Abstract
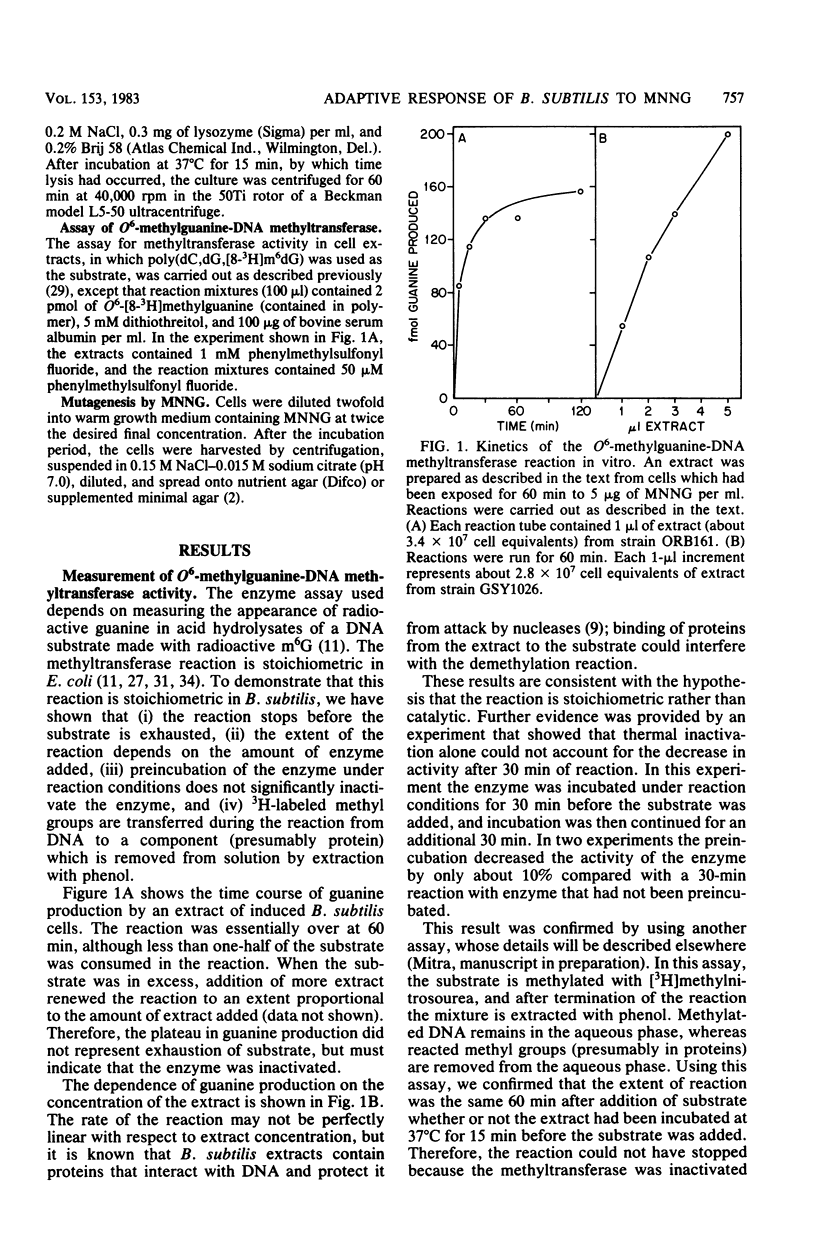
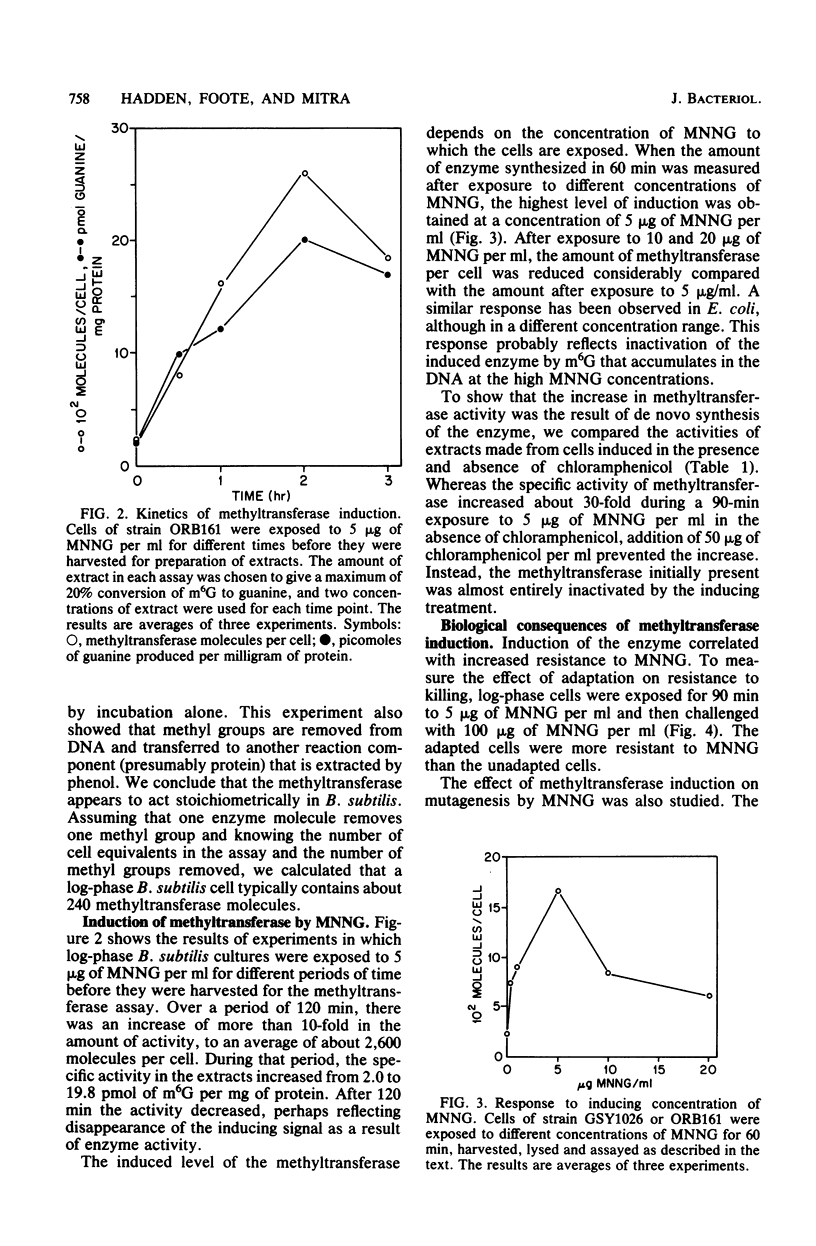
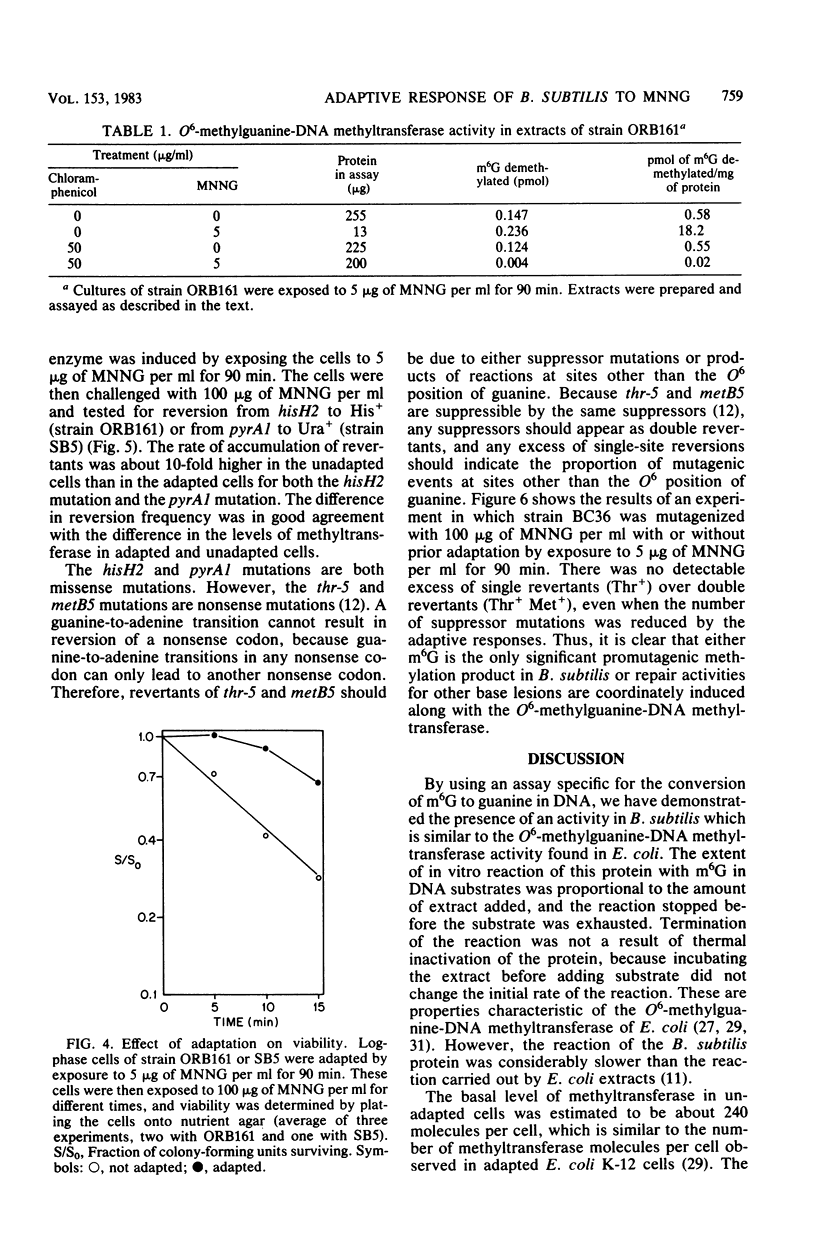
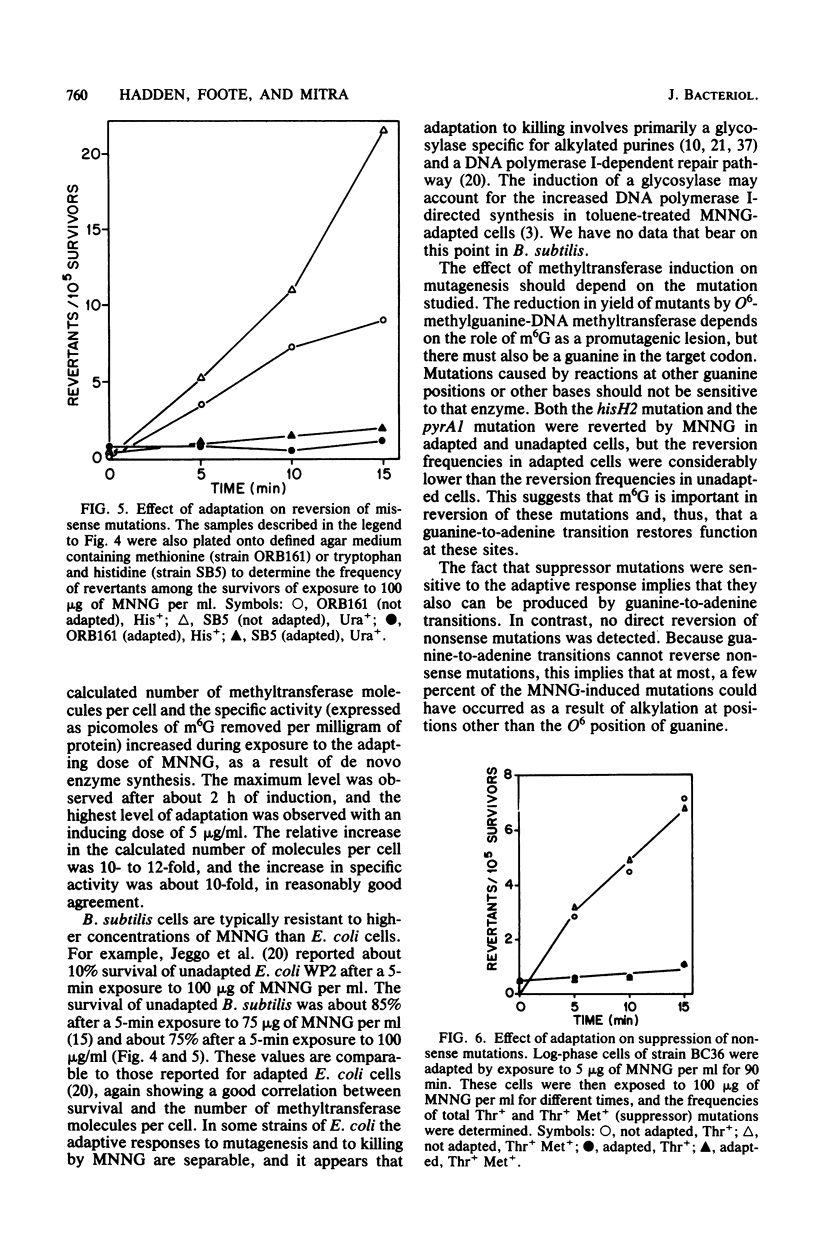
Cell extracts of Bacillus subtilis contain a methyltransferase that appears to remove the O6-methyl group from O6-methylguanine in DNA in situ. This reaction proceeds in a stoichiometric fashion, as in Escherichia coli. However, the basal level of the enzyme (approximately 240 molecules per cell) is significantly higher in B. subtilis than in E. coli. In addition, the methyltransferase level increases by an order of magnitude as a result of de novo protein synthesis after adaptive treatment with a low concentration of N-methyl-N'-nitro-N-nitrosoguanidine (MNNG), as in E. coli. Concomitant with adaptation, B. subtilis cells become more resistant to both killing and mutagenesis by a challenge dose of N-methyl-N'-nitro-N-nitrosoguanidine. We present evidence to support the hypothesis that the majority of N-methyl-N'-nitro-N-nitrosoguanidine-induced mutations in B. subtilis are of the guanine-to-adenine transition type.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beattie K. L., Kimball R. F. Involvement of DNA replication and repair in mutagenesis of Haemophilus influenzae induced by N-nitrosocarbaryl. Mutat Res. 1974 Aug;24(2):105–115. doi: 10.1016/0027-5107(74)90124-9. [DOI] [PubMed] [Google Scholar]
