Abstract
The xenoestrogen diethylstilbestrol (DES) is commonly believed to mimic the action of the natural estrogen 17β-estradiol (E2). To determine if these two estrogens exert similar actions in prostate carcinogenesis, we elevated circulating levels of estrogen in Noble (NBL) rats with E2/DES-filled implants, while maintaining physiological levels of testosterone (T) in the animals with T-filled implants. The two estrogens induced dysplasia in a lobe-specific manner, with E2 targeting only the lateral prostate (LP) and DES impacting only the ventral prostate (VP). Gene expression profiling identified distinct and common E2-disrupted versus DES-disrupted gene networks in each lobe. More importantly, hierarchical clustering analyses revealed that T + E2 treatment primarily affected the gene expression pattern in the LP, whereas T + DES treatment primarily affected the gene expression profile in the VP. Gene ontology analyses and pathway mapping suggest that the two hormone treatments disrupt unique and/or common cellular processes, including cell development, proliferation, motility, apoptosis, and estrogen signaling, which may be linked to dysplasia development in the rat prostate. These findings suggest that the effects of xenoestrogens and natural estrogens on the rat prostate are more divergent than previously suspected and that these differences may explain the lobe-specific carcinogenic actions of the hormones.
Introduction
Androgens are essential to the growth and functioning of the normal prostate and undoubtedly play a key role in prostate carcinogenesis [1]. Emerging evidence shows that estrogens also have a critical role in prostatic diseases, including prostate cancer (PCa) [2,3]. The incidence of PCa increases dramatically with age in human males, whose testosterone (T) levels in both the circulation and in prostate decline, whereas those of 17β-estradiol (E2) remain relatively stable [2–4]. Thus, the age-associated alterations in the sex hormone milieu toward an estrogen predominance have been proposed as an endogenous risk factor for prostate carcinogenesis. Dynamic changes in the expression of estrogen receptor-β during PCa progression in humans [5,6] also suggest the involvement of estrogen signaling in the malignant transformation of the prostate. Moreover, the findings of aberrant activation of aromatase expression in benign prostatic hyperplasia and PCa [3,7] further advance the concept that local production of estrogens is pivotal in the pathobiology of this gland.
Humans are exposed to xenoestrogens from various sources, including food, drugs, and other exogenous venues. Xenoestrogens are believed to cause endocrine disruptions that lead to pathogenesis in reproductive end organs [8,9]. One agent of public health concern is diethylstilbestrol (DES), a synthetic estrogen, which was widely used as a growth-promoting animal feed additive [10]. Epidemiological studies have established a strong association between maternal exposure to DES and increased risk of vaginal, cervical, and perhaps breast cancers in DES daughters [11,12]. It remains uncertain, however, whether prenatal exposure to DES elevates cancer risk in men [13]. Transient exposure of neonatal rodents to estrogen increases susceptibility of the prostate to inflammation and cancer development later in life [14]. Together, these findings strongly suggest that natural or synthetic estrogen-mediated endocrine disruption leads to the evolution of prostatic diseases, including PCa.
We have established the T plus estrogen-induced dysplasia/carcinoma model in Noble (NBL) rats as a robust study system for deciphering the contributions of estrogens, in adulthood, on the pathogenesis of dysplasia, a precancerous lesion, and adenocarcinoma of the prostate [15,16]. In this model, estrogens are administered through subcutaneous implants of hormone-filled Silastic capsules with the coimplantation of T-filled capsules to maintain physiological levels of this androgen in the circulation [17]. It is well known that increased exposure to estrogens leads to a decline in circulating levels of T through the hypothalamic-pituitary-gonadal axis. The topographical localization of premalignant/malignant lesions within the rat prostate gland has been found to be dependent on the type of estrogens used. Under the same exogenous androgen support, E2 induces epithelial dysplasia and adenocarcinoma in the lateral prostate (LP) but not in the ventral prostate (VP) [16–19], whereas synthetic DES specifically targets the VP but not the LP [19,20]. Why two different estrogens induce lobe-specific carcinogenic actions in the prostate remains unclear. To address this issue, we conducted gene expression profiling analyses on the LP and VP of NBL rats exposed to T + E2 or T + DES. The experimental design and the application of a bioinformatic tool of gene network mapping enabled us to identify lobe-specific and common estrogen-mediated disruptions in multiple biologic networks/pathways that may be linked to prostate carcinogenesis and explain the distinctive action of the two estrogens.
Materials and Methods
Animals and Hormonal Treatment
The animal usage protocol was approved by the Institutional Animal Care Committee at the University of Cincinnati. Male NBL rats (5–6 weeks old) were purchased from Charles River Laboratories (Kingston, NY), kept under standard conditions, and treated as previously reported [16,19,20]. Briefly, animals were randomized into three groups (n = 5 for each group). Rats in the T + E2 treatment group received subcutaneous implants of two pieces of 2-cm-long Silastic capsules containing T (Sigma, St Louis, MO) and one piece of 1-cm-long capsules packed with E2 (Sigma), whereas the T + DES treatment group received the same number of Silastic capsules of the same length filled with T and DES (Sigma). Age-matched untreated control rats were implanted with empty capsules. At the end of a 16-week treatment period, animals were sacrificed with an overdose of isoflurane, and VPs and LPs were excised. One half of each lobe was processed for histologic examination, and the other half was snap-frozen for RNA extraction.
Microarray Hybridization
The Atlas Glass Rat 3.8 I Microarray (Clontech, Palo Alto, CA) carrying 3800 named rat genes (spotted oligonucleotides) were used as the gene chip platform. The Atlas Glass Fluorescent Labeling Kit (Clontech) was used for synthesizing and purifying fluorescent-labeled cDNA probes for hybridization to glass microarrays. The labeling and hybridization procedures were performed in accordance with the manufacturer's instruction manual. In brief, amino-modified first-strand cDNA probes were synthesized with aminoallyl. 2′-deoxyuridine 5′-triphosphate incorporation. Then Cy3 fluorescent dye was coupled to the cDNAs derived from individual prostatic lobes, whereas Cy5 dye was conjugated to the universal rat reference RNA obtained from Stratagene (La Jolla, CA). Equal quantities of two labeled probes were mixed and hybridized in a Corning microarray hybridization chamber (Corning, Corning, NY) at 50°C overnight (≥16 hours). Spikes of positive control probes were also included as an internal control for the process of cDNA probe synthesis and the dye-coupled reaction. Finally, the signal was obtained using a microarray scanner (GenePix 4000B; Axon Instruments, Foster City, CA). A probe coverage of >90% was achieved for all arrays. Five animal replicates for each treatment/tissue group (T + E2-treated VP or LP, T + DES-treated VP and LP, and untreated VP and LP) were used to conduct a 30-microarray analysis to assess changes in the gene expression pattern due to treatment and lobe specificity.
Microarray Data Normalization and Analysis
The data were analyzed to identify differentially expressed genes in 1) the T + E2-treated LPs and VPs compared with untreated controls and 2) the T + DES-treated LPs and VPs compared with untreated controls. Five biologic replicate arrays for each experimental condition, all versus universal reference, were performed. R statistical software and the limma Bioconductor package [21] were used for analysis. Data normalization was performed in two steps separately for each microarray [22–24]. First, background-adjusted intensities were log-transformed, and the differences (M) and averages (A) of log-transformed values were calculated as M = log2(X1) - log2(X2) and A = [log2(X1) + log2(X2)]/2, where X1 and X2 denote the Cy5 and Cy3 intensities, respectively. Second, normalization was performed by fitting the array-specific local regression model of M as a function of A and obtaining residuals. The statistical analysis was performed for each gene separately by fitting a one-way analysis of variance (ANOVA) model with treatment. Estimated fold changes were calculated from the ANOVA model; an intensity-based empirical Bayes method was used to modify the resulting t-statistics from each comparison [25]. This method obtains more precise estimates of variance by pooling information across genes and by accounting for the dependency of variance on probe intensity level. Genes with a false discovery rate (FDR) <0.05 [26] were considered to be significantly differentially expressed. Clustering was performed using normalized, centered sample ratios. The gene list used for clustering consisted of all genes having an FDR <0.05 for at least one comparison (1063 genes). T + E2 and T + DES samples were each clustered using the Ward clustering method and Euclidean dissimilarity metric with 1063 genes.
Identification of Estrogen-Regulated Gene Expression
Genes differentially regulated in the different lobes with T + DES and T + E2 treatment were identified separately in each drug treatment. Gene lists were generated according to their expression signature in different gene expression clusters; each group was described in the Results section.
Pathway and Network Analysis
Biologic relationships between differentially expressed genes were mapped by Ingenuity Pathway Analysis (IPA) 3.1 software (www.ingenuity.com). Gene lists of different patterns of gene expression in response to T + E2 and T + DES treatments (as described in the Results section) were uploaded to the IPAWeb application in an Excel file format containing expression data and GenBank accession number as identifier. The biologic relationship of uploaded genes was mapped with IPA software into networks according to the published literature in the database. A score was assigned to each network in the data set to estimate the relevance of the network to the uploaded gene list. A higher score means that the network is more relevant to the gene list entered by the user [27]. The two highest score networks were selected in this study, and genes in these two networks were selected for further post hoc analysis.
Reverse Transcription and Real-Time Quantitative Polymerase Chain Reaction
The total RNA from each sample of prostate tissue was reverse-transcribed into cDNA using Superscript III (Invitrogen, Carlsbad, CA). Specific primers were designed using either Primer Express 3 (Applied Biosystems, Foster City, CA) or Primer3 software [28]; the sequence of the primer is listed in Table W1. Real-time quantitative polymerase chain reaction (q-PCR) was performed on the 7900HT Fast Real-Time PCR System (ABI Biosystems) using the Power SYBR Green PCR master mix (Applied Biosystems). Polymerase chain reaction was performed in a total volume of 10 µl containing 50 ng of total cDNA, 1x Power SYBR Green PCR master mix, and a final primer concentration of 800 nM. The relative expression level was analyzed by the ΔΔCt method [29] and one-way ANOVA followed by Tukey post hoc analysis, where P < .05 was considered statistically significant.
Results
T + E2 and T + DES Treatments Differentially Induced Dysplasia in LP and VP, Respectively
The expected physiological and histologic changes resulting from T + E2 and T + DES treatment were observed as reported previously [15,18–20]. Dysplasia was observed in the T + E2-treated LPs (100% incidence) and T + DES.treated VPs (100% incidence), whereas no preneoplastic lesions were observed in T + E2-treated VPs and T + DES LPs. In T + E2-treated LPs, the dysplastic lesions were often accompanied by inflammatory infiltrates [17].
Hierarchical Clustering Identified Differential Action of E2 and DES in the Two Prostate Lobes
Unsupervised hierarchical clustering was performed for each hormone treatment group (T + E2 or T + DES) to determine the relatedness of replicate LP and VP samples (n = 5 per group) according to similarity in gene expression patterns among the 1063 genes with a significant difference in expression across samples, without prior knowledge of gene and sample identity.
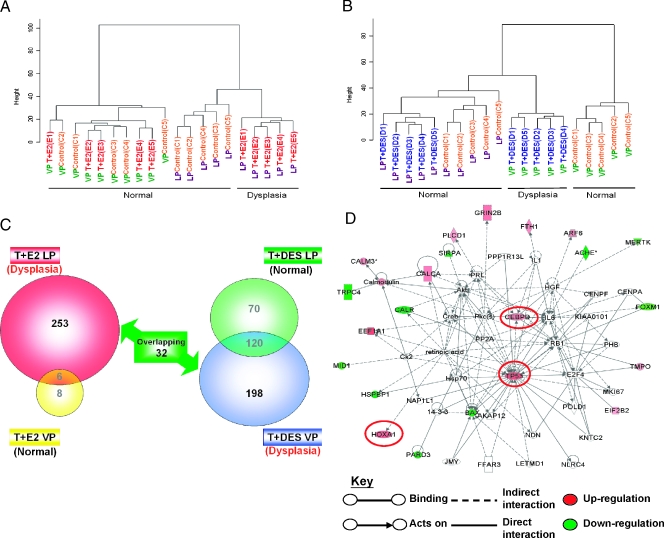
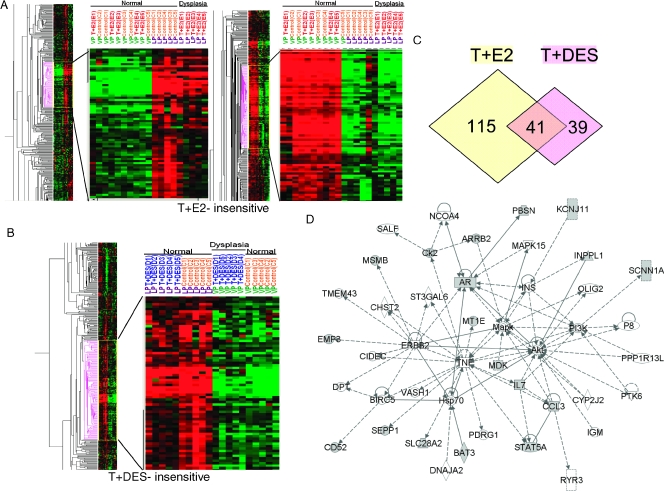
Hierarchical clustering of samples showed that all LPs treated with T + E2 formed one cluster distinct from the cluster containing all untreated LPs. The hormone treatment, however, did not partition the hormone-treated and -untreated VPs, which formed a single large cluster (Figure 1A). These results indicate that the T + E2 treatment induced changes in gene expression mainly in the LP and had little, if any, effect on the gene expression pattern in the VP.
Figure 1.
Hierarchical clustering analysis of T + E2 and T + DES gene expression data set. (A) Dendrogram of T + E2 expression data set in LP and VP. (B) Dendrogram of T + DES expression data set in LP and VP. (C) Venn diagram showing the number of genes differentially expressed in each treatment group compared with the respective untreated control. (D) Gene interaction network of a subset of differentially expressed genes that are common in both LP and VP dysplasia. Genes bordered with red were validated by quantitative real-time PCR.
