Abstract
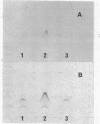
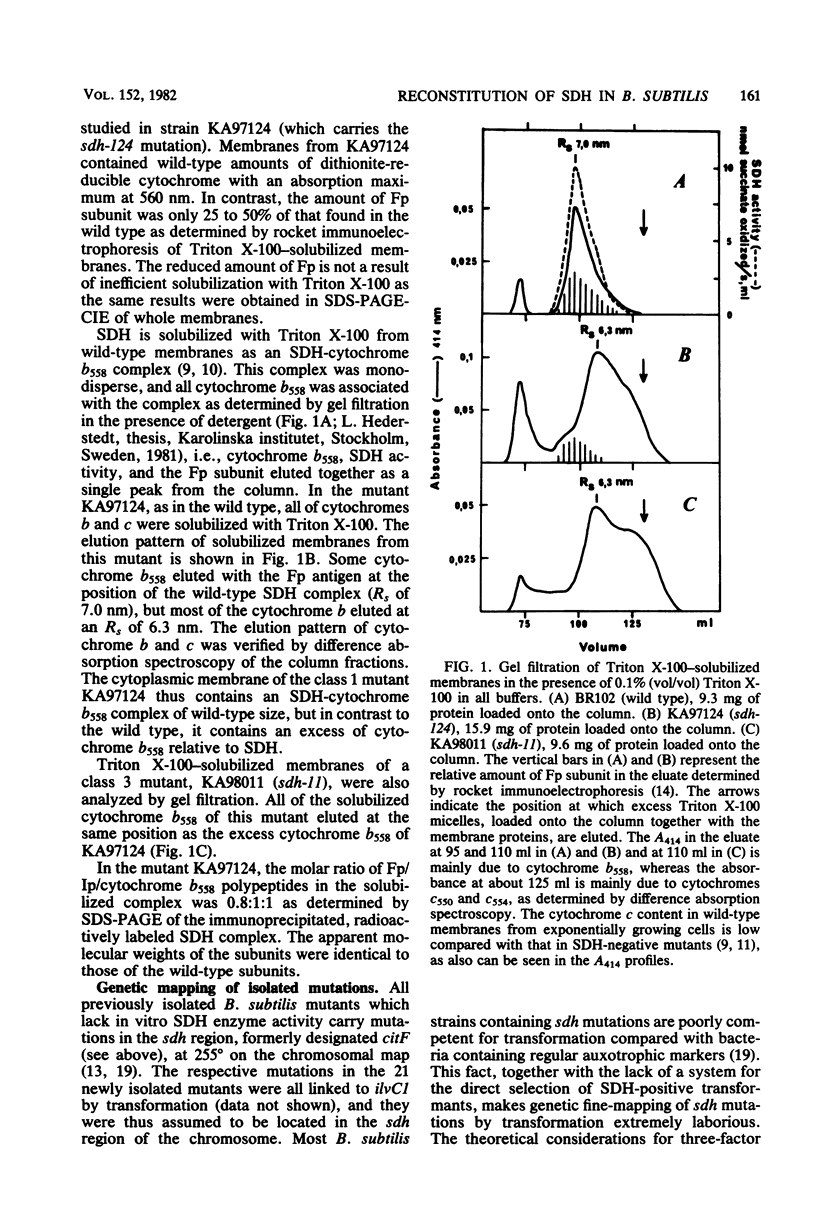
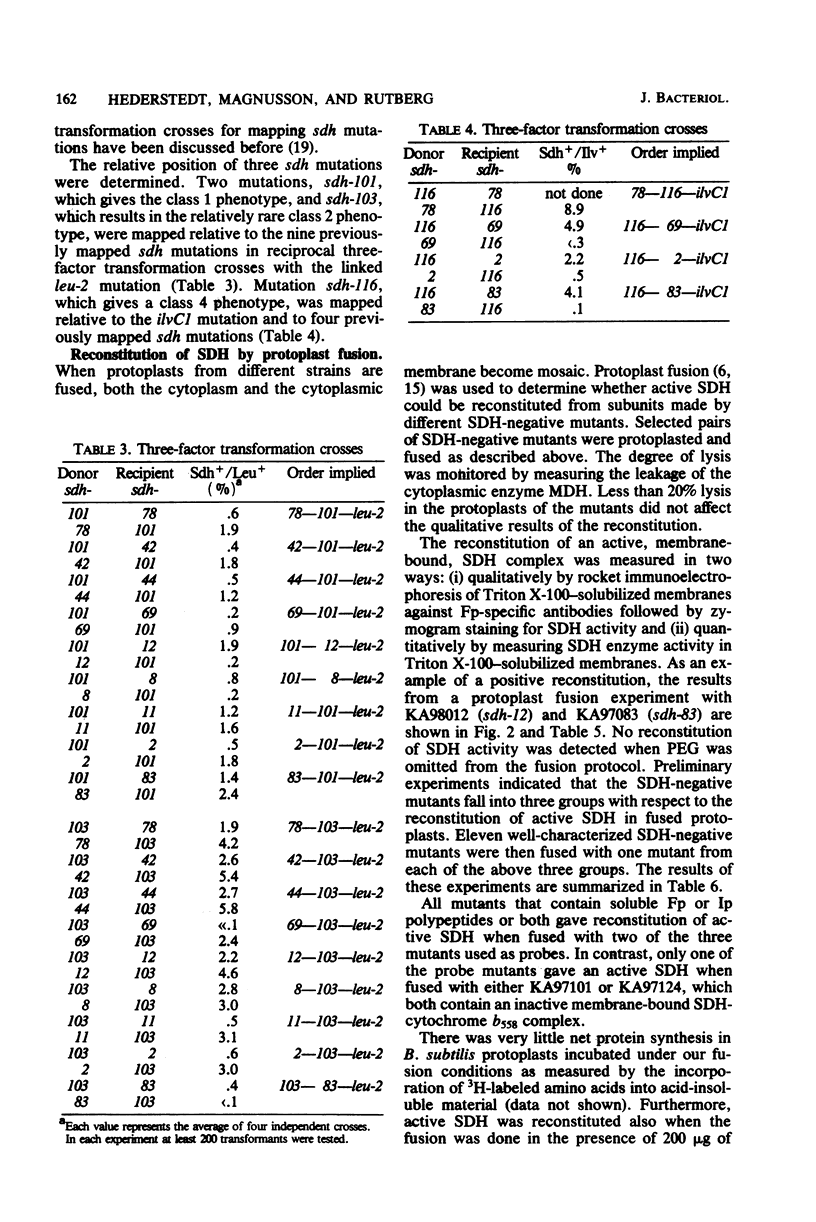
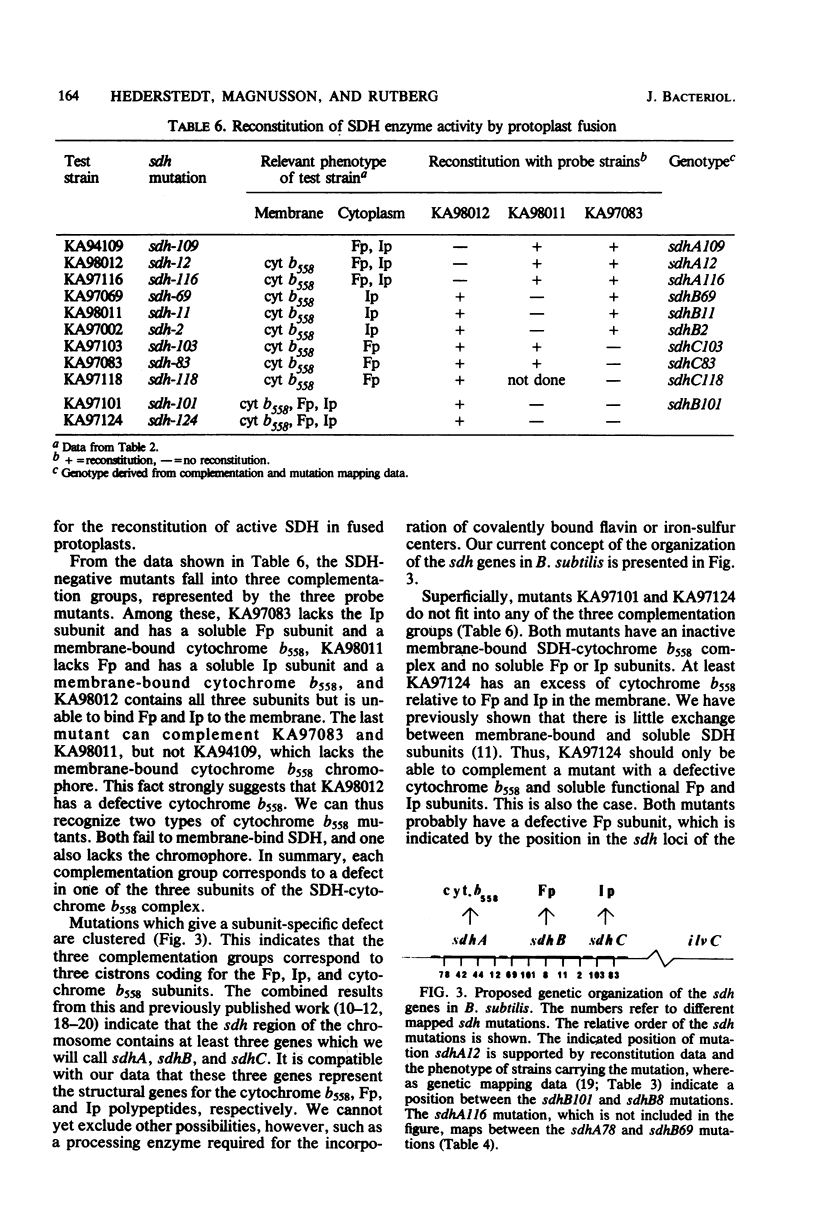
Bacillus subtilis succinate dehydrogenase (SDH) is composed of two unequal subunits designated Fp (Mr, 65,000) and Ip (Mr. 28,000). The enzyme is structurally and functionally complexed to cytochrome b 558 (Mr, 19,000) in the membrane. A total of 21 B. subtilis SDH-negative mutants were isolated. The mutants fall into five phenotypic classes with respect to the presence and localization of the subunits of the SDH-cytochrome b558 complex. One class contains mutants with an inactive membrane-bound complex. Membrane-bound enzymatically active SDH could be reconstituted in fused protoplasts of selected pairs of SDH-negative mutants. Most likely reconstitution is due to the assembly of preformed subunits in the fused cells. On the basis of the reconstitution data, the mutants tested could be divided into three complementation groups. The combined data of the present and previous work indicate that the complementation groups correspond to the structural genes for the three subunits of the membrane-bound SDH-cytochrome b558 complex. A total of 31 SDH-negative mutants of B. subtilis have now been characterized. The respective mutations all map in the citF locus at 255 degrees on the B. subtilis chromosomal map. In the present paper, we have revised the nomenclature for the genetics of SDH in B. subtilis. All mutations which give an SDH-negative phenotype will be called sdh followed by an isolation number. The designation citF will be omitted, and the citF locus will be divided into three genes: sdhA, sdhB, and sdhC. Mutations in sdhA affect cytochrome b558, mutations in sdhB affect Fp, and mutations in sdhC affect Ip.