Abstract
The structure of the subunit c oligomer of the H+-transporting ATP synthase of Escherichia coli has been modeled by molecular dynamics and energy minimization calculations from the solution structure of monomeric subunit c and 21 intersubunit distance constraints derived from cross-linking of subunits. Subunit c folds in a hairpin-like structure with two transmembrane helices. In the c12 oligomer model, the subunits pack to form a compact hollow cylinder with an outer diameter of 55–60 Å and an inner space with a minimal diameter of 11–12 Å. Phospholipids are presumed to pack in the inner space in the native membrane. The transmembrane helices pack in two concentric rings with helix 1 inside and helix 2 outside. The calculations strongly favor this structure versus a model with helix 2 inside and helix 1 outside. Asp-61, the H+-transporting residue, packs toward the center of the four transmembrane helices of two interacting subunits. From this position at the front face of one subunit, the Asp-61 carboxylate lies proximal to side chains of Ala-24, Ile-28, and Ala-62, projecting from the back face of a second subunit. These interactions were predicted from previous mutational analyses. The packing supports the suggestion that a c–c dimer is the functional unit. The positioning of the Asp-61 carboxyl in the center of the interacting transmembrane helices, rather than at the periphery of the cylinder, has important implications regarding possible mechanisms of H+-transport-driven rotation of the c oligomer during ATP synthesis.
H+-transporting F1Fo ATP synthases use the energy of a transmembrane electrochemical H+ gradient to catalyze formation of ATP. Closely related enzymes are found in the plasma membrane of eubacteria, the inner membrane of mitochondria, and the thylakoid membrane of chloroplasts (1). The enzyme is composed of distinct extramembranous and transmembrane sectors, termed F1 and Fo, respectively. Proton movement through Fo is reversibly coupled to ATP synthesis or hydrolysis in catalytic sites of F1. Each sector of the enzyme is composed of multiple subunits with the simplest composition being α3β3γδɛ for F1 and a1b2c12 for Fo in the Escherichia coli enzyme (2). Homologous subunits are found in mitochondria and chloroplasts. An atomic resolution x-ray structure of the α3β3γ portion of bovine F1 shows the three α and three β subunits alternating around a centrally located γ subunit, with the γ subunit interacting asymmetrically with the three β catalytic subunits (3). Subunit γ was subsequently shown to rotate with respect to the three β subunits during catalysis (4–6). Rotation of γ is thought to change the binding affinities in alternating catalytic sites to promote tight substrate binding and product release during catalysis (7). During ATP synthesis, the rotation of γ must be driven by proton translocation through Fo. The structure of Fo remains to be determined. Electron and atomic force microscopic studies suggest that the a and b subunits pack at the periphery of a ring of multiple subunit c (8–10). The subunit c oligomeric ring is proposed to rotate with respect to a static a1b2 subcomplex to drive rotation of subunit γ in F1 (4, 11–13). Proton binding and release from a conserved carboxylate side chain in the center of the membrane (Asp-61 in the case of E. coli subunit c) is proposed to drive rotation of the c oligomer.
A high-resolution NMR structure for monomeric subunit c was recently determined (14). The structural model shows that subunit c folds as a hairpin of two transmembrane α-helices with interacting residues in the two helices located at positions predicted from genetic and chemical studies of Fo in situ (14). Based on the positions of functionally interacting residues of the proton binding site, the protein was proposed to associate in the oligomer with the flattened “front” face of one subunit packed against the flattened “back” face of a second subunit to form a functional dimer. Such packing would maximize intersubunit contact. The prediction of front-to-back packing of subunit c in Fo was verified by an extensive cross-linking analysis with singly and doubly Cys-substituted variants of subunit c (15). The position of cross-linkable residues fit the packing predicted by the NMR model and provided support for the applicability of the model to the structural analysis of oligomeric subunit c in Fo. Several doubly Cys-substituted mutants formed extensive multimeric ladders on cross-linking with multimeric products extending to c10 and quite probably c12. The number of subunit c in the Fo ring could not be established with exactness from previous studies due to experimental uncertainties but is now proposed to be precisely 12 (16). Genetically fused dimers and trimers of E. coli subunit c are functional, indicating that the final stoichiometry must be a multiple of 2 and 3. On introduction of Cys at specific positions in the first and last helices of monomeric, dimeric, and trimeric subunit c, disulfide cross-linking led to multimers that maximized at the position equivalent to c12 for all three species (16).