In a similar manner, T + DES treatment segregated only the VPs into two distinct clusters (treated and untreated) and did not partition the LPs into distinctive groups (Figure 1B). These findings indicate that the hormone treatment altered primarily the gene expression pattern in the VP and had little effect on LP gene expression. It is interesting that the gene expression pattern in the VP after T + DES treatment appeared to more closely resemble that observed in the LPs, as the T + DES-treated VPs formed a cluster more closely linked to the LP cluster than the one comprising untreated VPs.
Identification of Estrogen-Induced Differentially Expressed Genes Related to Dysplasia in Rat LP or VP
We used the following criteria to identify two panels of dysplasia-related genes: 1) the T + E2-induced LP dysplasia panel contains genes whose expression changed following T + E2 treatment in the LPs harboring dysplasia but excludes those whose expression also changed in the VPs with no dysplastic changes (253 genes; Figure 1C, left; Table 1), and 2) the T + DES-induced VP dysplasia panel includes genes whose expression changed after T + DES treatment in the VPs harboring dysplasia but excludes those whose expression also changed in the LPs without dysplasia (198 genes; Figure 1C, right; Table 2).
Table 1.
T + E2-Induced LP Dysplasia Panel: Genes Whose Expression Selectively Changed following T + E2 Treatment in the LPs Harboring Dysplasia, but not in the VPs.
| Gene Names | Locus ID | Symbol | Fold Changes | Functions |
| prohibitin* | 25344 | PHB | 3.42 | Cell death; cell signaling; cellular growth and proliferation; gene transcription |
| nuclease-sensitive element binding protein 1* | 29206 | NSEP1 | 3.08 | Cell death; gene transcription |
| potassium channel, subfamily K, member 9* | 84429 | KCNK9 | 3.08 | Cell death |
| vesicle-associated membrane protein 2* | 24803 | VAMP2 | 2.97 | Cell death; cellular assembly and organization; cellular movement |
| cytochrome c oxidase subunit IV isoform 1* | 29445 | COX4I1 | 2.71 | Others/unclassified |
| PR-Vbeta1* | 498341 | PR-Vbeta1 | 2.68 | Others/unclassified |
| melanocortin 5 receptor* | 25726 | MC5R | 2.47 | Cell signaling |
| glutamate receptor, ionotropic, kainate 3* | 298521 | GRIK3 | 2.45 | Cell signaling |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4* | 54283 | PFKFB4 | 2.41 | Others/unclassified |
| potassium voltage-gated channel, Shaw-related subfamily, member 1* | 25327 | KCNC1 | 2.39 | Molecular transport |
| actin alpha cardiac 1* | 29275 | ACTC1 | 2.34 | Others/unclassified |
| eukaryotic translation elongation factor 1 alpha 1* | 171361 | EEF1A1 | 2.31 | Cell death, protein synthesis |
| ATPase, Na+/K+ transporting, alpha 3 polypeptide* | 24213 | ATP1A3 | 2.29 | Inflammation; molecular transport |
| gamma-aminobutyric acid A receptor, rho 1* | 29694 | GABRR1 | 2.25 | Cell death; cell signaling |
| guanylate cyclase 1, soluble, alpha 3* | 25201 | GUCY1A3 | 2.25 | Cellular movement |
| transforming growth factor alpha* | 24827 | TGFA | 2.24 | Biomolecule metabolism; cell cycle; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; gene transcription; molecular transport; protein synthesis |
| ribosomal protein S12* | 65139 | RPS12 | 2.20 | Protein synthesis |
| tumor protein p53* | 24842 | TP53 | 2.16 | Cell cycle; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; cellular response to therapeutics; DNA replication, recombination and repair; free radical scavenging; gene transcription; inflammation; molecular transport; post-translational modification; protein synthesis; RNA post-transcriptional modification |
| nuclear receptor subfamily 3, group C, member 2* | 25672 | NR3C2 | 2.13 | Biomolecule metabolism; cell signaling; gene transcription; molecular transport; protein synthesis |
| secretory carrier membrane protein 1* | 29521 | SCAMP1 | 2.09 | Cellular assembly and organization |
| CEA-related cell adhesion molecule 1* | 81613 | CEACAM1 | 2.08 | Cell death; cell signaling; cellular growth and proliferation; cellular movement; inflammation |
| olfactory marker protein* | 24612 | OMP | 2.08 | Cell signaling |
| guanine nucleotide binding protein, alpha z subunit* | 25740 | GNAZ | 2.05 | Cell signaling; cellular development; cellular movement; molecular transport; biomolecule metabolism |
| phosphodiesterase 1C* | 81742 | PDE1C | 2.04 | Others/unclassified |
| ribosomal protein S3a* | 29288 | RPS3A | 2.04 | Cell death; cellular development; cellular growth and proliferation; protein synthesis |
| insulin-like 6* | 50546 | IL1RAP | 1.95 | Cell signaling |
| A kinase (PRKA) anchor protein 14* | 60332 | AKAP14 | 1.94 | Others/unclassified |
| CD38 antigen* | 25668 | CD38 | 1.94 | Cell cycle; cell death; cell signaling; cellular development; cellular growth and proliferation; cellular movement; biomolecule metabolism; molecular transport; post-translational modification |
| p21 (CDKN1A)-activated kinase 1* | 29431 | PAK1 | 1.93 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular movement |
| regenerating islet-derived 3 alpha* | 171162 | REG3G | 1.93 | Others/unclassified |
| ubiquitin-conjugating enzyme E2I* | 25573 | UBE2I | 1.93 | Cell signaling; gene transcription; protein synthesis |
| ATPase, H+ transporting, V1 subunit F* | 116664 | ATP6V1F | 1.91 | Molecular transport |
| homeo box A1* | 25607 | HOXA1 | 1.89 | Cell death; cellular development; cellular movement; gene transcription |
| regenerating islet-derived 3 gamma* | 24620 | REG3G | 1.89 | Others/unclassified |
| Fas apoptotic inhibitory molecule 2* | 246274 | FAIM2 | 1.86 | Cell death |
| barrier to autointegration factor 1* | 114087 | BANF1 | 1.85 | DNA replication, recombination, and repair |
| thyroid hormone receptor alpha* | 81812 | THRA | 1.85 | Biomolecule metabolism; cell death; cell morphology; cellular development; free radical scavenging; gene transcription; protein synthesis |
| neuromedin B receptor* | 25264 | NMBR | 1.83 | Others/unclassified |
| ATPase, Na+/K+ transporting, beta 2 polypeptide* | 24214 | ATP1B2 | 1.82 | Others/unclassified |
| Chondroitin sulfate proteoglycan 5* | 50568 | CSPG5 | 1.81 | Cell signaling |
| MAD homolog 7 (Drosophila)* | 81516 | SMAD7 | 1.78 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; gene transcription |
| tachykinin receptor 2* | 25007 | TACR2 | 1.78 | Others/unclassified |
| alanyl (membrane) aminopeptidase* | 81641 | ANPEP | 1.76 | Cell death; cell morphology; cellular development; cellular movement; protein synthesis |
| growth hormone-releasing hormone* | 29446 | GHRH | 1.75 | Cell morphology; cell signaling; cellular growth and proliferation; biomolecule metabolism; molecular transport |
| protein kinase N1* | 29355 | PKN1 | 1.75 | Cell signaling; cellular growth and proliferation; cellular movement; gene transcription |
| slit homolog 3 (Drosophila)* | 83467 | SLIT3 | 1.74 | Others/unclassified |
| dopamine receptor 2* | 24318 | DRD2 | 1.72 | Cell death; cell signaling; cellular growth and proliferation; cellular movement; gene transcription; inflammation; biomolecule metabolism |
| glutathione peroxidase 3* | 64317 | GPX3 | 1.72 | Cellular growth and Proliferation; Post-translational modification; protein synthesis |
| vasoactive intestinal peptide receptor 1* | 24875 | VIPR1 | 1.71 | Cell death; cell signaling; cellular growth and proliferation; cellular movement; biomolecule metabolism; molecular transport |
| amphiphysin 1* | 60668 | AMPH | 1.70 | Cell signaling; cellular assembly and organization; cellular movement; gene transcription |
| G protein beta subunit-like* | 64226 | GBL | 1.70 | Others/unclassified |
| heat shock 70 kDa protein 5* | 25617 | HSPA5 | 1.69 | Cell death; cellular growth and proliferation; inflammation |
| hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1* | 29632 | HSD3B2 | 1.68 | Others/unclassified |
| POU domain, class 3, transcription factor 2* | 29588 | POU3F2 | 1.68 | Cellular development; cellular growth and proliferation; cellular movement; gene transcription |
| ribosomal protein S9* | 81772 | RPS9 | 1.64 | Protein synthesis |
| glucocorticoid modulatory element binding protein 2* | 83635 | GMEB2 | 1.63 | Gene transcription |
| glutamate receptor, ionotropic, NMDA2B* | 24410 | GRIN2B | 1.63 | Cell signaling |
| solute carrier family 8 (sodium/calcium exchanger), member 3* | 140448 | SLC8A3 | 1.63 | Cell death; cell signaling |
| ATPase, Ca2+ transporting, ubiquitous* | 25391 | ATP2A3 | 1.62 | Others/unclassified |
| CTD-binding SR-like rA1* | 56081 | SR-A1 | 1.60 | Others/unclassified |
| solute carrier family 2 (facilitated glucose transporter), member 2* | 25351 | SLC2A2 | 1.60 | Cell death |
| calcineurin binding protein 1* | 94165 | CABIN1 | 1.59 | Cell cycle; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair; gene transcription; inflammation; biomolecule metabolism; molecular transport |
| heterogeneous nuclear ribonucleoprotein A1* | 29578 | LOC144983 | 1.59 | Others/unclassified |
| casein kinase 1, gamma 1* | 64086 | CSNK1G1 | 1.57 | Others/unclassified |
| potassium inwardly rectifying channel, subfamily J, member 10* | 29718 | KCNJ10 | 1.56 | Cell morphology; cellular development; inflammation; molecular transport |
| potassium large conductance calcium-activated channel, subfamily M, beta member 1* | 29747 | KCNMB1 | 1.56 | Cell signaling; molecular transport |
| potassium voltage-gated channel, Shaw-related subfamily, member 3* | 117101 | KCNC3 | 1.56 | Molecular transport |
| CCAAT/enhancer binding protein (C/EBP), delta* | 25695 | CEBPD | 1.55 | Cell death; cellular development; cellular growth and proliferation; gene transcription; inflammation |
| guanine nucleotide binding protein, beta 3* | 60449 | GNB3 | 1.55 | Cell signaling |
| ribosomal protein S19* | 29287 | RPS19 | 1.54 | Cellular development; cellular growth and proliferation; cellular movement; protein synthesis |
| aquaporin 5* | 25241 | AQP5 | 1.53 | Inflammation |
| chloride channel Kb* | 79430 | CLCNKB | 1.53 | Others/unclassified |
| fatty acid binding protein 3* | 79131 | FABP3 | 1.52 | Cellular growth and proliferation; biomolecule metabolism; molecular transport |
| endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2* | 116744 | EDG2 | 1.51 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; gene transcription |
| killer cell lectin-like receptor subfamily B member 1B* | 25192 | KLRB1 | 1.51 | Cell death |
| suppression of tumorigenicity 18* | 266680 | ST18 | 1.50 | Gene transcription |
| pregnancy upregulated nonubiquitously expressed CaM kinase* | 29660 | PNCK | 1.49 | Others/unclassified |
| Arg/Abl-interacting protein ArgBP2* | 114901 | SORBS2 | 1.48 | Cell death; cell morphology; cell signaling |
| sodium channel, voltage-gated, type IV, alpha polypeptide* | 25722 | SCN4A | 1.48 | Molecular transport |
| calcitonin/calcitonin-related polypeptide, alpha* | 24241 | CALCA | 1.47 | Cell cycle; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair; gene transcription; inflammation; biomolecule metabolism; molecular transport |
| RASD family, member 2* | 171099 | RASD2 | 1.47 | Others/unclassified |
| spondin 1* | 64456 | SPON1 | 1.46 | Others/unclassified |
| sulfotransferase family, cytosolic, 1C, member 2* | 171072 | SULT1C2 | 1.46 | Others/unclassified |
| acyl-CoA synthetase long-chain family member 4* | 113976 | ACSL4 | 1.45 | Cell death |
| aldehyde dehydrogenase family 1, subfamily A2* | 116676 | ALDH1A2 | 1.45 | Cell death; cellular development; cellular growth and proliferation; biomolecule metabolism |
| CD3 antigen, zeta polypeptide* | 25300 | CD247 | 1.45 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; gene transcription; inflammation; post-translational modification |
| discs, large homolog 4 (Drosophila)* | 29495 | DLG4 | 1.45 | Cell signaling; cellular assembly and organization |
| calmodulin 3 | 24244 | CALM3 | 1.43 | Cell signaling; cellular growth and proliferation |
| carboxypeptidase E* | 25669 | CPE | 1.43 | Biomolecule metabolism; molecular transport |
| phosphofructokinase, muscle* | 65152 | PFKM | 1.42 | Others/unclassified |