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carls R. A., Hanson R. S. Isolation and characterization of tricarboxylic acid cycle mutants of Bacillus subtilis. J Bacteriol. 1971 Jun;106(3):848–855. doi: 10.1128/jb.106.3.848-855.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966 Jul;54(1):61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D. L., Belsole D. M., Guzik H. J., Unger B. W. Selection of succinic dehydrogenase mutants of Neurospora crassa. J Bacteriol. 1979 Feb;137(2):900–904. doi: 10.1128/jb.137.2.900-904.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortnagel P., Freese E. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol. 1968 Apr;95(4):1431–1438. doi: 10.1128/jb.95.4.1431-1438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frehel C., Lheritier A. M., Sanchez-Rivas C., Schaeffer P. Electron microscopic study of Bacillus subtilis protoplast fusion. J Bacteriol. 1979 Mar;137(3):1354–1361. doi: 10.1128/jb.137.3.1354-1361.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstedt L. Cytochrome b reducible by succinate in an isolated succinate dehydrogenase-cytochrome b complex from Bacillus subtilis membranes. J Bacteriol. 1980 Dec;144(3):933–940. doi: 10.1128/jb.144.3.933-940.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstedt L., Holmgren E., Rutberg L. Characterization of a succinate dehydrogenase complex solubilized from the cytoplasmic membrane of Bacillus subtilis with the nonionic detergent Triton X-100. J Bacteriol. 1979 May;138(2):370–376. doi: 10.1128/jb.138.2.370-376.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstedt L., Rutberg L. Biosynthesis and membrane binding of succinate dehydrogenase in Bacillus subtilis. J Bacteriol. 1980 Dec;144(3):941–951. doi: 10.1128/jb.144.3.941-951.