- Billen D., Carreira L. B., Hadden C. T., Silverstein S. J. Evidence suggestive of compartmentalization of deoxyribonucleic acid-synthesizing systems in freeze-treated Bacillus subtilis. J Bacteriol. 1971 Dec;108(3):1250–1256. doi: 10.1128/jb.108.3.1250-1256.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billen D., Hellerman G., Carreira L. Gene frequency analysis of chromosomal initiation sites in Bacillus subtilis after ultraviolet light or x-ray exposure. J Bacteriol. 1972 Jan;109(1):379–384. doi: 10.1128/jb.109.1.379-384.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billen D., Hellermann G. R. Enhancement of deoxyribonucleic acid polymerase I-directed repair synthesis in toluene-treated Escherichia coli after growth in the presence of low levels of N-methyl-N'-nitro-N-nitrosoguanidine. J Bacteriol. 1979 Mar;137(3):1439–1442. doi: 10.1128/jb.137.3.1439-1442.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdá-Olmedo E., Hanawalt P. C., Guerola N. Mutagenesis of the replication point by nitrosoguanidine: map and pattern of replication of the Escherichia coli chromosome. J Mol Biol. 1968 May 14;33(3):705–719. doi: 10.1016/0022-2836(68)90315-x. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Dawes I. W., Mackinnon D. A., Ball D. E., Hardie I. D., Sweet D. M., Ross F. M., Macdonald F. Identifying sites of simultaneous DNA replication in eukaryotes by N-methyl-N'-nitro-N-nitrosoguanidine multiple mutagenesis. Mol Gen Genet. 1977 Mar 28;152(1):53–57. doi: 10.1007/BF00264939. [DOI] [PubMed] [Google Scholar]
- Eisenstadt E., Lange R., Willecke K. Competent Bacillus subtilis cultures synthesize a denatured DNA binding activity. Proc Natl Acad Sci U S A. 1975 Jan;72(1):323–327. doi: 10.1073/pnas.72.