| translocase of outer mitochondrial membrane 20 homology (yeast)* | 266601 | TOMM20 | 1.42 | Others/unclassified |
| complement component 1, q subcomponent, beta polypeptide* | 29687 | C1QB | 1.40 | Others/unclassified |
| regulator of G-protein signaling 19* | 59293 | RGS19 | 1.40 | Cell signaling; cellular development; biomolecule metabolism; protein synthesis |
| Carcinoembryonic antigen gene family (CGM3)* | 24256 | PSG18 | 1.39 | Others/unclassified |
| gamma-glutamyl hydrolase* | 25455 | GGH | 1.39 | Others/unclassified |
| N-acetyltransferase 8 (camello like)* | 64570 | NAT8 | 1.37 | Others/unclassified |
| pyrimidinergic receptor P2Y, G-protein-coupled, 6* | 117264 | P2RY6 | 1.37 | Others/unclassified |
| protein tyrosine phosphatase, receptor type, K, extracellular region* | 360302 | PTPRK | 1.36 | Cellular growth and proliferation |
| cytochrome P450, 4a12* | 266674 | CYP4A22 | 1.35 | Others/unclassified |
| gamma-aminobutyric acid A receptor, alpha 1* | 29705 | GABRA1 | 1.35 | Cell morphology; cell signaling |
| ferritin, heavy polypeptide 1* | 25319 | FTH1 | 1.34 | Cell death; cell morphology; cellular growth and proliferation; DNA replication, recombination, and repair; free radical scavenging; cell death; cell morphology; cellular growth and proliferation; free radical scavenging |
| guanylate cyclase 1, soluble, beta 2* | 25206 | GUCY1B2 | 1.34 | Others/unclassified |
| proteasome (prosome, macropain) 28 subunit, beta* | 29614 | PSME2 | 1.31 | Cell signaling; cellular growth and proliferation |
| small inducible cytokine A4* | 116637 | CCL4 | 1.31 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; inflammation biomolecule metabolism |
| ADP-ribosylation factor 6* | 79121 | ARF6 | 1.30 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular movement; biomolecule metabolism; molecular transport; protein trafficking |
| mannan-binding lectin serine protease 2 | 64459 | MASP2 | 1.30 | Others/unclassified |
| dopamine receptor 4* | 25432 | DRD4 | 1.29 | Cell signaling; biomolecule metabolism; molecular transport |
| ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit* | 245965 | ATP5D | 1.26 | Energy production; molecular transport; biomolecule metabolism |
| calponin 1 | 65204 | CNN1 | 1.26 | Cellular assembly and organization; cellular growth and proliferation; cellular movement |
| hydroxyacid oxidase 2 (long chain) | 84029 | HAO2 | 1.25 | Others/unclassified |
| cytotoxic T-lymphocyte-associated protein 4 | 63835 | CTLA4 | 1.23 | Cell cycle; cell death; cell signaling; cellular development; cellular growth and proliferation; cellular movement; inflammation |
| potassium voltage-gated channel, subfamily H (eag-related), member 7* | 170739 | KCNH7 | 1.23 | Others/unclassified |
| Ras association (RalGDS/AF-6) domain family 5 | 54355 | RASSF5 | 1.23 | Cell cycle; cell death; cellular growth and proliferation |
| thymopoietin | 25359 | TMPO | 1.22 | Cell cycle; cellular assembly and organization; DNA replication, recombination, and repair; gene transcription |
| polypyrimidine tract binding protein 1 | 29497 | PTBP1 | 1.19 | Protein synthesis |
| protein kinase C and casein kinase substrate in neurons 2 | 124461 | PACSIN2 | 1.16 | Cell morphology; cell signaling; cellular assembly and organization |
| leptin receptor overlapping transcript | 56766 | LEPROT | 1.15 | Others/unclassified |
| phospholipase C, delta 1 | 24655 | PLCD1 | 1.15 | Biomolecule metabolism; cellular growth and proliferation |
| synaptonemal complex protein SC65 | 59101 | SC65 | 1.15 | Others/unclassified |
| ADP-ribosylation factor 5 | 79117 | ARF5 | 1.14 | Molecular transport; protein trafficking |
| crystallin, beta B2 | 25422 | CRYBB2 | 1.11 | Others/unclassified |
| prolactin-like protein L | 171556 | PRLPL | 1.10 | Others/unclassified |
| protein phosphatase 3, catalytic subunit, beta isoform | 24675 | PPP3CB | 1.09 | Cell death; cellular development; inflammation |
| calpain 3 | 29155 | CAPN3 | 1.07 | Protein synthesis |
| cathepsin D | 171293 | CTSD | 1.07 | Cell death; cellular growth and proliferation; free radical scavenging; inflammation; molecular transport; protein synthesis |
| eukaryotic translation initiation factor 2B, subunit 2 beta | 84005 | EIF2B2 | 1.07 | Cellular development; cellular growth and proliferation; protein synthesis |
| calcitonin/calcitonin-related polypeptide, alpha | 24241 | CALCA | -1.13 | Cell cycle; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair; gene transcription; inflammation; biomolecule metabolism; molecular transport |
| forkhead box M1 | 58921 | FOXM1 | -1.13 | Cell cycle; cell death; cell morphology; cellular growth and proliferation; gene transcription |
| adrenal secretory serine protease precursor | 64565 | TMPRSS11D | -1.14 | Others/unclassified |
| MAP-kinase activating death domain | 94193 | MADD | -1.19 | Cell death; cellular growth and proliferation |
| calmodulin 3 | 24244 | CALM3 | -1.22 | Cell signaling; cellular growth and proliferation |
| neural visinin-like Ca2+-binding protein type 3* | 50871 | HPCAL1 | -1.28 | Others/unclassified |
| fertility protein SP22 | 117287 | PARK7 | -1.30 | Cell death; cell signaling; cellular growth and proliferation |
| actin, beta* | 81822 | ACTB | -1.31 | Cellular growth and proliferation; cellular movement |
| G-protein-coupled receptor 37* | 117549 | GPR37 | -1.33 | Cell death; cell signaling |
| glycogen synthase kinase 3 alpha* | 50686 | GSK3A | -1.34 | Cellular movement |
| ribosomal protein L22 | 81768 | RPL22 | -1.34 | Protein synthesis |
| actinin alpha 4* | 63836 | ACTN4 | -1.35 | Cell death; cellular growth and proliferation; cellular movement |
| midline 1* | 54252 | MID1 | -1.35 | Cellular assembly and organization |
| dipeptidase 1 (renal)* | 94199 | DPEP1 | -1.36 | Others/unclassified |
| polymeric immunoglobulin receptor* | 25046 | PIGR | -1.36 | Cell signaling; cellular growth and proliferation; cellular movement |
| adducin 2 (beta)* | 24171 | ADD2 | -1.38 | Cellular development; cellular growth and proliferation; inflammation; molecular transport |
| c-mer protooncogene tyrosine kinase* | 65037 | MERTK | -1.38 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation |
| caspase 7* | 64026 | CASP7 | -1.39 | Cell death; protein synthesis |
| ddx5 gene | 287765 | DDX5 | -1.40 | Cell death; cellular growth and proliferation; gene transcription |
| vesicle docking protein* | 56042 | VDP | -1.40 | Cellular assembly and organization; molecular transport; protein trafficking |
| acetylcholinesterase* | 83817 | ACHE | -1.41 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; DNA replication, recombination, and repair; post-translational modification; protein synthesis |
| phenylalkylamine Ca2+ antagonist (emopamil) binding protein* | 117278 | EBP | -1.41 | Cellular development |
| phosphodiesterase 4C, cAMP-specific* | 290646 | PDE4C | -1.41 | Inflammation |
| transition protein 2* | 24840 | TNP2 | -1.44 | Others/unclassified |
| casein kinase 1, alpha 1* | 113927 | CSNK1A1 | -1.45 | Cell death |
| nuclear receptor subfamily 4, group A, member 3 | 58853 | NR4A3 | -1.46 | Cellular development; cellular growth and proliferation; gene transcription |
| phospholipase C, beta 3* | 29322 | PLCB3 | -1.46 | Biomolecule metabolism; cellular movement; molecular transport |
| myelin-associated oligodendrocytic basic protein* | 25037 | MOBP | -1.47 | Others/unclassified |
| retinoic acid receptor, alpha* | 24705 | RARA | -1.47 | Cell death; cell signaling; cellular development; cellular growth and proliferation; cellular movement; biomolecule metabolism; gene transcription; inflammation |
| putative pheromone receptor Go-VN13C* | 286986 | EG665255 | -1.48 | Others/unclassified |
| prolyl 4-hydroxylase, beta polypeptide* | 25506 | P4HB | -1.49 | Cell death |
| protein kinase, lysine-deficient 1* | 116477 | WNK1 | -1.49 | Molecular transport |
| Pyruvate carboxylase* | 25104 | PC | -1.49 | Others/unclassified |
| ribosomal protein S15* | 29285 | RPS15 | -1.49 | Others/unclassified |
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit* | 192241 | ATP5O | -1.50 | Others/unclassified |
| granzyme M (lymphocyte met-ase 1)* | 29252 | GZMM | -1.50 | Cell death |
| metastasis-associated 1* | 64520 | MTA1 | -1.50 | Cell morphology; cell signaling; cellular growth and proliferation; cellular movement; gene transcription |
| Rous sarcoma oncogene* | 83805 | SRC | -1.50 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; gene transcription; biomolecule metabolism; molecular transport; protein synthesis |
| neurexophilin 3* | 59315 | NXPH3 | -1.51 | Others/unclassified |
| phosphodiesterase 4A* | 25638 | PDE4A | -1.51 | Cell death; cell signaling; cellular development; inflammation; molecular transport; biomolecule metabolism |
| calcium channel, voltage-dependent, alpha 1I subunit* | 56827 | CACNA1I | -1.52 | Cell signaling |
| transient receptor potential cation channel, subfamily C, member 4* | 84494 | TRPC4 | -1.53 | Others/unclassified |
| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide* | 29753 | YWHAE | -1.53 | Cell cycle; cell signaling; cellular movement |
| polymerase (RNA) II (DNA-directed) polypeptide G* | 117017 | POLR2G | -1.54 | Gene transcription |
| F-box only protein 2* | 85273 | FBXO2 | -1.55 | Cellular growth and proliferation; protein synthesis |
| apolipoprotein C–I* | 25292 | APOC1 | -1.56 | Biomolecule metabolism; molecular transport |
| proprotein convertase subtilisin/kexin type 3* | 54281 | FURIN | -1.56 | Cell signaling; cellular growth and proliferation; cellular movement; protein synthesis |
| chemokine orphan receptor 1* | 84348 | CXCR7 | -1.57 | Cellular growth and proliferation; cellular movement |
| heat shock protein, alpha-crystallin-related, B6* | 192245 | HSPB6 | -1.57 | Others/unclassified |
| interleukin 5* | 24497 | IL5 | -1.57 | Cell cycle; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; gene transcription; molecular transport |
| interleukin enhancer binding factor 3* | 84472 | ILF3 | -1.57 | Cellular growth and proliferation; gene transcription |
| protein tyrosine phosphatase, receptor type, F* | 360406 | PTPRF | -1.57 | Cell cycle; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular growth and proliferation; cellular movement |
| par-3 (partitioning defective 3) homolog (C. elegans)* | 81918 | PARD3 | -1.58 | Cell morphology; cell signaling; cellular development; gene Transcription |
| ribosomal protein L27* | 64306 | RPL27 | -1.58 | Others/unclassified |
| linker for activation of T cells* | 81511 | LAT | -1.59 | Biomolecule metabolism; cell death; cell morphology; cell signaling; cellular development; gene transcription; molecular transport |
| nuclear factor I/C* | 29228 | NFIC | -1.59 | Gene transcription |
| potassium voltage-gated channel, shaker-related subfamily, beta member 1* | 29737 | KCNAB1 | -1.59 | Molecular transport |
| Bcl2-associated X protein* | 24887 | BAX | -1.60 | Cell cycle; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular response to therapeutics; DNA replication, recombination, and repair; free radical scavenging; inflammation; biomolecule metabolism; molecular transport |
| carboxylesterase 1* | 29225 | ES22 | -1.60 | Others/unclassified |
| ribosomal protein L10A* | 81729 | RPL10A | -1.60 | Others/unclassified |
| upstream of NRAS* | 117180 | CSDE1 | -1.60 | Others/unclassified |
| nucleoporin 62* | 65274 | NUP62 | -1.61 | Cell death; cell signaling; cellular growth and proliferation; gene transcription |
| eukaryotic translation initiation factor 2B, subunit 4 delta* | 117019 | EIF2B4 | -1.62 | Cellular development; protein synthesis |
| apolipoprotein A–V* | 140638 | APOA5 | -1.63 | Biomolecule metabolism; molecular transport |
| myosin IE* | 25484 | MYO1E | -1.64 | Others/unclassified |
| paired box gene 8* | 81819 | PAX8 | -1.64 | Cellular development; gene transcription |
| prion protein* | 24686 | PRNP | -1.64 | Cell death; cellular development; cellular growth and proliferation; cellular movement |
| amelogenin X chromosome* | 29160 | AMELX | -1.67 | Others/unclassified |