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstedt L., Rutberg L. Succinate dehydrogenase--a comparative review. Microbiol Rev. 1981 Dec;45(4):542–555. doi: 10.1128/mr.45.4.542-555.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Hoch J. A. The Bacillus subtilis chromosome. Microbiol Rev. 1980 Mar;44(1):57–82. doi: 10.1128/mr.44.1.57-82.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren E., Hederstedt L., Rutberg L. Role of heme in synthesis and membrane binding of succinic dehydrogenase in Bacillus subtilis. J Bacteriol. 1979 May;138(2):377–382. doi: 10.1128/jb.138.2.377-382.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A. Genetic studies with bacterial protoplasts. Annu Rev Microbiol. 1981;35:237–272. doi: 10.1146/annurev.mi.35.100181.001321. [DOI] [PubMed] [Google Scholar]
- Hotchkiss R. D., Gabor M. H. Biparental products of bacterial protoplast fusion showing unequal parental chromosome expression. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3553–3557. doi: 10.1073/pnas.77.6.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Spizizen J. Increased rate of asporogenous mutations following treatment of Bacillus subtilis spores with ethyl methanesulfonate. Mutat Res. 1971 Sep;13(1):93–96. doi: 10.1016/0027-5107(71)90130-8. [DOI] [PubMed] [Google Scholar]
- Ohné M. Regulation of the dicarboxylic acid part of the citric acid cycle in Bacillus subtilis. J Bacteriol. 1975 Apr;122(1):224–234. doi: 10.1128/jb.122.1.224-234.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohné M., Rutberg B., Hoch J. A. Genetic and biochemical characterization of mutants of Bacillus subtilis defective in succinate dehydrogenase. J Bacteriol. 1973 Sep;115(3):738–745. doi: 10.1128/jb.115.3.738-745.