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Foote R. S., Mitra S., Pal B. C. Demethylation of O6-methylguanine in a synthetic DNA polymer by an inducible activity in Escherichia coli. Biochem Biophys Res Commun. 1980 Nov 28;97(2):654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Suppressor system in Bacillus subtilis 168. J Bacteriol. 1969 Mar;97(3):1397–1402. doi: 10.1128/jb.97.3.1397-1402.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerola N., Ingraham J. L., Cerdá-Olmedo E. Induction of closely linked multiple mutations by nitrosoguanidine. Nat New Biol. 1971 Mar 24;230(12):122–125. doi: 10.1038/newbio230122a0. [DOI] [PubMed] [Google Scholar]
- Hill T., Prakash L., Strauss B. Mutagen stability of alkylation-sensitive mutants of Bacillus subtilis. J Bacteriol. 1972 Apr;110(1):47–55. doi: 10.1128/jb.110.1.47-55.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y., Jacob F., Ryter A., Buttin G., Nakai T. On the process of cellular division in Escherichia coli. I. Asymmetrical cell division and production of deoxyribonucleic acid-less bacteria. J Mol Biol. 1968 Jul 14;35(1):175–192. doi: 10.1016/s0022-2836(68)80046-4. [DOI] [PubMed] [Google Scholar]
- Hoch J. A., Barat M., Anagnostopoulos C. Transformation and transduction in recombination-defective mutants of Bacillus subtilis. J Bacteriol. 1967 Jun;93(6):1925–1937. doi: 10.1128/jb.93.6.1925-1937.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Nester E. W. Gene-enzyme relationships of aromatic acid biosynthesis in Bacillus subtilis. J Bacteriol. 1973 Oct;116(1):59–66. doi: 10.1128/jb.116.1.59-66.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P., Defais T. M., Samson L., Schendel P. An adaptive response of E. coli to low levels of alkylating agent: comparison with previously characterised DNA repair pathways. Mol Gen Genet. 