| Unc4.1 homeobox (C. elegans)* | 29375 | UNCX4.1 | -1.67 | Cell death |
| cofilin 1* | 29271 | CFL1 | -1.68 | Cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; molecular transport; protein trafficking |
| caudal type homeo box 1* | 171042 | CDX1 | -1.69 | Others/unclassified |
| phosphatidylinositol-4-phosphate 5-kinase, type II, alpha* | 116723 | PIP5K2A | -1.70 | Biomolecule metabolism; molecular transport |
| platelet-activating factor acetylhydrolase, isoform 1b, alpha1 subunit* | 114113 | PAFAH1B3 | -1.70 | Cell death; cellular development; biomolecule metabolism |
| protein tyrosine phosphatase, nonreceptor type substrate 1* | 25528 | SIRPA | -1.70 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; gene transcription; inflammation |
| acetylcholinesterase* | 83817 | ACHE | -1.71 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; DNA replication, recombination, and repair; post-translational modification; protein synthesis |
| ribosomal protein L36* | 58927 | RPL36 | -1.71 | Others/unclassified |
| G-protein-coupled receptor 24* | 83567 | MCHR1 | -1.72 | Cell signaling; molecular transport; biomolecule metabolism |
| epididymal retinoic acid-binding protein* | 29552 | LCN5 | -1.73 | Biomolecule metabolism |
| gap junction membrane channel protein beta 4* | 117055 | GJB4 | -1.75 | Others/unclassified |
| zinc finger protein 111* | 170849 | ZNF227 | -1.75 | Others/unclassified |
| cleavage and polyadenylation-specific factor 4* | 252943 | CPSF4 | -1.76 | Cellular growth and proliferation |
| ADP-ribosylation factor 1* | 64310 | ARF1 | -1.77 | Cellular assembly and organization; cellular growth and proliferation; biomolecule metabolism; molecular transport; protein trafficking |
| coagulation factor X* | 29243 | F10 | -1.77 | Cell signaling; cellular movement; inflammation; biomolecule metabolism; molecular transport |
| ephrin A1* | 94268 | EFNA1 | -1.77 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement |
| rabaptin 5* | 54190 | RABEP1 | -1.78 | Others/unclassified |
| allograft inflammatory factor 1* | 29427 | AIF1 | -1.79 | Cell death; cell morphology; cellular development; cellular growth and proliferation |
| Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1* | 64466 | CITED1 | -1.79 | Cellular development; cellular growth and proliferation; gene transcription |
| testis enhanced gene transcript* | 24822 | TEGT | -1.79 | Cell death |
| neurogenic differentiation 2* | 54276 | NEUROD2 | -1.80 | Cellular development; gene transcription |
| heat shock 10 kDa protein 1* | 25462 | HSPE1 | -1.83 | Cell death |
| hsp70-interacting protein* | 246146 | HSPBP1 | -1.83 | Others/unclassified |
| calreticulin* | 64202 | CALR | -1.84 | Cell death; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; gene transcription; molecular transport; protein trafficking |
| discoidin domain receptor family, member 1* | 25678 | DDR1 | -1.84 | Cell death; cellular development; cellular growth and proliferation; cellular movement |
| myosin ID* | 25485 | MYO1D | -1.84 | Others/unclassified |
| ribosomal protein L28* | 64638 | RPL28 | -1.84 | Others/unclassified |
| gap junction membrane channel protein alpha 3* | 79217 | GJA3 | -1.85 | Cell morphology; cell signaling; cellular development |
| hairy and enhancer of split 3 (Drosophila)* | 64628 | HES3 | -1.85 | Cellular development; gene transcription |
| ornithine decarboxylase antizyme 1* | 25502 | OAZ1 | -1.87 | Cell death; cellular growth and proliferation |
| a disintegrin and metallopeptidase domain 1a* | 56777 | ADAM1A | -1.89 | Cellular movement |
| mucosal vascular addressin cell adhesion molecule 1* | 54266 | MADCAM1 | -1.89 | Cell signaling; cellular development; cellular movement; inflammation |
| membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)* | 114202 | MPP3 | -1.90 | Others/unclassified |
| sulfotransferase family 4A, member 1* | 58953 | SULT4A1 | -1.90 | Others/unclassified |
| CCAAT/enhancer binding protein (C/EBP), alpha* | 24252 | CEBPA | -1.91 | Cell cycle; cell death; cellular development; cellular growth and proliferation; cellular response to therapeutics; gene transcription; biomolecule metabolism; molecular transport |
| SNF-related kinase* | 170837 | SNRK | -1.92 | Cellular development |
| ADP-ribosylation factor 3* | 140940 | ARF3 | -1.94 | Molecular transport; protein trafficking |
| aspartyl-tRNA synthetase* | 116483 | DARS | -1.94 | Cell cycle; cell signaling; protein synthesis |
| glycine cleavage system protein H (aminomethyl carrier)* | 171133 | GCSH | -1.94 | Biomolecule metabolism; post-translational modification |
| 3-hydroxyisobutyrate dehydrogenase* | 63938 | HIBADH | -1.96 | Others/unclassified |
| preoptic regulatory factor-2* | 286903 | KIAA1688 | -1.96 | Others/unclassified |
| amino-terminal enhancer of split* | 29466 | AES | -1.97 | Cell death; gene transcription |
| ribosomal protein L29* | 29283 | RPL29 | -1.99 | Others/unclassified |
| quinoid dihydropteridine reductase* | 64192 | QDPR | -2.01 | Others/unclassified |
| complexin 2* | 116657 | CPLX2 | -2.02 | Cellular assembly and organization; cellular movement |
| hairy and enhancer of split 2 (Drosophila)* | 29567 | HES2 | -2.02 | Gene transcription |
| transmembrane 4 superfamily member 11* | 64364 | PLLP | -2.02 | Molecular transport |
| ferritin light chain 1* | 29292 | FTL | -2.03 | Cellular growth and proliferation |
| growth hormone-releasing hormone receptor* | 25321 | GHRHR | -2.04 | Cell signaling; cellular growth and proliferation; molecular transport; biomolecule metabolism |
| membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)* | 85275 | MPP2 | -2.04 | Cell signaling |
| ribosomal protein S27* | 94266 | RPS27 | -2.05 | Cell signaling; cellular growth and proliferation; protein synthesis |
| translocase of inner mitochondrial membrane 22 homolog (yeast)* | 79463 | TIMM22 | -2.06 | Others/unclassified |
| sec22 homolog* | 117513 | SEC22A | -2.08 | Molecular transport; protein trafficking |
| transmembrane protein with EGF-like and two follistatin-like domains 1* | 63845 | TMEFF1 | -2.11 | Cellular growth and proliferation |
| paired-like homeodomain transcription factor 3* | 29609 | PITX3 | -2.12 | Cellular development; gene transcription |
| fibrinogen, gamma polypeptide* | 24367 | FGG | -2.25 | Cell signaling |
| neurogenic differentiation 1* | 29458 | NEUROD1 | -2.26 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; gene transcription |
| steroid-sensitive gene 1* | 64387 | CCDC80 | -2.31 | Others/unclassified |
| ubiquitin A-52 residue ribosomal protein fusion product 1* | 64156 | UBA52 | -2.33 | Gene transcription; protein synthesis |
| synuclein, gamma* | 64347 | SNCG | -2.42 | Cell death; cell morphology; cellular growth and proliferation; cellular movement |
The change in expression was significant compared with untreated control (P < .05). Gene names in italics are common in both panels of T + E2-treated LP and T + DES-treated VP.
Table 2.
T + DES-Induced VP Dysplasia Panel: Genes Whose Expression Selectively Changed following T + DES Treatment in the VPs Harboring Dysplasia, but not in the LPs.
| Gene Names | Locus ID | Symbol | Fold Changes | Functions |
| glutathione peroxidase 3* | 64317 | GPX3 | 17.188 | Cellular development; cellular growth and proliferation; post-translational modification; protein synthesis |
| ferritin, heavy polypeptide 1* | 25319 | FTH1 | 6.304 | Cell death; cell morphology; cellular growth and proliferation; DNA replication, recombination, and repair; free radical scavenging; post-translational modification |
| protein phosphatase 2a, catalytic subunit, alpha isoform* | 24672 | PPP2CA | 3.388 | Biomolecule metabolism; cell death; cellular growth and proliferation; post-translational modification |
| monocarboxylate transporter* | 80878 | SLC16A3 | 2.263 | Others/unclassified |
| nuclease-sensitive element binding protein 1* | 29206 | NSEP1 | 1.898 | Cell death; gene transcription |
| thymopoietin* | 25359 | TMPO | 1.814 | Cell cycle; DNA replication, recombination, and repair; gene transcription |
| homeo box A1* | 25607 | HOXA1 | 1.586 | Cell death; cellular development; cellular movement; gene transcription |
| cystatin C* | 25307 | CST3 | 1.585 | Cell death; cellular development; cellular growth and proliferation |
| thyroid hormone receptor alpha* | 81812 | THRA | 1.583 | Cell death; cell morphology; cellular development; free radical scavenging; gene transcription |
| tubulin, alpha 1* | 64158 | TUBA1A | 1.580 | Inflammation |
| phospholipase C, delta 1* | 24655 | PLCD1 | 1.532 | Cellular growth and proliferation |
| glutamate receptor, metabotropic 7* | 81672 | GRM7 | 1.481 | Cell signaling |
| potassium voltage-gated channel, subfamily H (eag-related),member 7* | 170739 | KCNH7 | 1.481 | Others/unclassified |
| plasminogen activator, urokinase* | 25619 | PLAU | 1.461 | Cell cycle; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; inflammation |
| vesicle-associated membrane protein 2 | 24803 | VAMP2 | 1.452 | Cell signaling; cellular assembly and organization; cellular movement; molecular transport |
| glyceraldehyde-3-phosphate dehydrogenase* | 24383 | GAPDH | 1.401 | Others/unclassified |
| protein phosphatase 1F (PP2C domain containing)* | 287931 | PPM1F | 1.392 | Biomolecule metabolism; cell death; post-translational modification |
| tumor protein p53* | 24842 | TP53 | 1.379 | Cell cycle; cell death; cell morphology; cell signaling; cellular assembly and; organization; cellular development; cellular growth and proliferation; cellular movement; cellular response to therapeutics; DNA replication, recombination, and repair; free radical scavenging; gene transcription; inflammation; post-translational modification; protein synthesis |
| opioid receptor, mu 1* | 25601 | OPRM1 | 1.375 | Cell death; cell signaling; cellular growth and proliferation; cellular movement; inflammation |
| calcitonin/calcitonin-related polypeptide, alpha* | 24241 | CALCA | 1.361 | Cell cycle; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair; gene transcription; inflammation |
| testis-specific protein* | 192229 | C3ORF34 | 1.357 | Others/unclassified |
| eukaryotic translation initiation factor 2B, subunit 2 beta* | 84005 | EIF2B2 | 1.334 | Cellular development; cellular growth and proliferation |
| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide* | 25576 | YWHAH | 1.333 | Cellular assembly and organization; gene transcription |
| glucagon-like peptide 1 receptor | 25051 | GLP1R | 1.328 | Cell death; gene transcription; inflammation |
| transient receptor potential cation channel, subfamily C, member 2* | 64573 | TRPC2 | 1.322 | Others/unclassified |
| dopamine receptor 4* | 25432 | DRD4 | 1.311 | Cell signaling |
| early growth response 3* | 25148 | EGR3 | 1.289 | Cell death; cell signaling; cellular development; cellular growth and proliferation; gene transcription |
| glutamate receptor, ionotropic, NMDA2B* | 24410 | GRIN2B | 1.248 | Cell morphology; cell signaling |
| chemokine (C-X-C motif) ligand 5* | 60665 | CXCL6 | 1.236 | Cell signaling; cellular movement |
| myosin IC | 65261 | MYO1C | 1.190 | Cell morphology |
| breast cancer anti-estrogen resistance 1 | 25414 | BCAR1 | 1.190 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement |
| aryl hydrocarbon receptor nuclear translocator 2 | 25243 | ARNT2 | 1.172 | Gene transcription |
| CCAAT/enhancer binding protein (C/EBP), delta | 25695 | CEBPD | 1.151 | Cell cycle; cell death; cellular development; cellular growth and proliferation; gene transcription; inflammation; biomolecule metabolism; molecular transport |
| CTD-binding SR-like rA1 | 56081 | SR-A1 | 1.148 | Others/unclassified |
| ADP-ribosylation factor 6 | 79121 | ARF6 | 1.142 | Cell death; cell morphology; cellular assembly and organization; cellular movement |
| calmodulin 3 | 24244 | CALM3 | 1.138 | Cell death; cellular growth and proliferation; post-translational modification |
| synaptotagmin 3 | 25731 | SYT3 | 1.115 | Cell signaling; cellular assembly and organization; cellular movement; molecular transport |