In sum, the studies above indicate that the c subunits in the Fo oligomer interact with the front face of one subunit packed against the back face of the next. The oligomer is packed so that the α-helical segments form two concentric rings with the N- and C-terminal helices located to the inner and outer ring, respectively. In this paper, we have used the NMR model of monomeric subunit c and distance restraints from the aforementioned cross-linking studies to model the oligomeric ring by molecular dynamics and energy minimization. In the molecular model of the c oligomer, the side chain of Asp-61 packs toward the center of the four transmembrane helices of two interacting subunit c in a position shielded from the lipid environment that is expected to surround the surface of the cylinder. The important functional implications of this placement of Asp-61 to the mechanism of rotary motion are considered.
MATERIALS AND METHODS
Rationale.
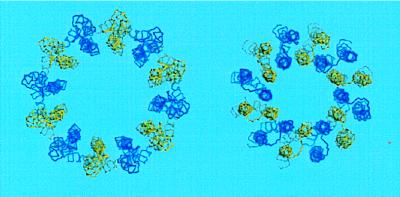
Based on the structural considerations described in the Introduction, the input model for calculation of structure was a ring of 12 c subunits positioned with the N-terminal helices closer to the central axis and the C-terminal helices on the outside. The distance between the center of mass of each monomer and the central axis was varied over a range of 20–40 Å, the distance not proving to be critical, 25 Å being used in the final simulation (Fig. 1). The monomeric structure used was the lowest energy model from the ensemble deposited at Protein Data Bank as 1A91 (model 1). The defined elements of secondary structure in the NMR model are likely to remain unperturbed in the oligomer in that the lipid environment should stabilize the hydrogen bonding characteristic of an α-helix. On the other hand, the NMR-derived distance and angle restraints allow for significant variation in the overall bending and crossing angle of the α-helices. Accordingly, two kinds of intramolecular restraints were used in the modeling calculations made in this paper: (i) Relatively strong distance restraints were imposed to maintain proper geometry for (i, i + 4) hydrogen bonding. The distances between the carbonyl oxygen of residue i and backbone nitrogen or amide proton of residue i+4 were restrained to a range of 2.7–3.2 Å and 1.8–2.3 Å, respectively, for the segments of α-helix i = residues 3–35, i = residues 47–59, and i = residues 61–74. (ii) Backbone angles were more weakly restrained to their values in the NMR structure. This created a bias in the folding of the monomeric units in the model toward the solution structure but permitted variation in the bending and packing of helices.
Figure 1.
Backbone trace of input model (Left) and final model of subunit c oligomer (Right). Blue and yellow colors distinguish individual monomers.
Intermolecular distance restraints were derived from the extensive data on Cys–Cys cross-linking (15). Restraints were imposed on the distances between the α carbons of the cross-linkable Cys–Cys pairs based on the distances observed between the α carbons of Cys residues forming disulfide bridges in natural proteins, the distance ranging from 4 to 7.5 Å (17). A systematic procedure was devised for incorporating cross-linking data into distance restraints. First, the cross-linking data used came only from mutants showing reasonably robust growth via oxidative phosphorylation on succinate (i.e., colony size ≥1.2 mm versus 2.5 mm for wild type) to avoid structural distortions that were more likely to occur in nonfunctional or poorly functioning ATP synthase mutants. A number of double Cys substitutions in the cross-linking study generated high-yield cross-link products composed of multiple subunit c up to a maximum of 12. In these cases, the initial cross-link is likely to have had minimal perturbing effects on the structure because additional identical cross-links could be formed. For these cross-links the α carbon distances were restrained to a range of 4–8 Å (Table 1). For double Cys mutants forming dimers only, the range was restrained to 4–11 Å, because in these cases the initial cross-link is more likely to perturb structure and/or may have resulted initially from larger thermal motions. The data from single Cys substitutions forming dimers were used if the yield of cross-link product was ≥50% and the distance was restrained to 4–11 Å. The α carbon distances for residue 8–8′, 11–11′, 15–15′, 26–26′, and 30–30′ were restrained to 4–8 Å because each residue when combined with one of the other formed a multimeric ladder (15), which indicated that the initial cross-link had minimal perturbing effects on structure.‡ Finally, the restraints for residues 74–74′, 75–75′, and 78–78′ were set as high as 14 Å because of indications that this terminal region of the protein is more mobile, perhaps because it extends from the lipid bilayer. In this region, a continuous stretch of singly substituted residues form dimers that must indicate rotation or mobility of the observed α-helix (15).
