-RNase toward malignant cells can be transferred to a monomeric, noncytotoxic RNase of human origin by converting monomeric human pancreatic RNase into a dimeric enzyme. This was achieved through the substitution of a few of its residues with the corresponding key residues of BS-RNase, a homologous dimeric antitumor protein (12).
The residues of BS-RNase found to be determinant for its dimeric structure—hence, its selective cytotoxicity—are Pro-19, Leu-28, and Cys-31 and Cys-32 (11, 12, 34). They are respectively responsible for (i) determining the correct conformations in the peptide loop connecting the N-terminal α-helix to the main subunit body (10, 35); (ii) a hydrophobic interaction between the two subunits; and (iii) the formation of the two intersubunit disulfides.
Because a proline residue is already present at position 19 of wild-type HP-RNase, the engineering of the mutant was confined to the putative intesubunit interface of the programmed dimer. In this region, the HP-RNase sequence differs from that of BS-RNase at four positions (see Fig. 1). Three (Gln-28, and Arg-31 and -32) were mutated to insert the structural determinants indicated above as essential for the antitumor action of BS-RNase. The fourth mutation (a Lys for Asn-34) was inserted to endow the mutant with an interface identical with that of BS-RNase and to increase the positive net charge at a position adjacent to the intersubunit disulfides. This has been shown to increase reactivity of adjacent disulfides (36).
The effects on cell growth of HHP-RNase first were tested by an assay system with a built-in control: i.e., the same cell line before and after malignant transformation. The dimer exerted a severe cytotoxic action on malignant cells and had no effects on normal cells. The selectivity of HHP-RNase cytotoxic action was also evident when normal human fibroblasts, and cell lines derived from thyroid tumors with different degrees of malignancy, were tested. When the nature of cell death was tested on the most responsive cell line (NPA), evident symptoms of apoptotic cell death were evidenced whereas for normal fibroblasts, no effects were detected. Furthermore, a series of malignant cell lines derived from human tumors of different histogenesis were all inhibited in their growth by HHP-RNase, although with varying degrees of sensitivity.
Two structural requirements appear to be crucial for the antitumor action of dimeric human RNase: its dimeric structure and the availability of its original N terminus. In fact, (i) monomeric wild-type HP-RNase, as well as the monomeric mutant LCCK-HP-RNase, which carries two molecules of glutathione linked at Cys-31 and -32, were totally ineffective on tumor cells, and (ii) the removal of the methionine residue at position −1 was found to be essential for HHP-RNase antitumor action. It should be noted that also recombinant onconase has no antitumor activity when the N terminus is not the original pyroglutamate (37). These results may suggest that the N termini of these antitumor RNases are involved in the molecular interactions that eventually lead to cell death.
The molecular mechanistic bases are not clear for the cytotoxic action of HHP-RNase nor for its selective action on tumor cells. But then those for the very similar antitumor action of BS-RNase still remain unclear, even after intense scrutiny by different laboratories (5, 7, 9, 38). Here, we report that the action of an antitumor RNase on malignant cells is based on induction of apoptosis. This finding may help to design new experimental approaches to elucidate the mechanism of antitumor action of dimeric RNases.
This report on the construction from a human protein of a cytotoxic agent selective for malignant cells may have a special interest. The only RNase to have entered clinical trials so far with cancer patients is Onconase, an RNase from amphibia (3). Onconase has been found to be well tolerated by Phase I patients, compared with most chemotherapeutic agents (39). However, we may presume that a human protein should be decisively less toxic, and much less immunogenic, if at all, compared with an amphibian protein.
Acknowledgments
The authors thank Drs. Giancarlo Vecchio, Alberto Di Donato, and Valeria Cafaro for precious advise and helpful discussions, Dr. Piero Pucci for mass spectrometric analyses, Dr. Antimo Di Maro for protein sequence determinations, and Dr. Corrado Garbi for immunofluorescence analyses. Dr. Cinátl acknowledges Hilfe für krebskranke Kinder, Frankfurt am Main, and Frankfurter Stiftung für krebskranke Kinder. This work was financed by grants from the European Union (INCO-Copernicus, contract no. IC15CT 960903), Associazione Italiana per la Ricerca sul Cancro, Ministero dell’Università e della Ricerca Scientifica e Tecnologica, and Consiglio Nazionale delle Ricerche, Italy. Dr. Spalletti-Cernia was supported by a fellowship from Federazione Italiana per la Ricerca sul Cancro, Italy.