| dynein, cytoplasmic, light chain 1 | 58945 | DYNLL1 | 1.090 | Cell death; cell morphology; cellular assembly and organization |
| MAD homolog 7 (Drosophila) | 81516 | SMAD7 | 1.062 | Cell cycle; cell death; cell morphology; cellular development; cellular growth and proliferation; cellular movement; gene transcription |
| v-maf musculoaponeurotic fibrosarcom oncogene family, protein B (avian) | 54264 | MAFB | 1.062 | Cell death; cellular development; cellular movement; gene transcription |
| calmodulin 3 | 24244 | CALM3 | 1.060 | Cellular growth and proliferation; post-translational modification |
| thymosin, beta 10 | 50665 | TMSB10 | 1.059 | Cell death; cellular assembly and organization; cellular growth and proliferation |
| aldehyde dehydrogenase family 1, subfamily A2 | 116676 | ALDH1A2 | 1.035 | Cell death; cellular development; cellular growth and proliferation |
| syntaxin 1B2 | 24923 | STX1B2 | 1.032 | Others/unclassified |
| aldolase A | 24189 | ALDOA | -1.013 | Others/unclassified |
| prosaposin | 25524 | PSAP | -1.031 | Cell cycle; cell death; cellular growth and proliferation; biomolecule metabolism; molecular transport; post-translational modification |
| vesicle-associated membrane protein 2 | 24803 | VAMP2 | -1.031 | Cell death; cell signaling; cellular assembly and organization; cellular movement; molecular transport |
| Rous sarcoma oncogene | 83805 | SRC | -1.070 | Biomolecule metabolism; cell cycle; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair; gene transcription; molecular transport; post-translational modification |
| acetylcholinesterase | 83817 | ACHE | -1.072 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; DNA replication, recombination, and repair; post-translational modification; protein synthesis |
| Max interacting protein 1 | 25701 | MXI1 | -1.120 | Cell morphology; cellular growth and proliferation; gene transcription |
| betacellulin | 64022 | BTC | -1.137 | Cell cycle; cell death; cell signaling; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair |
| argininosuccinate synthetase | 25698 | ASS1 | -1.165 | Others/unclassified |
| nuclear receptor subfamily 1, group D, member 2 | 259241 | NR1D2 | -1.170 | Gene transcription |
| stannin | 29140 | SNN | -1.242 | Cell death |
| phosphatase and tensin homolog* | 50557 | PTEN | -1.272 | Cell cycle; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; gene transcription inflammation; biomolecule metabolism; molecular transport; post-translational modification |
| solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7* | 116638 | SLC17A7 | -1.287 | Cell signaling |
| carnitine palmitoyltransferase 1, liver* | 25757 | CPT1A | -1.291 | Biomolecule metabolism |
| isopentenyl-diphosphate delta isomerase* | 89784 | IDI1 | -1.294 | Others/unclassified |
| LIM homeobox protein 5 | 124451 | LHX5 | -1.317 | Others/unclassified |
| calpain, small subunit 1* | 29156 | CAPNS1 | -1.318 | Cell death; cellular assembly and organization; cellular growth and proliferation; cellular movement |
| filaggrin | 24641 | FLG | -1.325 | Cellular assembly and organization |
| microsomal glutathione S-transferase 1 | 171341 | MGST1 | -1.329 | Biomolecule metabolism |
| protein tyrosine phosphatase, nonreceptor type substrate 1* | 25528 | SIRPA | -1.330 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; gene transcription; inflammation |
| neurofibromatosis 2 | 25744 | NF2 | -1.333 | Cell death; cellular growth and proliferation; cellular movement |
| Notch gene homolog 3 (Drosophila)* | 56761 | NOTCH3 | -1.335 | Cellular development; gene transcription |
| killer cell lectin-like receptor subfamily B member 1B* | 25192 | KLRB1 | -1.338 | Cell death |
| basic transcription element binding protein 1* | 117560 | KLF9 | -1.339 | Gene transcription |
| regulating synaptic membrane exocytosis 1* | 84556 | RIMS1 | -1.353 | Cell morphology; cell signaling; cellular assembly and organization; cellular movement; molecular transport |
| transglutaminase 1* | 60335 | TGM1 | -1.376 | Biomolecule metabolism; cell death; cellular development; post-translational modification |
| translocase of inner mitochondrial membrane 23 homolog (yeast)* | 54312 | IMM23 | -1.393 | Others/unclassified |
| lysozyme | 25211 | LYZ | -1.399 | Others/unclassified |
| solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11* | 64201 | SLC25A11 | -1.407 | Others/unclassified |
| hydroxysteroid 11-beta dehydrogenase 1* | 25116 | HSD11B1 | -1.407 | Cellular development; cellular growth and proliferation |
| potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3* | 54263 | KCNN3 | -1.412 | Cell signaling |
| B-cell leukemia/lymphoma 2* | 24224 | BCL2 | -1.413 | Cell cycle; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; cellular response to therapeutics; DNA replication, recombination, and repair; biomolecule metabolism; gene transcription; inflammation; molecular transport; post-translational modification |
| hyperpolarization-activated, cyclic nucleotide-gated K+ 4* | 59266 | HCN4 | -1.415 | Cell signaling |
| perforin 1 (pore-forming protein)* | 50669 | PRF1 | -1.417 | Cell death; cell morphology; cell signaling; DNA replication, recombination, and repair; inflammation |
| calcitonin/calcitonin-related polypeptide, alpha* | 24241 | CALCA | -1.433 | Cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair; gene transcription; inflammation |
| nuclear RNA export factor 1* | 59087 | NXF1 | -1.436 | Protein synthesis |
| fibroblast growth factor 14* | 63851 | FGF14 | -1.439 | Others/unclassified |
| solute carrier family 7 (cationic amino acid transporter,y+ system), member 3* | 29485 | SLC7A3 | -1.455 | Others/unclassified |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit b, isoform 1* | 171375 | ATP5F1 | -1.456 | Cellular growth and proliferation |
| septin 3* | 56003 | SEPT3 | -1.468 | Others/unclassified |
| cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal)* | 54239 | CHRNB2 | -1.469 | Cell signaling |
| wild-type p53-induced gene 1* | 64394 | ZMAT3 | -1.480 | Cell death; cellular growth and proliferation; DNA replication, recombination, and repair |
| glutamate receptor, ionotropic, kainate 4* | 24406 | GRIK4 | -1.482 | Cell signaling |
| Kruppel-like factor 15* | 85497 | KLF15 | -1.488 | Gene transcription |
| aquaporin 6* | 29170 | AQP6 | -1.496 | Others/unclassified |
| unc-5 homolog A (C. elegans)* | 60629 | UNC5A | -1.498 | Cell death |
| integrin beta 4* | 25724 | ITGB4 | -1.502 | Cell death; cell morphology; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; gene transcription |
| peptidylprolyl isomerase A* | 25518 | PPIA | -1.526 | Cell death; cellular development; cellular growth and proliferation; cellular movement; DNA replication, recombination, and repair; inflammation; post-translational modification |
| FXYD domain-containing ion transport regulator 6* | 63847 | FXYD6 | -1.562 | Others/unclassified |
| ribosomal protein L37* | 81770 | RPL37 | -1.579 | Others/unclassified |
| RT1 class II, locus Da* | 294269 | HLA-DRA | -1.580 | Cell signaling; inflammation |
| FK506 binding protein 12-rapamycin-associated protein 1* | 56718 | FRAP1 | -1.596 | Biomolecule metabolism; cell cycle; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; gene transcription; inflammation; post-translational modification; protein synthesis |
| eukaryotic translation elongation factor 1 alpha 1* | 171361 | EEF1A1 | -1.618 | Cell death; protein synthesis |
| glutathione S-transferase theta 1* | 25260 | GSTT1 | -1.622 | Biomolecule metabolism |
| solute carrier family 4, member 1* | 24779 | SLC4A1 | -1.630 | Cell death; cellular growth and proliferation |
| fibroblast growth factor 17* | 29368 | FGF17 | -1.642 | Cellular growth and proliferation |
| forkhead box M1* | 58921 | FOXM1 | -1.646 | Cell cycle; cell death; cell morphology; cellular growth and proliferation; gene transcription |
| ATPase, class II, type 9A* | 84011 | ATP9A | -1.649 | Others/unclassified |
| phosphorylase kinase, gamma 2 (testis)* | 140671 | PHKG2 | -1.679 | Biomolecule metabolism; post-translational modification |
| arrestin, beta 1* | 25387 | ARRB1 | -1.683 | Cellular movement |
| rhoB gene* | 64373 | RHOB | -1.684 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; gene transcription |
| par-3 (partitioning defective 3) homolog (C. elegans)* | 81918 | PARD3 | -1.686 | Cell morphology; cell signaling; cellular assembly and organization; cellular development; gene transcription |
| ribosomal protein L21* | 79449 | RPL21 | -1.687 | Others/unclassified |
| RAS protein-specific guanine nucleotide-releasing factor 1* | 192213 | RASGRF1 | -1.715 | Cell morphology; cell signaling; cellular growth and proliferation; gene transcription |
| acetylcholinesterase* | 83817 | ACHE | -1.741 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; DNA replication, recombination, and repair; post-translational modification; protein synthesis |
| ribosomal protein L18* | 81766 | RPL18 | -1.741 | Others/unclassified |
| granzyme M (lymphocyte met-ase 1)* | 29252 | GZMM | -1.745 | Cell death |
| Bcl2-associated X protein* | 24887 | BAX | -1.746 | Cell cycle; cell death; cell morphology; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular response to therapeutics; DNA replication, recombination, and repair; free radical scavenging; inflammation; biomolecule metabolism; molecular transport; protein synthesis |
| legumain* | 63865 | LGMN | -1.749 | Cellular movement |
| RAB6A, member RAS oncogene family* | 84379 | RAB6A | -1.752 | Cellular assembly and organization |
| matrix metalloproteinase 16* | 65205 | MMP16 | -1.763 | Cellular movement; inflammation |
| olfactory receptor 226* | 65140 | OR6A2 | -1.770 | Others/unclassified |
| mitogen-activated protein kinase 14* | 81649 | MAPK14 | -1.775 | Biomolecule metabolism; cell cycle; cell death; cell morphology; cellular development; cellular growth and proliferation; cellular movement; gene transcription; inflammation; post-translational modification |
| c-mer protooncogene tyrosine kinase* | 65037 | MERTK | -1.780 | Biomolecule metabolism; cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; post-translational modification |
| clathrin, heavy polypeptide (Hc)* | 54241 | CLTC | -1.786 | Cell morphology; cellular growth and proliferation |
| transient receptor potential cation channel, subfamily C, member 4* | 84494 | TRPC4 | -1.793 | Others/unclassified |
| carnitine palmitoyltransferase 1b* | 25756 | CPT1B | -1.795 | Biomolecule metabolism |
| PDZ and LIM domain 1 (elfin)* | 54133 | PDLIM1 | -1.796 | Gene transcription |
| fatty acid amide hydrolase* | 29347 | FAAH | -1.798 | Cell death; inflammation |
| fatty acid binding protein 4, adipocyte* | 79451 | FABP4 | -1.820 | Gene transcription |
| protein tyrosine phosphatase, nonreceptor type 12* | 117255 | PTPN12 | -1.826 | Biomolecule metabolism; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular movement; post-translational modification |
| potassium voltage-gated channel, Shab-related subfamily, member 2* | 117105 | KCNB2 | -1.834 | Others/unclassified |
| glutathione S-transferase, pi 2* | 29438 | GSTP1 | -1.856 | Cell death; cellular growth and proliferation; biomolecule metabolism; gene transcription |
| Fc receptor, IgG, alpha chain transporter* | 29558 | FCGRT | -1.870 | Cell signaling; molecular transport |
| voltage-gated channel like 1* | 266760 | VGCNL1 | -1.871 | Others/unclassified |
| prostaglandin D2 synthase* | 25526 | PTGDS | -1.888 | Cell death; cell morphology |
| MAP-kinase activating death domain* | 94193 | MADD | -1.934 | Cell death; cellular growth and proliferation |
| neogenin* | 81735 | NEO1 | -1.935 | Cellular growth and proliferation; cellular movement; gene transcription |
| calbindin 1* | 83839 | CALB1 | -1.950 | Cell death; cell morphology; cell signaling |
| guanylate cyclase 2e* | 79222 | GUCY2D | -1.957 | Others/unclassified |
| adrenal secretory serine protease precursor* | 64565 | TMPRSS11D | -1.971 | Others/unclassified |
| ubc2e ubiquitin-conjugating enzyme* | 641452 | Ube2d2 | -1.986 | Others/unclassified |