Table 1.
Cross-linking distance constraints used in modeling the c oligomer
| Residue pair | Cross-linking properties | Distance constraint range, Å | Final distance, Å |
|---|---|---|---|
| Helix 1 to helix 1 | |||
| 8–8′ | Dimer; multimers with 30 | 4–8 | 7.2–7.8 |
| 10–10′ | Dimer | 4–11 | 9.0–10.3 |
| 11–11′ | Dimer; multimers with 15 | 4–8 | 7.2–7.8 |
| 15–15′ | Dimer; multimers with 11, 26, 30 | 4–8 | 6.4–7.1 |
| 26–26′ | Dimer; multimers with 15, 30 | 4–8 | 6.5–6.8 |
| 30–30′ | Dimer; multimers with 15, 26 | 4–8 | 6.8–7.2 |
| 14–16′ | High-yield multimers | 4–8 | 5.6–5.8 |
| 16–18′ | High-yield multimers | 4–8 | 4.1–4.3 |
| Helix 1 to helix 2 | |||
| 13–68′ | Dimer | 4–11 | 10.1–10.9 |
| 14–72′ | High-yield multimers | 4–8 | 7.0–7.6 |
| 16–68′ | Dimer | 4–11 | 10.5–11.0 |
| 20–62′ | Dimer | 4–11 | 9.2–10.1 |
| 21–65′ | High-yield multimers | 4–8 | 5.4–5.9 |
| Helix 2 to helix 2 | |||
| 53–53′ | Dimer | 4–11 | 11.3–14.8 |
| 63–63′ | Dimer | 4–11 | 11.9–13.2 |
| 66–66′ | Dimer | 4–11 | 11.3–11.4 |
| 68–71′ | Dimer | 4–11 | 11.4–11.5 |
| 70–72′ | High-yield multimers | 4–8 | 8.1–8.2 |
| 74–74′ | Dimer at C-terminus | 4–14 | 13.7–14.0 |
| 75–75′ | Dimer at C terminus | 4–14 | 14.1–14.4 |
| 78–78′ | Dimer at C terminus | 4–14 | 14.1–14.4 |
Calculations.
The structure calculations were done with the insightii/discover-3 suite (Biosym/MSI, San Diego) using the Consistent Valence Forcefield (CVFF). All the ionizable groups were modeled in their uncharged forms. All the restraints were quadratic; i.e., the energy penalty was proportional to the square of the deviation from the target value. The restraint function force constants for intramolecular hydrogen bonds were 100 times those used for the upper limit of intermolecular distance restraints set from cross-linking data. In the final model, the energy contribution of the distance restraint violations was small compared with dispersive and repulsive components of the van der Waals potential and covalent geometry terms. After the initial energy minimization, the input structure was subjected to 5 ps of molecular dynamics simulation at 1,000 K with subsequent slow cooling to 100 K, followed by the steepest descents and conjugate gradients minimization. Inspections of the trajectory during this simulation showed that the thermal fluctuations of the protein molecules at 1,000 K are large enough to sample the conformational space of all NMR structures reported in the Protein Data Bank file. The resulting structure was cloned to 12 identical structures, which were then best-fitted in a circular shift fashion; i.e., if the oligomeric structures are lettered A to L and monomers in the ring are numbered 1 to 12, then structure B would be fitted to structure A so that c monomers are aligned in the order A1 to B2, A2 to B3, … , A12 to B1 and structure C would be fitted to structure A so that the monomers are aligned in the order A1 to C3, A2 to C4, … , A11 to C1, A12 to C2, and so on. At this stage the rms deviation (rmsd) for backbone atoms was 2.3 Å. A mean structure of the oligomer was then calculated. This procedure is similar to symmetrization of an image by calculating an average of 12 superpositions related to each other by rotation in 30° increments. The whole cycle including molecular dynamics, gradient minimization, and averaging was repeated on the mean structure, except that molecular dynamics simulation was run at 600 K. The best fit rmsd for backbone atoms for a circular shift alignment was 1.1 Å. The cycle was repeated again on the mean structure with the molecular dynamics simulation at 300 K. No averaging was done after this cycle. The resulting structure was minimized by using a conjugate gradient procedure to a maximum derivative of 0.1 kcal/mol⋅Å2. The final structural model was evaluated by using procheck (18) and what_check (19) and shown to meet normal standards of structure determination. Because no explicit symmetry constraints were imposed in the procedure and because the final structures were not averaged, the method allows for structural variability within an oligomer meeting the imposed experimental restraints. The procedure is similar to that used in solving NMR structures with a final product being an ensemble of 12 monomeric structures packed in a low-energy oligomeric complex. The molmol program (20) was used for visual analysis of the structure and in preparation of molecular graphics figures.