ABBREVIATIONS
- BS-RNase
bull semen RNase
- HP-RNase
human pancreas RNase
- HDF
human diploid fibroblasts
References
- 1.D’Alessio G. Trends Cell Biol. 1993;3:106–109. doi: 10.1016/0962-8924(93)90166-x. [DOI] [PubMed] [Google Scholar]
- 2.Youle R J, D’Alessio G. In: Ribonucleases: Structures and Functions. D’Alessio G, Riordan J F, editors. San Diego: Academic; 1997. pp. 491–514. [Google Scholar]
- 3.Juan G, Ardelt B, Li X, Mikulski S M, Shogen K, Ardelt W, Mittelman A, Darzynkiewicz Z. Leukemia. 1998;12:1241–1248. doi: 10.1038/sj.leu.2401100. [DOI] [PubMed] [Google Scholar]
- 4.Cinátl J, Matoušek J, Stanek R. Folia Biol (Prague) 1977;23:235–242. [PubMed] [Google Scholar]
- 5.Vescia S, Tramontano D, Augusti Tocco G, D’Alessio G. Cancer Res. 1980;40:3740–3744. [PubMed] [Google Scholar]
- 6.Laccetti P, Portella G, Mastronicola M R, Russo A, Piccoli R, D’Alessio G, Vecchio G. Cancer Res. 1992;52:4582–4586. [PubMed] [Google Scholar]
- 7.Mastronicola M R, Piccoli R, D’Alessio G. Eur J Biochem. 1995;230:242–249. doi: 10.1111/j.1432-1033.1995.tb20557.x. [DOI] [PubMed] [Google Scholar]
- 8.Poucková P, Soucek J, Jelínek J, Zadinová M, Hlous̆ková D, Polívková J, Navrátil L, Cinátl J, Matoušek J. Neoplasma. 1998;45:30–34. [PubMed] [Google Scholar]
- 9.Wu Y, Saxena S K, Ardelt W, Gadina M, Mikulski S M, De Lorenzo C, D’Alessio G, Youle R J. J Biol Chem. 1995;270:17476–17481. doi: 10.1074/jbc.270.29.17476. [DOI] [PubMed] [Google Scholar]
- 10.Mazzarella L, Capasso S, Demasi D, Di Lorenzo G, Mattia C A, Zagari A. Acta Crystallogr D. 1993;49:389–402. doi: 10.1107/S0907444993003403. [DOI] [PubMed] [Google Scholar]
- 11.Di Donato A, Cafaro V, D’Alessio G. J Biol Chem. 1994;269:17394–17396. [PubMed] [Google Scholar]
- 12.Di Donato A, Cafaro V, Romeo I, D’Alessio G. Protein Sci. 1995;4:1470–1477. doi: 10.1002/pro.5560040804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weickmann J L, Elson M, Glitz D G. Biochemistry. 1981;20:1272–1278. doi: 10.1021/bi00508a035. [DOI] [PubMed] [Google Scholar]
- 14.Beintema J J, Wietzes P, Weickmann J L, Glitz D G. Anal Biochem. 1984;136:48–64. doi: 10.1016/0003-2697(84)90306-3. [DOI] [PubMed] [Google Scholar]
- 15.Tamburrini M, Piccoli R, De Prisco R, Di Donato A, D’Alessio G. Ital J Biochem (Engl Ed) 1986;35:22–32. [PubMed] [Google Scholar]
- 16.Dostál J, Matoušek J. J Reprod Fertil. 1973;33:263–274. doi: 10.1530/jrf.0.0330263. [DOI] [PubMed] [Google Scholar]
- 17.Russo N, de Nigris M, Ciardiello A, Di Donato A, D’Alessio G. FEBS Lett. 1993;333:233–237. doi: 10.1016/0014-5793(93)80660-m. , and erratum (1995) 369, 352. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;76:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Nigris M, Russo N, Piccoli R, D’Alessio G, Di Donato A. Biochem Biophys Res Commun. 1993;193:155–160. doi: 10.1006/bbrc.1993.1603. [DOI] [PubMed] [Google Scholar]