| neuroblastoma, suppression of tumorigenicity 1* | 50594 | NBL1 | -2.003 | Cellular movement |
| collagen, type 1, alpha 1* | 29393 | COL1A1 | -2.023 | Cell morphology; cellular growth and proliferation; cellular movement |
| homeodomain interacting protein kinase 3* | 83617 | HIPK3 | -2.034 | Biomolecule metabolism; cell signaling; post-translational modification |
| intercellular adhesion molecule 1* | 25464 | ICAM1 | -2.052 | Cell death; cell morphology; cell signaling; cellular development; cellular growth and proliferation; cellular movement; inflammation |
| cyclin-dependent kinase 5* | 140908 | CDK5 | -2.134 | Biomolecule metabolism; cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; molecular transport; post-translational modification |
| ATPase, Cu2+ transporting, beta polypeptide* | 24218 | ATP7B | -2.137 | Cell death |
| Jun D protooncogene* | 24518 | JUND | -2.159 | Cell death; cell morphology; cellular growth and proliferation; gene transcription |
| neurexophilin 3* | 59315 | NXPH3 | -2.168 | Others/unclassified |
| heterogeneous nuclear ribonucleoprotein methyltransferase-like 3 (S. cerevisiae)* | 89820 | PRMT3 | -2.171 | Biomolecule metabolism; post-translational modification |
| midline 1* | 54252 | MID1 | -2.180 | Cellular assembly and; organization |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 1* | 24565 | ABCC1 | -2.191 | Cell death; cellular movement; inflammation; biomolecule metabolism; molecular transport |
| prothymosin alpha* | 29222 | PTMA | -2.313 | Cell cycle; cell death; cellular development; cellular growth and proliferation; gene transcription |
| ubiquilin 1* | 114590 | UBQLN1 | -2.322 | Others/unclassified |
| similar to Leydig cell tumor 10 kDa protein* | 288913 | C19ORF53 | -2.346 | Others/unclassified |
| Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein* | 25587 | ID2 | -2.357 | Cell cycle; cell death; cell morphology; cellular development; cellular growth and proliferation; cellular movement; gene transcription |
| lectin, galactose binding, soluble 1* | 56646 | LGALS1 | -2.385 | Cell cycle; cell death; cell morphology; cellular development; cellular growth and proliferation; cellular movement gene transcription; inflammation; post-translational modification |
| calreticulin* | 64202 | CALR | -2.401 | Cell cycle; cell death; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement; gene transcription; post-translational modification |
| carbonic anhydrase 5* | 54233 | CA5A | -2.427 | Inflammation |
| mitogen-activated protein kinase 13* | 29513 | MAPK13 | -2.438 | Cell death; cell morphology; gene transcription; inflammation |
| cytochrome P450, family 19, subfamily a, polypeptide 1* | 25147 | CYP19A1 | -2.440 | Cell death; cell morphology; cellular development; cellular growth and proliferation; cellular movement; gene transcription; biomolecule metabolism |
| nucleobindin 1* | 84595 | NUCB1 | -2.513 | Others/unclassified |
| tropomodulin 2* | 58814 | TMOD2 | -2.563 | Cell signaling; cellular assembly and organization |
| apolipoprotein B editing complex 1* | 25383 | APOBEC1 | -2.655 | Others/unclassified |
| solute carrier family 7 (cationic amino acid transporter, y+ system), member 7* | 83509 | SLC7A7 | -2.720 | Cell signaling; cellular growth and proliferation |
| hsp70-interacting protein* | 246146 | HSPBP1 | -2.772 | Post-translational modification |
| Rsec5 protein* | 171455 | EXOC2 | -2.779 | Cell signaling; molecular transport |
| MAD homolog 9 (Drosophila)* | 85435 | SMAD9 | -2.815 | Others/unclassified |
| ERM-binding phosphoprotein* | 59114 | SLC9A3R1 | -2.851 | Cell signaling; cellular growth and proliferation |
| low-density lipoprotein receptor-related protein 3* | 89787 | LRP3 | -2.869 | Others/unclassified |
| Rab geranylgeranyl transferase, a subunit* | 58983 | RABGGTA | -2.890 | Biomolecule metabolism; post-translational modification |
| cadherin EGF LAG seven-pass G-type receptor 2* | 83465 | CELSR2 | -2.923 | Others/unclassified |
| tropomyosin 4* | 24852 | TPM4 | -3.019 | Cellular movement |
| cytochrome P450, family 2, subfamily e, polypeptide 1* | 25086 | CYP2E1 | -3.161 | Cell death; biomolecule metabolism |
| CEA-related cell adhesion molecule 9* | 116711 | CEACAM9 | -3.207 | Others/unclassified |
| discoidin domain receptor family, member 2* | 83573 | DDR2 | -3.253 | Others/unclassified |
| phosphoglucomutase 1* | 24645 | PGM1 | -3.308 | Others/unclassified |
| gamma-glutamyl carboxylase* | 81716 | GGCX | -3.455 | Post-translational modification |
| Cplx1 complexin 1* | 64832 | CPLX1 | -3.464 | Cell signaling; cellular assembly; and organization; cellular movement; molecular transport |
| septin 9* | 83788 | SEPT9 | -3.781 | Cell cycle; protein synthesis |
| matrix metallopeptidase 8* | 63849 | MMP8 | -3.785 | Cell death, cellular movement |
| thyroid hormone receptor interactor 10* | 116717 | TRIP10 | -4.232 | Cell death |
| carboxylesterase 3* | 113902 | CES1 | -4.325 | Others/unclassified |
| cyclin-dependent kinase 2* | 362817 | CDK2 | -4.458 | Biomolecule metabolism; cell cycle; cell death; cell morphology; cellular development; cellular growth and proliferation; DNA replication, recombination, and repair; gene transcription; post-translational modification |
| monoamine oxidase B* | 25750 | MAOB | -4.873 | Cell death |
| solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12* | 50676 | SLC6A12 | -5.287 | Others/unclassified |
| peroxiredoxin 1* | 117254 | PRDX1 | -5.360 | Cell death; cell morphology; cellular growth and proliferation; gene transcription; post-translational modification |
| ADP-ribosylation factor 4* | 79120 | ARF4 | -5.374 | Others/unclassified |
| glypican 3* | 25236 | GPC3 | -5.638 | Cell death; cellular growth and proliferation |
| protein kinase C, delta binding protein* | 85332 | PRKCDBP | -6.059 | Others/unclassified |
| cd86 antigen* | 56822 | CD86 | -6.118 | Cell signaling; cellular development; cellular growth and proliferation; cellular movement; gene transcription; inflammation |
| Sjogren syndrome antigen B* | 81783 | SSB | -6.131 | Gene transcription; protein synthesis |
| secretoglobin, family 2A, member 1* | 25010 | PSBP1 | -6.227 | Others/unclassified |
| procollagen, type I, alpha 2* | 84352 | COL1A2 | -7.076 | Cell morphology |
| A kinase (PRKA) anchor protein 1* | 114124 | AKAP1 | -8.502 | Cell death |
| methionine adenosyltransferase II, alpha* | 171347 | MAT2A | -9.474 | Others/unclassified |
| FXYD domain-containing ion transport regulator 1* | 58971 | FXYD1 | -9.644 | Cellular growth and proliferation; gene transcription |
| CD24 antigen* | 25145 | CD24 | -11.299 | Cell death; cell morphology; cell signaling; cellular assembly and organization; cellular development; cellular growth and proliferation; cellular movement |
| nuclear receptor subfamily 1, group I, member 2* | 84385 | NR1I2 | -11.958 | Biomolecule metabolism; gene transcription; inflammation; molecular transport; post-translational modification |
| glial cell line derived neurotrophic factor family receptor alpha 1* | 25454 | GFRA1 | -17.708 | Cell death; cell morphology; cellular development; cellular; growth and proliferation; cellular movement |
| cell division cycle 25 homolog A (S. cerevisiae)* | 171102 | CDC25A | -19.859 | Cell cycle; cell death; cell morphology; cellular growth and proliferation; dna replication, recombination, and repair |
| 5-hydroxytryptamine (serotonin) receptor 5B* | 79247 | HTR5B | -22.285 | Others/unclassified |
| NSFL1 (p97) cofactor (p47)* | 83809 | NSFL1C | -22.400 | Others/unclassified |
| membrane and microfilament-associated protein p58* | 207121 | RGD: 727794 | -49.533 | Cellular assembly and organization |
The change in expression was significant compared with untreated control (P < .05). Gene names in italics are common in both panels of T + E2-treated LP and T + DES-treated VP.
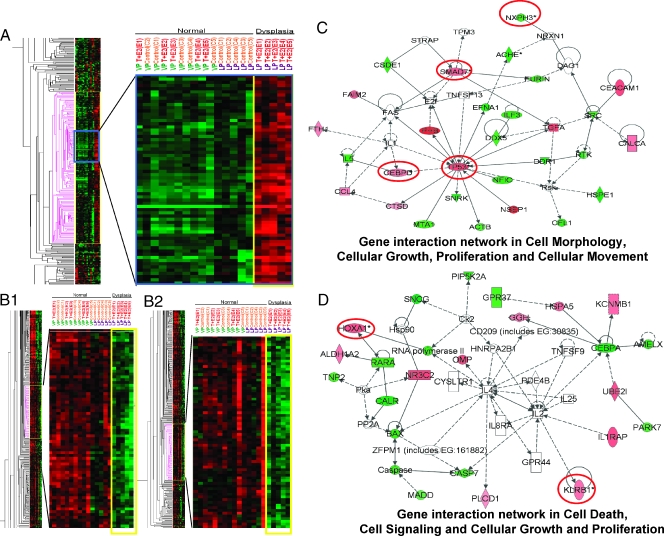
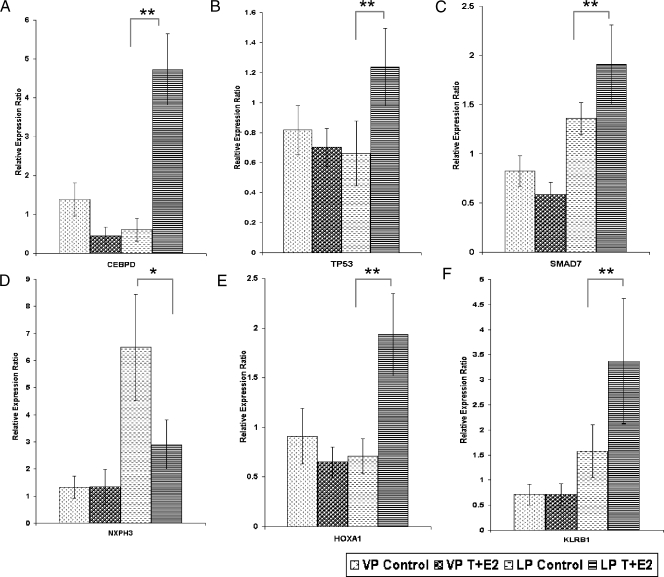
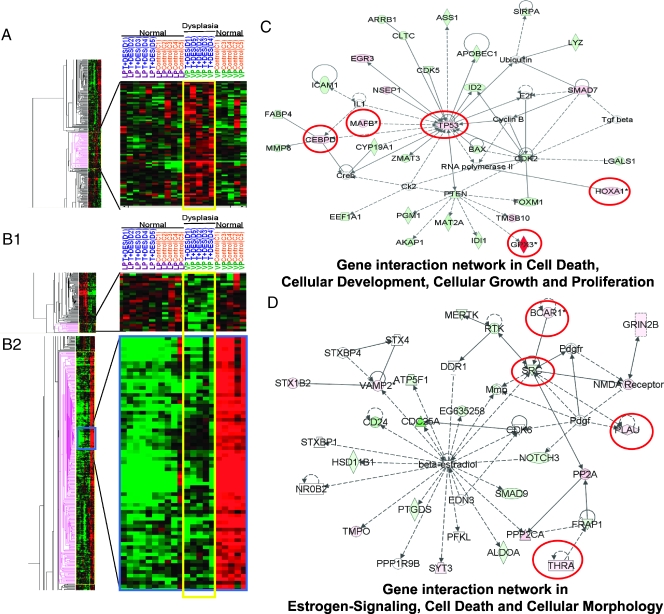
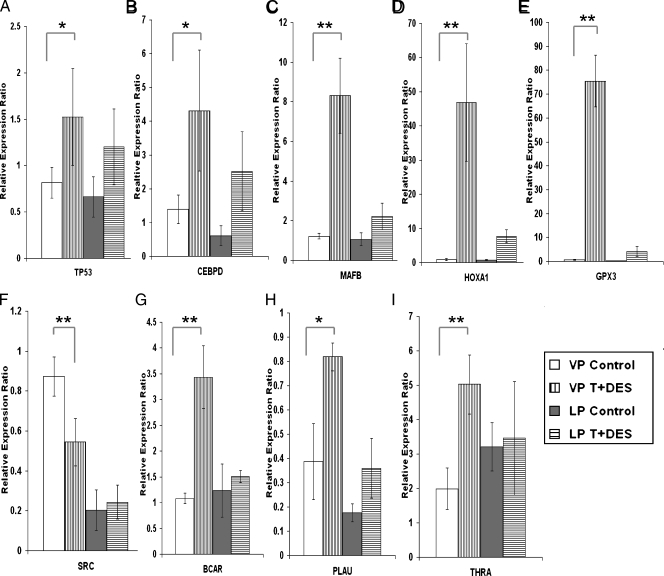
Image plots (Figure 2, A, and B1 and B2) showed the up- and downregulated genes in the T + E2-induced LP dysplasia panel (253 genes), respectively. Although genes in this panel were distributed among 27 IPA networks, two major networks with the highest relevancy scores were identified: one related to cell morphology, cellular growth, proliferation, and movement (Figure 2C), and the other related to apoptosis and cell signaling (Figure 2D). From these two networks, six genes were selected for post hoc confirmation by real-time q-PCR (Figure 3). All six genes showed the predicted expression patterns. Significant differences (P < .01/.05) were observed between the transcript levels in the T + E2-treated and -untreated LPs, whereas no significant differences (P > .05) in the transcript levels were observed between the treated and untreated VP groups. These genes are the CCAAT/enhancer binding protein-delta (Cebpd), the tumor protein Tp53, MAD homolog 7 (Smad7), homeobox A1 (Hoxa1), killer cell lectin-like receptor subfamily B member 1 (Klrb1), and neurexophilin 3 (Nxph3). Notably, the q-PCR data correlated well with those obtained by microarray analyses (Figure W1A). T + E2-response genes found in VP, in contrast to those found in the LP, were not mapped to any particular IPA networks (data not shown).
Figure 2.
Heat maps and gene interaction networks of differentially expressed genes found exclusively in the LP dysplasia following T + E2 treatment. Red and green denote upregulated and downregulated expression, respectively, as compared with the overall gene's mean value normalized to the universal rat reference RNA. Columns represent data from a single prostate sample, and rows correspond to a single gene probe. (A) A single cluster of upregulated genes identified in the T + E2 LP dysplasia, marked with pink (left panel); a selected region (blue box) of this cluster is enlarged (right panel). (B1 and B2) Two separate downregulated gene clusters observed only in T + E2 LP dysplasia, marked with pink (left panels); selected clusters are magnified (right panels). (C and D) Two representative gene interaction networks (with the highest relevancy scores) generated by IPA analysis from the differentially expressed genes in the T + E2 LP dysplasia panel. Green indicates downregulated; red, upregulated. Genes bordered in red were validated by real-time q-PCR. See Figure 1 for key to IPA network.
Figure 3.
Post hoc real-time q-PCR analyses of selected genes in the T + E2 LP dysplasia panel. Data were normalized to the levels of Rpl19. Bars indicate standard deviations (SD) of three to five animals in each treatment group. *P < .05, **P < .01 by one-way ANOVA with Tukey post hoc analysis.
Image plots illustrated distinctive upregulated (Figure 4A) and downregulated (Figure 4, B1 and B2) genes in the T + DES-induced VP dysplasia panel. In addition, a unique set of genes was exclusively repressed by T + DES in the VP harboring dysplasia (Figure 4B2). These genes were also underexpressed in LPs compared with the untreated VPs. Genes in the T + DES-treated VP panel (198 genes) mapped primarily to two major networks related to 1) apoptosis, cellular development, growth, and proliferation and 2) estrogen signaling, as they exhibited the highest relevancy scores (Figure 4, C and D, respectively). From the two networks, nine genes were selected for post hoc confirmation by q-PCR analyses that correlated well with those obtained by microarrays (Figure W1B). All nine genes showed significant increases in transcript levels (P < .01/.05) in the VP with T + DES treatment and exhibited little or no change in expression (P > .05) in the LPs of the treated group compared with untreated controls (Figure 5). They include Cebpd, Tp53, v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (Mafb), Hoxa1, breast cancer anti-estrogen resistance 1 (Bcar1), plasminogen activator urokinase (Plau), thyroid hormone receptor alpha (Thra), glutathione peroxidase 3 (Gpx3), and v-src sarcoma viral oncogene homolog (Src).
Figure 4.