RESULTS AND DISCUSSION
General Features of the Model.
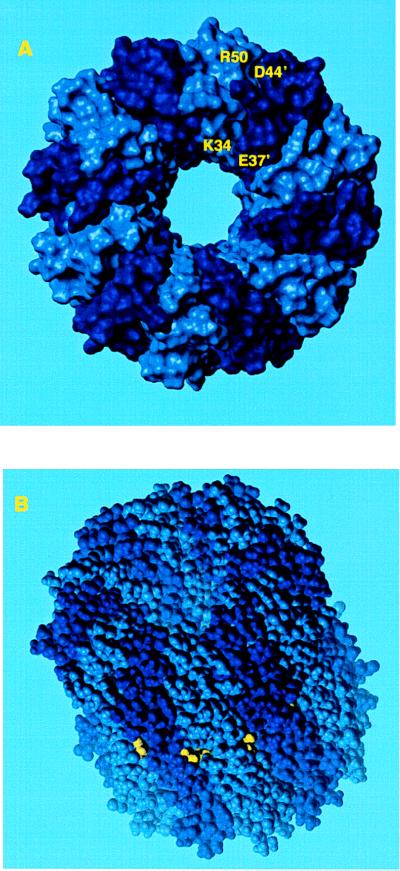
In the final model the c12 complex is a compact hollow cylinder with an outer diameter of 55–60 Å and inner cylindrical space with a minimum diameter of 11–12 Å (Fig. 2).§ In comparison, Birkhenhäger et al. (8) have estimated the diameter of the Fo complex, in perpendicular view to the plane of the membrane, to be 60–75 Å by electron microscopy. Most of the intersubunit cross-linking distance restraints were satisfied by the model (Table 1), the exceptions being those from residues 53 to 53′ and residues 63 to 63′. Both exceptions involve residues with bulky side chains (i.e., Phe and Ile), the replacement of which may result in altered local packing, abnormal thermal motions and perhaps aberrant cross-linking. We should emphasize that cross-linking between identical residues in helix 2 cannot in any case be explained by the simple ring model of Jones et al. (15). These cross-links are suggested to result from structural flexibility in helix 2 that may relate to changing the protonation/deprotonation accessibility of Asp-61, perhaps as a result of interactions with subunit a (15).
Figure 2.
Space-filling model of subunit c oligomer with shaded colors distinguishing individual subunits. (A) View looking down from the polar loop end of the molecule. (B) View from the side with polar loops at the top of the cylinder. The Asp-61 residue is shown in yellow.
A ring model of the subunit c oligomer was suggested by Groth and Walker (21) where, in contrast to our model, the C-terminal helices are located in the inner ring and the N-terminal helices on the outside. We attempted to apply our modeling procedure to an input structure where subunit c monomers were oriented in such way. A 1,000 K molecular dynamics/minimization cycle (see Materials and Methods) produced a highly asymmetric structure. The nonbond energy was +1,348 kcal/mol versus −287 kcal/mol in a comparable simulation performed with our input structure. The energy contribution from violated distance restraints was +4,637 kcal/mol in Groth–Walker type of the model compared with +545 kcal/mol in our study. Clearly, an orientation of the c monomers with the N-terminal helices forming the inner ring provides the best fit for the monomeric structure resolved by NMR, as well as the experimental cross-linking data. The placement of helix 2 on the outside is supported by the cross-linking experiments of Jiang and Fillingame (22), who have shown that multiple Cys residues in helix 4 of subunit a were cross-linked to multiple sites in helix 2 of subunit c, but no cross-links from helix 4 were formed to multiple Cys residues introduced in helix 1 of the subunit c.