- 21.Adinolfi B S, Cafaro V, D’Alessio G, Di Donato A. Biochem Biophys Res Commun. 1995;213:525–532. doi: 10.1006/bbrc.1995.2163. [DOI] [PubMed] [Google Scholar]
- 22.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Nature (London) 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 23.Cinátl J, Jr, Cinátl J, Mainke M, Weissflog A, Rabenau H, Kornhuber B, Doerr H W. Cancer Lett. 1993;70:15–24. doi: 10.1016/0304-3835(93)90069-l. [DOI] [PubMed] [Google Scholar]
- 24.Friedman H S, Burger P C, Bigner S H, Trojanowski J Q, Wikstrand C J, Halperin E C, Bigner D D. J Neuropathol Exp Neurol. 1985;44:592–605. doi: 10.1097/00005072-198511000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Khatib Z A, Matsushime H, Valentine M, Shapiro D N, Sherr C J, Look A T. Cancer Res. 1995;53:5535–5541. [PubMed] [Google Scholar]
- 26.Moore L, Sabachewsky L, Toolan H W. Cancer Res. 1955;15:598–602. [PubMed] [Google Scholar]
- 27.Tanaka J, Ogura T, Sato H, Hatano M. Virology. 1987;161:62–72. doi: 10.1016/0042-6822(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 28.Pang X J, Hershman M, Chung M, Pekary A E. Endocrinology. 1989;125:1783–1788. doi: 10.1210/endo-125-4-1783. [DOI] [PubMed] [Google Scholar]
- 29.Kantor G J, Mulkie J R, Hull D R. Exp Cell Res. 1978;113:283–294. doi: 10.1016/0014-4827(78)90368-3. [DOI] [PubMed] [Google Scholar]
- 30.Alley M C, Scudiero D A, Monks A, Hursey M L, Czerwinski M J, Fine D L, Abbott B J, Mayo J G, Shoemacker R H, Boyd M R. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 31.Kunitz M. J Biol Chem. 1946;164:563–568. [PubMed] [Google Scholar]
- 32.Laemmli U. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Cerrutti J, Trapasso F, Battaglia C, Zhang L, Martelli M L, Visconti R, Berlingieri M T, Fagin J A, Santoro M, Fusco A. Clin Cancer Res. 1996;2:119–126. [PubMed] [Google Scholar]
- 34.Ciglic M I, Jackson P J, Raillard S A, Haugg M, Jermann T M, Opitz J G, Trabesinger-Ruf N, Benner S A. Biochemistry. 1998;37:4008–4022. doi: 10.1021/bi972203e. [DOI] [PubMed] [Google Scholar]
- 35.Mazzarella L, Vitagliano L, Zagari A. Proc Natl Acad Sci USA. 1995;92:3799–3803. doi: 10.1073/pnas.92.9.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parente A, Merrifield B, Geraci G, D’Alessio G. Biochemistry. 1985;24:1098–1104. doi: 10.1021/bi00326a005. [DOI] [PubMed] [Google Scholar]
- 37.Boix E, Wu Y, Vasadani V M, Saxena S, Ardelt W, Ladner J, Youle R J. J Mol Biol. 1996;258:1–16. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 38.Kim J-S, Soucek J, Matoušek J, Raines R T. J Biol Chem. 1995;270:31097–31102. doi: 10.1074/jbc.270.52.31097. [DOI] [PubMed] [Google Scholar]
- 39.Mikulski S M, Grossman A M, Carter P W, Shogen K, Costanzi P W. Int J Oncol. 1993;3:57–64. doi: 10.3892/ijo.3.1.57. [DOI] [PubMed] [Google Scholar]