Heat maps and gene interaction networks of differentially expressed genes found uniquely in the VP dysplasia following T + DES treatment. (A) A single cluster of upregulated genes identified only in the T + DES VP dysplasia, marked with pink (left panel), and the selected cluster enlarged (right panel). (B1 and B2) Clusters of downregulated genes in the VP but not in the LP following T + DES exposure. Interestingly, a single cluster (B2, left panel) corresponds to the downregulated genes in the T + DES-treated VP and is also underexpressed in both untreated and treated LPs. A selected region (blue box) of this cluster is enlarged (right panel). (C and D) Two representative gene interaction networks (with the highest relevancy scores) generated by IPA analysis from the differentially expressed genes in the T + DES VP dysplasia panel. Green indicates downregulated; red, upregulated. Genes bordered with red were validated by real-time q-PCR. See Figure 1 for key to IPA network.
Figure 5.
Post hoc validation of selected genes in the T + DES VP dysplasia panel by real-time q-PCR. The data were normalized to the levels of Rpl19. Bars indicated standard deviations (SD) of three to five animals in each treatment group. *P < .05, **P < .01 by one-way ANOVA with Tukey post hoc analysis.
Only 32 genes were found to be common in both dysplasia-related panels (Figure 1C), and 23 (of 32) genes formed a unique gene network when mapped by IPA (Figure 1D). We postulate that this gene network is central to E2- or DES-induced dysplasia in the LP or VP.
Identification of Estrogen-induced Differentially Expressed Genes That Are Not Related to Dysplasia in Rat LP or VP
We also identified two sets of hormone-induced genes that are not related to dysplasia. These genes exhibited differential expression in the VP following T + E2 treatment (14 genes; Figure 1C, left bottom) and in the LP following T + DES treatment (190 genes; Figure 1C, right bottom). They are published in Tables W2 and W3.
Genes Insensitive to the T + E2 and T + DES Treatments
Two sets of 156 and 80 genes were identified as insensitive in the LP and/or VP to T + E2 or T + DES treatment, respectively (Figure 6, A–C; Tables W4 and W5). Forty-one genes were found in both hormone-insensitive panels and mapped to an IPA network (Figure 6D) that includes the androgen receptor (Ar), a key regulator of prostate function.
Figure 6.
Heat maps of genes insensitive to (A) T + E2 or (B) T + DES. (C) Venn diagram showing the number of genes insensitive to T + E2 and/or T + DES treatment. Note that 41 genes are found in common in both hormone-insensitive panels and mapped to an IPA network (D) that includes the Ar, a key regulator of prostate function. Gray indicates no change in gene expression levels compared with those in untreated counterparts. See Figure 1 for key to IPA network.
Discussion
Exposure of the human prostate to estrogen may increase as men age [2–4] or with increased exposure to dietary estrogens such as DES or zeranol residues in meat, bisphenol A from food containers, or phytoestrogens [9]. The NBL rat provides a relevant model for elucidating mechanisms underlying carcinogenesis caused by such exposures, such as dysplasia and PCa, that consistently developed in the LP or the VP following chronic T-supported treatment with E2 or DES, respectively [15,18–20]. In the present study, we assessed transcriptional profiles elicited by the xenoestrogen DES in comparison with those of the endogenous hormone E2 and observed prostatic lobe-specific differential responses to the two estrogens in NBL rats. Hierarchical clustering revealed that the T + E2 treatment principally affects the LP gene expression profile whereas the T + DES targets that of the VP. Two distinct panels of dysplasia-related genes were identified: the T + E2 LP panel (253 genes) and the T + DES VP panel (198 genes), with only 32 overlapping genes. These findings indicate that the two estrogens alter gene expression profiles only in the prostatic lobe that develops dysplasia following each of the hormonal treatments. These findings imply a functional divergence of the two estrogens with respect to their oncogenic actions in the rat prostate. Thus, endocrine disruption of gene expression and dysplasia induction of the two estrogens are categorically different [9], although they may share some biologic convergence [30].
The IPA analysis identified distinct gene interaction networks based on differential expression profiles. It classified genes into functional categories and suggested a possible mechanistic linkage among the differentially expressed genes. We also used this tool to help prioritize genes for confirmation. The T + E2 treatment altered the expression of LP genes primarily in two major networks: one related to cell morphology, cellular growth, proliferation, and movement, and the other related to apoptosis and cell signaling. Exposure to T + DES also affected expression of genes in a similar network associated with apoptosis, cellular development, growth, and proliferation. Intriguingly, it also altered the expression of a set of genes linked to E2, a finding that may explain why, after T + DES treatment, gene expression profiles in the VP shifted closer to those in the treated and untreated LPs, which are under the influence of natural estrogen. By first mapping genes to the IPA network before selecting genes for confirmation, we were able to validate most of the expression changes in the microarrays with real-time q-PCR. In this regard, we confirmed three genes (Cebpd, Hoxa1, and Tp53) common in the two dysplasia panels. Importantly, these three genes were upregulated only in the prostatic lobes that developed dysplasia, i.e., the T + E2-treated LP and the T + DES-treated VP. Other lobe-specific genes that were confirmed were Smad7, Klb1, and Nxph3 in the T + E2-treated LP and Mafb, Src, Bcar1, Gpx3, Plau, and Thra in T + DES-treated VP. Our findings suggest that these gene expression changes induced by T + E2/DES treatment could be causative factors of dysplasia development. However, it remains possible that these gene expression changes are simply part of the neoplastic transformation process. Future studies employing laser capture-microdissected samples to establish the temporal relationship between change in gene expression pattern and evolution of dysplasia may shed new light on the cause-effect link between the two.
Among all these validated genes, Cebpd, Hoxa1, and Tp53 are the ones upregulated in common in the two dysplasia panels. CCAAT/enhancer binding proteins (C/EBP) are a highly conserved family of basic leucine zipper transcription factors; six members (C/EBP-alpha to -zeta) have been identified to date. This protein family plays a critical role in cell proliferation, apoptosis, and inflammation, depending on the cell type and specific physiological stress [31]. The precise functional role of C/EBP-delta, in particular, in normal prostate biology and PCa remains poorly understood. Cebpd was shown to be an androgen-repressed gene in the normal rat prostate [32,33], supporting its role as an apoptotic mediator and/or a negative regulator of cell proliferation. In a human androgen-dependent PCa xenograft, androgen withdrawal, however, resulted in a decline in the expression of C/EBP-delta [32]. Most published studies focus on the androgenic regulation of C/EBP-delta; this is, therefore, the first report of the association of estrogen-mediated upregulation of C/EBP-delta gene expression with the early phase of prostate carcinogenesis. A recent study showed overexpression of C/EBP-beta, another member of the family, in proliferative inflammatory atrophy in human prostate specimens, suggesting its role in the inflammation-associated carcinogenesis in the prostate [34]. In microarray data, we observed downregulation of C/EBP-alpha (Cebpa) only in T + E2-treated dysplastic LPs, corroborating its putative suppressive function in epithelial tumorigenesis [35]. Together, these findings raise the possibility that an orchestrated deregulation of C/EBP-family transcripts is involved in the pathogenesis of PCa in humans and rodents.
Homeobox gene (HOX) is a family of transcription factors related to growth and development. In rodents and humans, 39 members of the HOX gene family have been assigned to four gene clusters (A–D) and are numbered according to their expression along the anterior- posterior axis. Hox13 was shown to be involved in cell differentiation during normal prostate development [36], whereas upregulations of HOXC have been observed in PCa cells [37,38]. Knockdown of HOXC6 gene expression led to apoptosis of PCa cells [39], and overexpression of the HOXC8 gene correlated with the loss of tumor differentiation in human PCa [37]. Increased expression of the HOXA1 transcript was detected in human clinical cervical cancer samples and in several cervical cell lines compared with expression in normal tissues [40,41]. Our data suggest that ectopic expression of Hoxa1 is related to the evolution of dysplasia in both LP and VP and that the detailed roles of Hoxa1 in PCa require further study.
Tp53 is a well-known tumor-suppressive transcription factor and acts as a gatekeeper in signaling pathways involved in monitoring cellular stress such as DNA damage and in the determination of congruous responses (DNA repair, cell growth arrest, or apoptosis) to specific physiological stress. Previous studies demonstrated that mutation of Tp53 is not necessarily as late an event in prostate carcinogenesis as previously reported; it is also frequently found in high-grade prostate intraepithelial neoplasia, a precursor lesion of PCa [42,43]. In this regard, we observed upregulation of Tp53 gene expression in dysplastic rat prostate glands treated with sex hormone, implying a role in early prostate tumorigenesis, but whether these glands express wild-type or mutated Tp53 is not known. Alternatively, the induction of Tp53 expression may be a compensatory response to cellular stress imposed by hormonal changes. We recently demonstrated that T + E2 treatment induced oxidative and nitrosative stress, accompanied by DNA and protein damages, specifically in the LP, which is susceptible to dysplasia/cancer induction [17].
Smad7, an inhibitory Smad, is rapidly induced by TGF-β, thereby creating a negative feedback loop to a variety of TGF-β signaling responses, including proliferation, differentiation, apoptosis, inflammation, tissue remodeling, angiogenesis, and cell adhesion [44]. Disruption of the TGF-β signaling cascade by aberrant overexpression of this negative modulator leads to increased tumorigenicity in colon cancer cells by blocking TGF-β-mediated growth inhibition and apoptosis [45]. In contrast, Smad7 has been shown to mediate apoptosis induced by TGF-β in PCa cells [46], perhaps through crosstalking with other cellular signaling cascades such as the p38 mitogen-activated protein kinase [47] and β-catenin/Wnt [48] pathways. In our study, upregulation of Smad7 in the dysplastic LP (also in the T + DES VP microarray data) suggests perturbations of TGF-β feedback regulation and thus the loss of balance between cell proliferation and apoptosis in the early phase of prostate carcinogenesis.
A particularly interesting gene network found in the T + DES-treated VP (Figure 4, B1 and B2) is related to E2 signaling. Members include Src, Bcar1, and Plau. Src, a nonreceptor protein tyrosine kinase, triggers the nongenomic estrogen signaling leading to activation of ERK1/2 and cell proliferation in prostate epithelial cells [49,50]. It remains to be determined if the downregulation of Src by DES represents a classic negative feedback loop. The Bcar1 interacts and modulates Src activity [51]. An increase in Bcar1 gene expression, as observed in the rat dysplastic VP, has also been reported in human PCa [52]. The coordinated dysregulation of both Src (downregulation) and Bcar1 (upregulation) may reflect a disruption of the Src-dependent signaling pathway during the early development of PCa, a notion further supported by the altered expression of Plau, a downstream target of Src [53]. Interestingly, both Bcar1 and Plau are involved in cell survival and migration [54,55].
Our group recently demonstrated significant oxidative stress and inflammation in the T + E2-treated NBL rat LP [17,56]. The microarray data show a relatively small fraction of genes related to redox homeostasis and immune response, possibly due to inherent limitations of the type and coverage size (about 3800 genes) of the array chips we selected. However, T + E2 treatment was found to upregulate Klrb1 in the LP. This gene, also known as NKR-P1A/CD161 in humans, encodes a natural killer (NK) cell C-type lectin-like receptor that regulates NK cell functions. KLRB1 is expressed not only in NK cells but also in immune cell types, such as T cells, monocytes, and dendritic cells [57]. Thus, it is likely that increased expression of Klrb1 in LP with T + E2 treatment is due to the infiltration of immune cells [17].
The observation of a dramatic induction of Gpx3 in the VP dysplasia panel, as shown by microarray (>17-fold) and q-PCR (>70-fold), is consistent with our previous report of a marked elevation of GPX enzyme activity and lipid peroxidation in this lobe after T + DES treatment [56]. Hence, the activation in this glutathione-associated detoxification enzyme might be a response to hormone-induced oxidative stress in the gland or represent a cytoprotective mechanism, adapted by dysplastic cells, against oxidative insults. An elevation of GPX3 expression, but not of GPX1 and GPX2, was found in precancerous lesions of NKX3.1 mutant mouse prostate [58]. Taken together, these findings suggest that GPX3 is specifically involved in early prostatic transformation that has a mechanistic link to oxidative stress.
In summary, we showed that genes such as Cebpd, Tp53, and Hoxa1 are at the core of disrupted biologic networks related to hormoneinduced dysplasia. Unbiased gene profiling clearly demonstrated differential susceptibility of the LP and VP to natural and xenoestrogen with regard to alterations in gene expression and induction of dysplasia. Our data suggest more functional divergence between DES and E2 in the disruption of prostatic functions than that previously suspected. Methodologically, the combined utility of expression profiling and gene network mapping provides an instrumental platform for transcriptome studies.