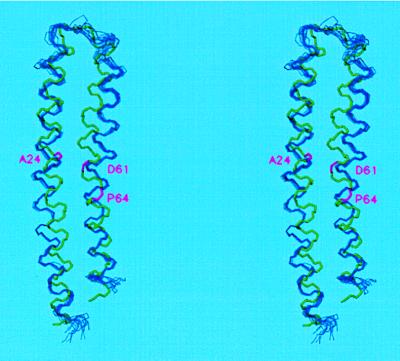
The average rmsd to mean structure for the backbone atoms (of residues 3 to 77) of the 12 monomers in the ring was 0.58 ± 0.10 Å. Not surprisingly, the general area of the polar loop (residues 39 to 49) where distance restraints were absent had a higher rmsd of 1.01 ± 0.27 Å than the rest of molecule (0.46 ± 0.08 Å). Superposition of the solution NMR structure on the best-fit ensemble of the 12 c monomers from the oligomeric model is shown in Fig. 3. Backbone rmsd between the mean structure in the oligomeric model and the NMR structure was 2.6 Å. This value is accounted for primarily by differences in the overall curvature of the molecule and changes in orientation of the N- and C-terminal helical segments as they pack against each other. Such changes in packing were anticipated due to the relatively weak restraints on backbone angles. Because the NMR-derived distance constraints between helices were not explicitly applied during the simulation, the final model is largely independent of the NMR monomeric structure. Alignment of the N-terminal helices (residues 3–38) or C-terminal helices (residues 50–77) separately results in lower backbone rmsds of 1.9 Å in both cases. The most notable difference in structure of the monomer versus oligomer is found in the bending of the C-terminal helix around Pro-64. The angle between the helical segments of residues 48–62 and 65–78, calculated by fitting cylinders to the α carbon trace, is 27° in the initial NMR structure. It is reduced to 16° ± 3° (mean ± SEM) in the oligomeric model. The change is not surprising in that new residue–residue interactions introduced by the packing of subunits in the oligomer would be expected to change the orientation of optimal helix packing from that calculated with the monomer, where protein–protein interactions are minimal.
Figure 3.
Superposition of the backbones of the 12 individual subunits of the c oligomer (blue) and of the input NMR structure of monomeric subunit c (green) in stereo. The mean structure in the c oligomer, calculated from the best fit of the 12 structures, was aligned with the NMR structure by best-fit calculations. Backbone positions of Ala-24, Asp-,61 and Pro-64 are magenta.
Structural Features of the Oligomer.
The van der Waals interactions achieved through packing of complementary surfaces are believed to play a major role in maintaining the structure of oligomeric membrane proteins (23). A pattern of interdigitation of long and short side chains is observed between interacting c monomers. Examples of such pairs include Gly-18/Met-16′, Gly-18/Leu-19′, Gly-23/Ile-22′, Gly-27/Ile-26′, Gly-29/Leu-31′, Gly-32/Phe-54′, Gly-33/Phe-35′, Gly-33/Lys-34′, Gly-38/Glu-37′, Gly-58/Ile-28′, and Gly-69/Met-17′. It is not certain whether salt bridges play a major role in maintaining the structure of membrane proteins. In our model, side chains of Lys-34 and Glu-37′ on the neighboring subunits are found in close proximity to each other, so that hydrogen-bond or salt-bridge formation is possible (Figs. 2 and 4A). The calculations were carried out with ionizable groups in their uncharged forms to prevent artifacts arising from strong unscreened electrostatic interactions. In a simulation where final energy minimization was carried out with the charged species of Lys-34 and Glu-37, in 10 of 12 neighbor pairs, the minimal distance between the ɛ-amino proton of Lys-34 and a side-chain oxygen of Glu-37′ was in the range of 1.6–2.0 Å, a distance expected in an ionic interaction. Another intersubunit interaction that may contribute to the stabilization of c oligomer is between Asp-44′ and Arg-50 (Figs. 2A and 4B). Hydrogen bonds between the uncharged Asp-44 and Arg-50 side chains were found for 10 of 12 neighbor pairs in the model, and distances consistent with ionic bonds were found in 10 pairs when the simulation was done with charged residues.
Figure 4.
Electrostatic potential surfaces in the polar loop region of two subunit c of the oligomer. Subunits have been manually moved apart at the plain indicated by the black line to emphasize their complementary interacting surfaces and individual electrostatic potentials. (A) Two subunits viewed from the inside of the oligomeric ring. (B) Two subunits viewed from the outside of the oligomeric ring after 180° rotation of the image shown in A.