1977 Nov 29;157(1):1–9. doi: 10.1007/BF00268680. [DOI] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Griffin B. Adaptive response to alkylating agents involves alteration in situ of O6-methylguanine residues in DNA. Nature. 1979 Jul 5;280(5717):76–77. doi: 10.1038/280076a0. [DOI] [PubMed] [Google Scholar]
- Kimball R. F. Further studies on the induction of mutation in Haemophilus influenzae by N-methyl-N'-nitro-N-nitrosoguanidine: lack of an inducible error-free repair system and the effect of exposure medium. Mutat Res. 1980 Aug;72(3):361–372. doi: 10.1016/0027-5107(80)90111-6. [DOI] [PubMed] [Google Scholar]
- Kimball R. F., Setlow J. K. Mutation fixation in MNNG-treated Haemophilus influenzae as determined by transformation. Mutat Res. 1974 Jan;22(1):1–14. doi: 10.1016/0027-5107(74)90002-5. [DOI] [PubMed] [Google Scholar]
- Kimball R. F., Setlow J. K. Mutations induced in Haemophilus influenzae by transformation with nitrosoguanidine-treated DNA. Mutat Res. 1972 Feb;14(2):137–146. doi: 10.1016/0027-5107(72)90042-5. [DOI] [PubMed] [Google Scholar]
- Kimball R. F. Studies on the mutagenic action of N-methyl-N'-nitro-N-nitrosoguanidine in Paramecium aurella with emphasis on repair processes. Mutat Res. 1970 Mar;9(3):261–271. doi: 10.1016/0027-5107(70)90128-4. [DOI] [PubMed] [Google Scholar]