Supplementary Material
Abbreviations
- C/EBP
CCAAT/enhancer binding protein
- DES
diethylstilbestrol
- E2
17β-estradiol
- LP
lateral prostate
- NBL
Noble
- PCa
prostate cancer
- PCR
polymerase chain reaction
- VP
ventral prostate
Footnotes
References
- 1.Taplin ME, Ho SM. Clinical review 134: the endocrinology of prostate cancer. J Clin Endocrinol Metab. 2001;86:3467–3477. doi: 10.1210/jcem.86.8.7782. [DOI] [PubMed] [Google Scholar]
- 2.Ho SM, Leung YK, Chung I. Estrogens and antiestrogens as etiological factors and therapeutics for prostate cancer. Ann NY Acad Sci. 2006;1089:177–193. doi: 10.1196/annals.1386.005. [DOI] [PubMed] [Google Scholar]
- 3.Carruba G. Estrogens and mechanisms of prostate cancer progression. Ann NY Acad Sci. 2006;1089:201–217. doi: 10.1196/annals.1386.027. [DOI] [PubMed] [Google Scholar]
- 4.Bosland MC. Sex steroids and prostate carcinogenesis: integrated, multifactorial working hypothesis. Ann NY Acad Sci. 2006;1089:168–176. doi: 10.1196/annals.1386.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leav I, Lau KM, Adams JY, McNeal JE, Taplin ME, Wang J, Singh H, Ho SM. Comparative studies of the estrogen receptors β and α and the androgen receptor in normal human prostate glands, dysplasia, and in primary and metastatic carcinoma. Am J Pathol. 2001;159:79–92. doi: 10.1016/s0002-9440(10)61676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu X, Leav I, Leung YK, Wu M, Liu Q, Gao Y, McNeal JE, Ho SM. Dynamic regulation of estrogen receptor-β expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004;164:2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellem SJ, Risbridger GP. Aromatase and prostate cancer. Minerva Endocrinol. 2006;31:1–12. [PubMed] [Google Scholar]
- 8.Singleton DW, Khan SA. Xenoestrogen exposure and mechanisms of endocrine disruption. Front Biosci. 2003;8:s110–s118. doi: 10.2741/1010. [DOI] [PubMed] [Google Scholar]
- 9.Leffers H, Naesby M, Vendelbo B, Skakkebaek NE, Jorgensen M. Oestrogenic potencies of zeranol, oestradiol, diethylstilboestrol, bisphenol-A and genistein: implications for exposure assessment of potential endocrine disrupters. Hum Reprod. 2001;16:1037–1045. doi: 10.1093/humrep/16.5.1037. [DOI] [PubMed] [Google Scholar]
- 10.Epstein SS. The chemical jungle: today's beef industry. Int J Health Serv. 1990;20:277–280. doi: 10.2190/3MFD-569U-K87C-P8CM. [DOI] [PubMed] [Google Scholar]
- 11.Herbst AL, Ulfelder H, Poskanzer DC. Adenocarcinoma of the vagina. Association of maternal stilbestrol therapy with tumor appearance in young women. N Engl J Med. 1971;284:878–881. doi: 10.1056/NEJM197104222841604. [DOI] [PubMed] [Google Scholar]
- 12.Troisi R, Hatch EE, Titus-Ernstoff L, Hyer M, Palmer JR, Robboy SJ, Strohsnitter WC, Kaufman R, Herbst AL, Hoover RN. Cancer risk in women prenatally exposed to diethylstilbestrol. Int J Cancer. 2007;121:356–360. doi: 10.1002/ijc.22631. [DOI] [PubMed] [Google Scholar]
- 13.Strohsnitter WC, Noller KL, Hoover RN, Robboy SJ, Palmer JR, Titus-Ernstoff L, Kaufman RH, Adam E, Herbst AL, Hatch EE. Cancer risk in men exposed in utero to diethylstilbestrol. J Natl Cancer Inst. 2001;93:545–551. doi: 10.1093/jnci/93.7.545. [DOI] [PubMed] [Google Scholar]
- 14.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leav I, Ho SM, Ofner P, Merk FB, Kwan PW, Damassa D. Biochemical alterations in sex hormone-induced hyperplasia and dysplasia of the dorsolateral prostates of Noble rats. J Natl Cancer Inst. 1988;80:1045–1053. doi: 10.1093/jnci/80.13.1045. [DOI] [PubMed] [Google Scholar]
- 16.Leav I, Merk FB, Kwan PW, Ho SM. Androgen-supported estrogenenhanced epithelial proliferation in the prostates of intact Noble rats. Prostate. 1989;15:23–40. doi: 10.1002/pros.2990150104. [DOI] [PubMed] [Google Scholar]
- 17.Tam NN, Leav I, Ho SM. Sex hormones induce direct epithelial and inflammation-mediated oxidative/nitrosative stress that favors prostatic carcinogenesis in the Noble rat. Am J Pathol. 2007;171:1334–1341. doi: 10.2353/ajpath.2007.070199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson CJ, Tam NN, Joyce JM, Leav I, Ho SM. Gene expression profiling of testosterone and estradiol-17β-induced prostatic dysplasia in Noble rats and response to the antiestrogen ICI 182,780. Endocrinology. 2002;143:2093–2105. doi: 10.1210/endo.143.6.8846. [DOI] [PubMed] [Google Scholar]
- 19.Bosland MC, Ford H, Horton L. Induction at high incidence of ductal prostate adenocarcinomas in NBL/Cr and Sprague-Dawley Hsd:SD rats treated with a combination of testosterone and estradiol-17β or diethylstilbestrol. Carcinogenesis. 1995;16:1311–1317. doi: 10.1093/carcin/16.6.1311. [DOI] [PubMed] [Google Scholar]
- 20.Ofner P, Bosland MC, Vena RL. Differential effects of diethylstilbestrol and estradiol-17β in combination with testosterone on rat prostate lobes. Toxicol Appl Pharmacol. 1992;112:300–309. doi: 10.1016/0041-008x(92)90200-c. [DOI] [PubMed] [Google Scholar]
- 21.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 22.Guo J, Sartor M, Karyala S, Medvedovic M, Kann S, Puga A, Ryan P, Tomlinson CR. Expression of genes in the TGF-β signaling pathway is significantly deregulated in smooth muscle cells from aorta of aryl hydrocarbon receptor knockout mice. Toxicol Appl Pharmacol. 2004;194:79–89. doi: 10.1016/j.taap.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Karyala S, Guo J, Sartor M, Medvedovic M, Kann S, Puga A, Ryan P, Tomlinson CR. Different global gene expression profiles in benzo[a] pyrene- and dioxin-treated vascular smooth muscle cells of AHR-knockout and wild-type mice. Cardiovasc Toxicol. 2004;4:47–73. doi: 10.1385/ct:4:1:47. [DOI] [PubMed] [Google Scholar]
- 24.Sartor M, Schwanekamp J, Halbleib D, Mohamed I, Karyala S, Medvedovic M, Tomlinson CR. Microarray results improve significantly as hybridization approaches equilibrium. Biotechniques. 2004;36:790–796. doi: 10.2144/04365ST02. [DOI] [PubMed] [Google Scholar]
- 25.Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 27.Daly-Burns B, Alam TN, Mackay A, Clark J, Shepherd CJ, Rizzo S, Tatoud R, O'Hare MJ, Masters JR, Hudson DL. A conditionally immortalized cell line model for the study of human prostatic epithelial cell differentiation. Differentiation. 2007;75:35–48. doi: 10.1111/j.1432-0436.2006.00113.x. [DOI] [PubMed] [Google Scholar]
- 28.Rozen S, Skaletsky HJ. Primer3. 1998 Available at: http://www-genome.wi.mit.edu/genome_software/other/primer3.html.
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Buterin T, Koch C, Naegeli H. Convergent transcriptional profiles induced by endogenous estrogen and distinct xenoestrogens in breast cancer cells. Carcinogenesis. 2006;27:1567–1578. doi: 10.1093/carcin/bgi339. [DOI] [PubMed] [Google Scholar]
- 31.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang G, Gregory CW, Shang Q, O'Brien DA, Zhang YL. Differential expression of CCAAT/enhancer-binding protein-delta (C/EBPdelta) in rat androgen-dependent tissues and human prostate cancer. J Androl. 2001;22:471–480. [PubMed] [Google Scholar]
- 33.Desai KV, Michalowska AM, Kondaiah P, Ward JM, Shih JH, Green JE. Gene expression profiling identifies a unique androgen-mediated inflammatory/immune signature and a PTEN (phosphatase and tensin homolog deleted on chromosome 10)-mediated apoptotic response specific to the rat ventral prostate. Mol Endocrinol. 2004;18:2895–2907. doi: 10.1210/me.2004-0033. [DOI] [PubMed] [Google Scholar]
- 34.Wang GL, Shi X, Salisbury E, Sun Y, Albrecht JH, Smith RG, Timchenko NA. Growth hormone corrects proliferation and transcription of phosphoenolpyruvate carboxykinase in livers of old mice via elimination of CCAAT/enhancer-binding protein α-Brm complex. J Biol Chem. 2007;282:1468–1478. doi: 10.1074/jbc.M608226200. [DOI] [PubMed] [Google Scholar]
- 35.Loomis KD, Zhu S, Yoon K, Johnson PF, Smart RC. Genetic ablation of CCAAT/enhancer binding protein α in epidermis reveals its role in suppression of epithelial tumorigenesis. Cancer Res. 2007;67:6768–6776. doi: 10.1158/0008-5472.CAN-07-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L, Pu Y, Hepps D, Danielpour D, Prins GS. Posterior Hox gene expression and differential androgen regulation in the developing and adult rat prostate lobes. Endocrinology. 2007;148:1235–1245. doi: 10.1210/en.2006-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waltregny D, Alami Y, Clausse N, de Leval J, Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 38.Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, Lucia MS, Nordeen SK. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 39.Ramachandran S, Liu P, Young AN, Yin-Goen Q, Lim SD, Laycock N, Amin MB, Carney JK, Marshall FF, Petros JA, et al. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- 40.Shim C, Zhang W, Rhee CH, Lee JH. Profiling of differentially expressed genes in human primary cervical cancer by complementary DNA expression array. Clin Cancer Res. 1998;4:3045–3050. [PubMed] [Google Scholar]
- 41.Hung YC, Ueda M, Terai Y, Kumagai K, Ueki K, Kanda K, Yamaguchi H, Akise D, Ueki M. Homeobox gene expression and mutation in cervical carcinoma cells. Cancer Sci. 2003;94:437–441. doi: 10.1111/j.1349-7006.2003.tb01461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Maghrabi J, Vorobyova L, Chapman W, Jewett M, Zielenska M, Squire JA. p53 Alteration and chromosomal instability in prostatic high-grade intraepithelial neoplasia and concurrent carcinoma: analysis by immunohistochemistry, interphase in situ hybridization, and sequencing of laser-captured microdissected specimens. Mod Pathol. 2001;14:1252–1262. doi: 10.1038/modpathol.3880471. [DOI] [PubMed] [Google Scholar]
- 43.Downing SR, Russell PJ, Jackson P. Alterations of p53 are common in early stage prostate cancer. Can J Urol. 2003;10:1924–1933. [PubMed] [Google Scholar]
- 44.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr, Wrana JL, et al. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 45.Halder SK, Beauchamp RD, Datta PK. Smad7 induces tumorigenicity by blocking TGF-β-induced growth inhibition and apoptosis. Exp Cell Res. 2005;307:231–246. doi: 10.1016/j.yexcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Landstrom M, Heldin NE, Bu S, Hermansson A, Itoh S, ten Dijke P, Heldin CH. Smad7 mediates apoptosis induced by transforming growth factor β in prostatic carcinoma cells. Curr Biol. 2000;10:535–538. doi: 10.1016/s0960-9822(00)00470-x. [DOI] [PubMed] [Google Scholar]
- 47.Edlund S, Bu S, Schuster N, Aspenstrom P, Heuchel R, Heldin NE, ten Dijke P, Heldin CH, Landstrom M. Transforming growth factor-β1 (TGF- β)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-β-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell. 2003;14:529–544. doi: 10.1091/mbc.02-03-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edlund S, Lee SY, Grimsby S, Zhang S, Aspenstrom P, Heldin CH, Landstrom M. Interaction between Smad7 and β-catenin: importance for transforming growth factor β-induced apoptosis. Mol Cell Biol. 2005;25:1475–1488. doi: 10.1128/MCB.25.4.1475-1488.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Migliaccio A, Castoria G, Di Domenico M, de Falco A, Bilancio A, Lombardi M, Barone MV, Ametrano D, Zannini MS, Abbondanza C, et al. Steroid-induced androgen receptor-oestradiol receptor β-Src complex triggers prostate cancer cell proliferation. EMBO J. 2000;19:5406–5417. doi: 10.1093/emboj/19.20.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chieffi P, Kisslinger A, Sinisi AA, Abbondanza C, Tramontano D. 17β-Estradiol-induced activation of ERK1/2 through endogenous androgen receptor-estradiol receptor α-Src complex in human prostate cells. Int J Oncol. 2003;23:797–801. [PubMed] [Google Scholar]
- 51.Burnham MR, Bruce-Staskal PJ, Harte MT, Weidow CL, Ma A, Weed SA, Bouton AH. Regulation of c-SRC activity and function by the adapter protein CAS. Mol Cell Biol. 2000;20:5865–5878. doi: 10.1128/mcb.20.16.5865-5878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fromont G, Vallancien G, Validire P, Levillain P, Cussenot O. BCAR1 expression in prostate cancer: association with 16q23 LOH status, tumor progression and EGFR/KAI1 staining. Prostate. 2007;67:268–273. doi: 10.1002/pros.20516. [DOI] [PubMed] [Google Scholar]
- 53.Kleiner S, Faisal A, Nagamine Y. Induction of uPA gene expression by the blockage of E-cadherin via Src- and Shc-dependent Erk signaling. FEBS J. 2007;274:227–240. doi: 10.1111/j.1742-4658.2006.05578.x. [DOI] [PubMed] [Google Scholar]
- 54.Gaylis FD, Keer HN, Wilson MJ, Kwaan HC, Sinha AA, Kozlowski JM. Plasminogen activators in human prostate cancer cell lines and tumors: correlation with the aggressive phenotype. J Urol. 1989;142:193–198. doi: 10.1016/s0022-5347(17)38709-8. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Fei Z, Bu X, Zhen H, Zhang Z, Gu J, Chen Y. Expression and significance of urokinase type plasminogen activator gene in human brain gliomas. J Surg Oncol. 2000;74:90–94. doi: 10.1002/1096-9098(200006)74:2<90::AID-JSO2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 56.Tam NN, Ghatak S, Ho SM. Sex hormone-induced alterations in the activities of antioxidant enzymes and lipid peroxidation status in the prostate of Noble rats. Prostate. 2003;55:1–8. doi: 10.1002/pros.10169. [DOI] [PubMed] [Google Scholar]
- 57.Poggi A, Costa P, Zocchi MR, Moretta L. Phenotypic and functional analysis of CD4+ NKRP1A+ human T lymphocytes. Direct evidence that the NKRP1A molecule is involved in transendothelial migration. Eur J Immunol. 1997;27:2345–2350. doi: 10.1002/eji.1830270932. [DOI] [PubMed] [Google Scholar]
- 58.Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. 2005;65:6773–6779. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.