Groth et al. (24) have recently constructed single Trp substitutions in residues 61–72 of helix 2 in an attempt to probe the arrangement of c subunits. The activities of most of the substitutions were severely compromised or nil. Some activity was observed with substitutions centered around Pro-64, i.e., at residues 62, 63, and 65, which may reflect structural flexibility in this region. Because tryptophan residues in membrane proteins are typically observed at the hydrocarbon–polar head group interface of the lipid bilayer (25), the explanation for some of these substitutions may be more complex than simple steric hindrance. The effects of substitutions at positions 69–72, which are expected to be located near the hydrocarbon–head group interface, are consistent with an α-helical periodicity and the model presented herein. The G71W mutant exhibits a wild-type phenotype and the Trp side chain would be expected to project directly from the periphery of the ring into the phospholipid bilayer. The G69W mutant is completely inactivated and in the model Gly-69 lies in a tightly packed interface between subunits, neighboring Met-17′. The L70W and L72W substitutions retain partial activity and the bulkier Trp side chains would be expected to project to the inside or outside of the ring.
The front-to-back packing of flattened surfaces of c subunits in a cylindrical ring inevitably leads to a central hole, with a minimal diameter of 10–12 Å, extending the length of the cylinder. A closure of the hole is obviously necessary to maintain the semipermeable properties of the E. coli inner membrane. Similar holes are seen in the crystal structure of the Rhodopseudomonas acidophilia LH2 light-harvesting complex (minimal diameter = approximately 24 Å) (26) and in the lattice of bacteriorhodopsin trimers in the purple membrane of Halobacterium halobium (minimal diameter = 8–9 Å) (27). The holes of both structures are presumed to be filled with lipid in the native membrane environment (26–28); lipid has been visualized by high-resolution electron or x-ray crystallography in the purple membrane (27, 28). In the bacteriorhodopsin trimer, three ether-linked glycerol lipids with isoprenoid hydrocarbon chains are packed into the hole from each membrane surface. Based on the similar dimensions of the hole in the modeled subunit c oligomer, we conclude that least three, and perhaps as many as six, E. coli phospholipid molecules with unbranched acyl chains could pack into the hole from each membrane surface. It is also possible that a portion of the γ subunit of F1, which is not resolved in the crystal structure (3), could extend into a part of the hole.
Functional Implications of the Model.
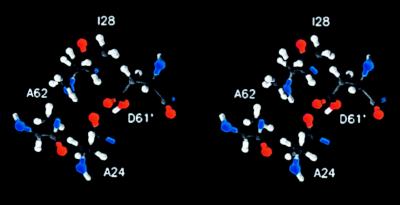
The packing of helices in the oligomer allows close contact between the “front” face of one monomer and the “back” face of its neighbor. As shown in Fig. 5, Asp-61′ at the front face of one monomer packs in close proximity to Ala-24, Ile-28, and Ala-62 of the neighboring monomer with the side chains of these residues facing toward each other (Fig. 5). The proximal positioning of residues 24 and 61′ provides an explanation for the observed function in the A24D/D61G and A24D/D61N aspartate-interchange mutants (29, 30) in that the H+ transporting carboxylate could pack in essentially the same position whether attached to the β carbon of either residue 24 or 61′. The proximal positioning of residues 24, 28, and 61′, as shown in Fig. 5, was also expected from the dicyclohexylcarbodiimide resistance of the A24S and I28T mutants (31). Replacement modeling indicates that a hydroxyl from Ser-24 could potentially hydrogen bond with the Asp-61 carboxylate to change its pKa and reactivity, although purely steric explanations for the dicyclohexylcarbodiimide resistance are also possible. The predicted structure is also consistent with the proposed role of Gln-32 of subunit c in the Na+-translocating F1Fo of Propionigenium modestum in Na+ binding (32). P. modestum Gln-32 lies at a position equivalent to Ile-28 in E. coli subunit c, a position from which it could easily participate in binding Na+ with the essential carboxylate. Finally, Ala-62 is located in close proximity to Asp-61′ of the neighboring subunit in the oligomeric model. Ala-62 is the equivalent of residue Ser-66 in P. modestum subunit c, the Ser-66 residue being necessary for binding of Na+ or Li+. For E. coli subunit c, the A62S mutation confers Li+ sensitivity to the enzyme when it is made in conjunction with other enabling substitutions in this region, including an Asp-61→ Glu substitution (33). A seryl hydroxyl at this position could easily participate in binding of the cation.