- Loveless A. Possible relevance of O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969 Jul 12;223(5202):206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- Mitra S., Pal B. C., Foote R. S. O6-methylguanine-DNA methyltransferase in wild-type and ada mutants of Escherichia coli. J Bacteriol. 1982 Oct;152(1):534–537. doi: 10.1128/jb.152.1.534-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeschger M. P., Berlyn M. K. A simple procedure for localized mutagenesis using nitrosoguanidine. Mol Gen Genet. 1974;134(1):77–83. doi: 10.1007/BF00332814. [DOI] [PubMed] [Google Scholar]
- Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980 Nov 25;255(22):10569–10571. [PubMed] [Google Scholar]
- Robins P., Cairns J. Quantitation of the adaptive response to alkylating agents. Nature. 1979 Jul 5;280(5717):74–76. doi: 10.1038/280074a0. [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Schendel P. F., Robins P. E. Repair of O6-methylguanine in adapted Escherichia coli. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6017–6020. doi: 10.1073/pnas.75.12.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siccardi A. G., Ferrari F. A., Mazza G., Galizzi A. Identification of coreplicating chromosomal sectors in Bacillus subtilis by nitrosoguanidine-induced comutation. J Bacteriol. 1976 Mar;125(3):755–761. doi: 10.1128/jb.125.3.755-761.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar R. Enchancement of nitrosoguanidine mutagenesis by chloramphenicol in Escherichia coli K-12. J Bacteriol. 1978 Oct;136(1):460–462. doi: 10.1128/jb.136.1.460-462.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L., Yang C. H., Goldthwait D. A. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982 Mar 16;21(6):1162–1169. doi: 10.1021/bi00535a009. [DOI] [PubMed] [Google Scholar]