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg B., Hederstedt L., Holmgren E., Rutberg L. Characterization of succinic dehydrogenase mutants of Bacillus subtilis by crossed immunoelectrophoresis. J Bacteriol. 1978 Oct;136(1):304–311. doi: 10.1128/jb.136.1.304-311.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg B., Hoch J. A. Citric acid cycle: gene-enzyme relationships in Bacillus subtilis. J Bacteriol. 1970 Nov;104(2):826–833. doi: 10.1128/jb.104.2.826-833.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rivas C., Garro A. J. Bacterial fusion assayed by a prophage complementation test. J Bacteriol. 1979 Mar;137(3):1340–1345. doi: 10.1128/jb.137.3.1340-1345.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Cami B., Hotchkiss R. D. Fusion of bacterial protoplasts. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2151–2155. doi: 10.1073/pnas.73.6.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Hotchkiss R. D. Fusion of bacterial protoplasts. Methods Cell Biol. 1978;20:149–158. doi: 10.1016/s0091-679x(08)62017-8. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unden G., Hackenberg H., Kröger A. Isolation and functional aspects of the fumarate reductase involved in the phosphorylative electron transport of Vibrio succinogenes. Biochim Biophys Acta. 1980 Jul 8;591(2):275–288. doi: 10.1016/0005-2728(80)90159-0. [DOI] [PubMed] [Google Scholar]
- Weiss H., Kolb H. J. Isolation of mitochondrial succinate: ubiquinone reductase, cytochrome c reductase and cytochrome c oxidase from Neurospora crassa using nonionic detergent. Eur J Biochem. 1979 Aug 15;99(1):139–149. doi: 10.1111/j.1432-1033.1979.tb13240.x. [DOI] [PubMed] [Google Scholar]
- YOSHIDA A. PURIFICATION AND CHEMICAL CHARACTERIZATION OF MALATE DEHYDROGENASE OF BACILLUS SUBTILIS. J Biol Chem. 1965 Mar;240:1113–1117. [PubMed] [Google Scholar]