Figure 5.
Stereo representation of orientation of Asp-61′ of one subunit relative to Ala-24, Ile-28, and Ala-62 of a neighboring subunit in the c oligomeric ring.
The proximal positioning of residues 24, 28, and 62 with 61′ in the model is somewhat remarkable in that there were no distance constraints imposed between these residues in the calculation procedure. On the other hand, one of the strong distance constraints (range = 4–8 Å) imposed was between the α carbons of residues 21 and 65′ (Table 1). To test whether the proximity of residues shown in Fig. 5 was dictated largely by this constraint, the calculations were repeated with omission of this one constraint. No differences were obvious on comparing the backbone packing of the oligomer in the two models. The packing of side chains in the region of Asp-61′ and residues Ala-24, Ile-28, and Ala-62 in the neighboring monomer were compared in detail, and again no major differences were apparent. The average distances between the Asp-61′ γ carbon and the β carbons of Ala-24, Ile-28, and Ala-62 for the 12 interacting pairs were 4.22 ± 0.04 Å, 4.46 ± 0.26 Å, and 5.50 ± 0.50 Å for the original model and 4.17 ± 0.17 Å, 5.10 ± 0.25 Å, and 5.29 ± 0.25 Å for the model derived with omission of the 21–65′ restraint.
Rotational models of Fo function suggest that the transmembrane proton path may be formed by an interaction between the Asp-61 side chain carboxylate and inlet and outlet channels formed at the interface of subunits a and c (4, 11–13). This necessitates an access to the Asp-61 carboxylate from the outer surface of the cylinder, where interaction with subunit a is known to take place (21). The positions of the Asp-61 side chains within the oligomer are shown in Fig. 6. Asp-61 was modeled in the protonated form in this structure since the NMR structure was solved under these conditions. The protonated carboxyl is clearly lodged at the center of the four α-helices of a functional dimeric unit. The modeling indicates that the protonated state could be stabilized by hydrogen bonding between the protonated Asp-61′ carboxyl and backbone carbonyl of Ala-24 or Ala-21 of the neighboring c subunit. In the rotational model, the state shown may correspond to that in which the outer surface of the oligomeric cylinder is rotating through and exposed to the hydrocarbon milieu of the lipid bilayer. During the proposed interaction with subunit a, in which deprotonation and reprotonation takes place, the carboxyl group might be expected to move more toward the periphery of the ring by the opening of the c–c interface by the swiveling of adjacent helices 1 at the center of the ring. Such a swiveling and opening of the c–c interface was predicted from the cross-linking studies of helix 4 of subunit a with helix 2 of subunit c (22). An opening of the c–c interface would also be required for interaction of subunit a with the functional Asp-24 carboxylate in the A24D/D61G and A24D/D61N aspartyl-interchange mutants. The predicted swiveling and repositioning of helices during the stepwise movement of the rotor may prove to be an important facet of the rotary mechanism. The structural model presented herein contrasts with the previous hypotheses, which place the essential carboxylate at the periphery of the ring (4, 11–13).
Figure 6.
Cross section of the contact surface in the c oligomer showing orientation of the Asp-61 side chains (yellow) with their carboxyl oxygens (red).
Acknowledgments
We thank Dr. Mark Girvin (Albert Einstein College of Medicine) for his thoughtful advice on modeling and critique of the manuscript. This work was supported by U. S. Public Health Service Grant GM23105 from the National Institutes of Health and a grant from the Human Frontiers Science Program.
ABBREVIATION
- rmsd
rms deviation
Footnotes
The residues cross-linked are designated with and without prime (′) to indicate that the cross-links are between different subunit c.
A coordinate file of the final model is available by e-mail at dmitriev@iris.bmolchem.wisc.edu.
References
- 1.Senior A E. Physiol Rev. 1988;68:177–231. doi: 10.1152/physrev.1988.68.1.177. [DOI] [PubMed] [Google Scholar]
- 2.Fillingame R H. J Exp Biol. 1997;200:217–224. doi: 10.1242/jeb.200.2.217. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams J P, Leslie A G W, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 4.Duncan T M, Bulygin V V, Zhou Y, Hutcheon M L, Cross R L. Proc Natl Acad Sci USA. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabbert D, Engelbrecht S, Junge W. Nature (London) 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 6.Noji H, Yasuda R, Yoshida M, Kiniosita K., Jr Nature (London) 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 7.Boyer P D. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 8.Birkenhäger R, Hoppert M, Deckers-Hebestreit G, Mayer F, Altendorf K. Eur J Biochem. 1995;230:58–67. [PubMed] [Google Scholar]
- 9.Takeyasu K, Omote H, Nettikadan S, Tokumasu F, Iwamoto-Kihara A, Futai M. FEBS Lett. 1996;392:110–113. doi: 10.1016/0014-5793(96)00796-x. [DOI] [PubMed] [Google Scholar]
- 10.Singh S, Turina P, Bustamante C J, Keller D J, Capaldi R A. FEBS Lett. 1996;397:30–34. doi: 10.1016/s0014-5793(96)01127-1. [DOI] [PubMed] [Google Scholar]
- 11.Vik S B, Antonio B J. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 12.Engelbrecht S, Junge W. FEBS Lett. 1997;414:485–491. doi: 10.1016/s0014-5793(97)00997-6. [DOI] [PubMed] [Google Scholar]
- 13.Elston T, Wang H, Oster G. Nature (London) 1998;391:510–513. doi: 10.1038/35185. [DOI] [PubMed] [Google Scholar]
- 14.Girvin M E, Rastogi V K, Abildgaard F, Markley J L, Fillingame R H. Biochemistry. 1998;37:8817–8824. doi: 10.1021/bi980511m. [DOI] [PubMed] [Google Scholar]
- 15.Jones P C, Jiang W, Fillingame R H. J Biol Chem. 1998;273:17178–17185. doi: 10.1074/jbc.273.27.17178. [DOI] [PubMed] [Google Scholar]
- 16.Jones P C, Fillingame R H. J Biol Chem. 1998;273:29701–29705. doi: 10.1074/jbc.273.45.29701. [DOI] [PubMed] [Google Scholar]
- 17.Richardson J S, Richardson D C. In: Prediction of Protein Structure and the Principles of Protein Conformation. Fasman G, editor. New York: Plenum; 1989. pp. 1–98. [Google Scholar]
- 18.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:946–950. [Google Scholar]
- 19.Hooft R W W, Vriend G, Sander C, Abola E E. Nature (London) 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 20.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:283–298. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 21.Groth G, Walker J E. FEBS Lett. 1997;410:117–123. doi: 10.1016/s0014-5793(97)00529-2. [DOI] [PubMed] [Google Scholar]
- 22.Jiang W, Fillingame R H. Proc Natl Acad Sci USA. 1998;95:6607–6661. doi: 10.1073/pnas.95.12.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemmon M A, Engelman D E. Q Rev Biophys. 1994;27:157–218. doi: 10.1017/s0033583500004522. [DOI] [PubMed] [Google Scholar]
- 24.Groth G, Tilg Y, Schirwitz K. J Mol Biol. 1998;281:49–59. doi: 10.1006/jmbi.1998.1936. [DOI] [PubMed] [Google Scholar]
- 25.Yau W-M, Wimley W C, Gawrisch K, White S H. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 26.Prince S M, Papiz M Z, Freer A A, McDermott G, Hawthornwaite-Lawless A M, Cogdell R J, Isaacs N W. J Mol Biol. 1997;268:412–423. doi: 10.1006/jmbi.1997.0966. [DOI] [PubMed] [Google Scholar]
- 27.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 28.Essen L O, Siegert R, Lehmann W D, Oesterhelt D. Proc Natl Acad Sci USA. 1998;95:11673–11678. doi: 10.1073/pnas.95.20.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller M J, Oldenburg M, Fillingame R H. Proc Natl Acad Sci USA. 1990;87:4900–4904. doi: 10.1073/pnas.87.13.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Fillingame R H. J Biol Chem. 1994;269:5473–5479. [PubMed] [Google Scholar]
- 31.Fillingame R H, Oldenburg M, Fraga D. J Biol Chem. 1991;266:20934–20939. [PubMed] [Google Scholar]
- 32.Kaim G, Wehrle F, Gerike U, Dimroth P. Biochemistry. 1997;36:9185–9194. doi: 10.1021/bi970831q. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Fillingame R H. J Biol Chem. 1995;270:87–93. doi: 10.1074/jbc.270.1.87. [DOI] [PubMed] [Google Scholar]