Abstract
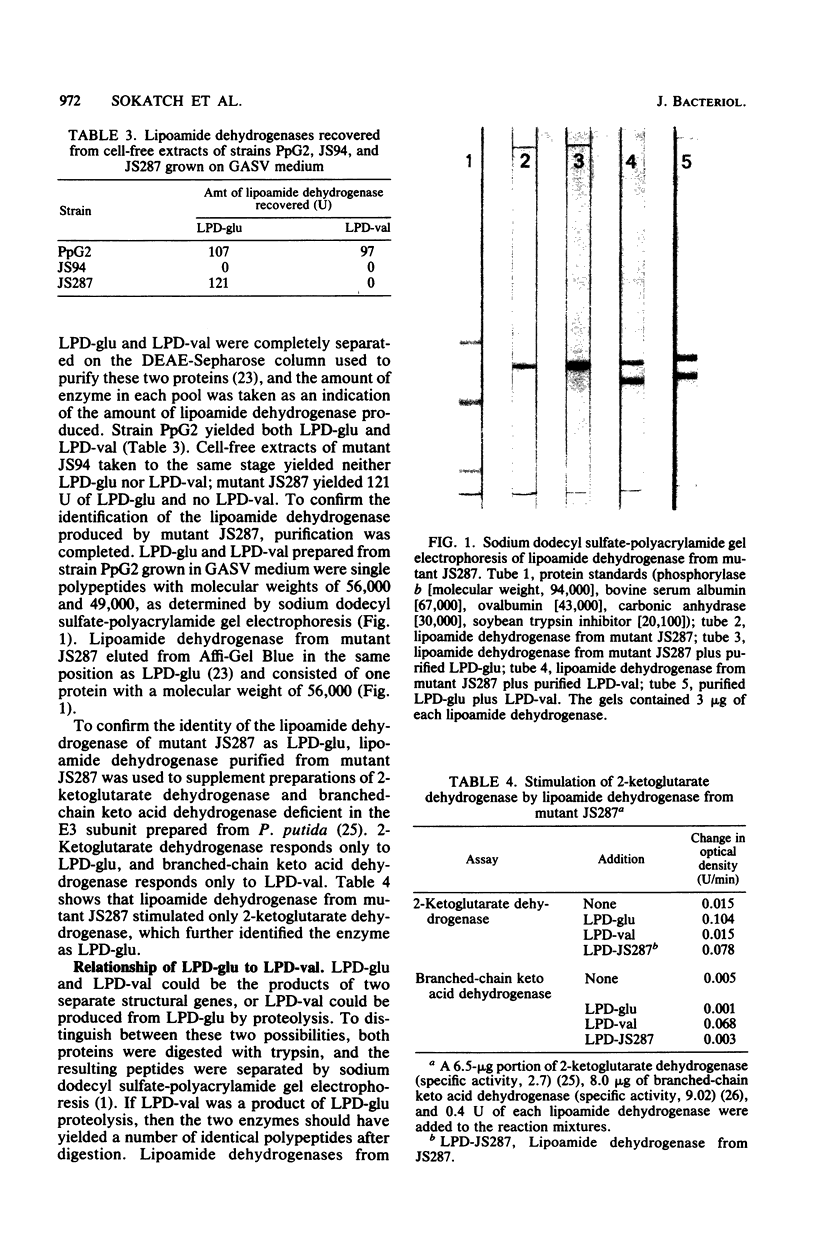
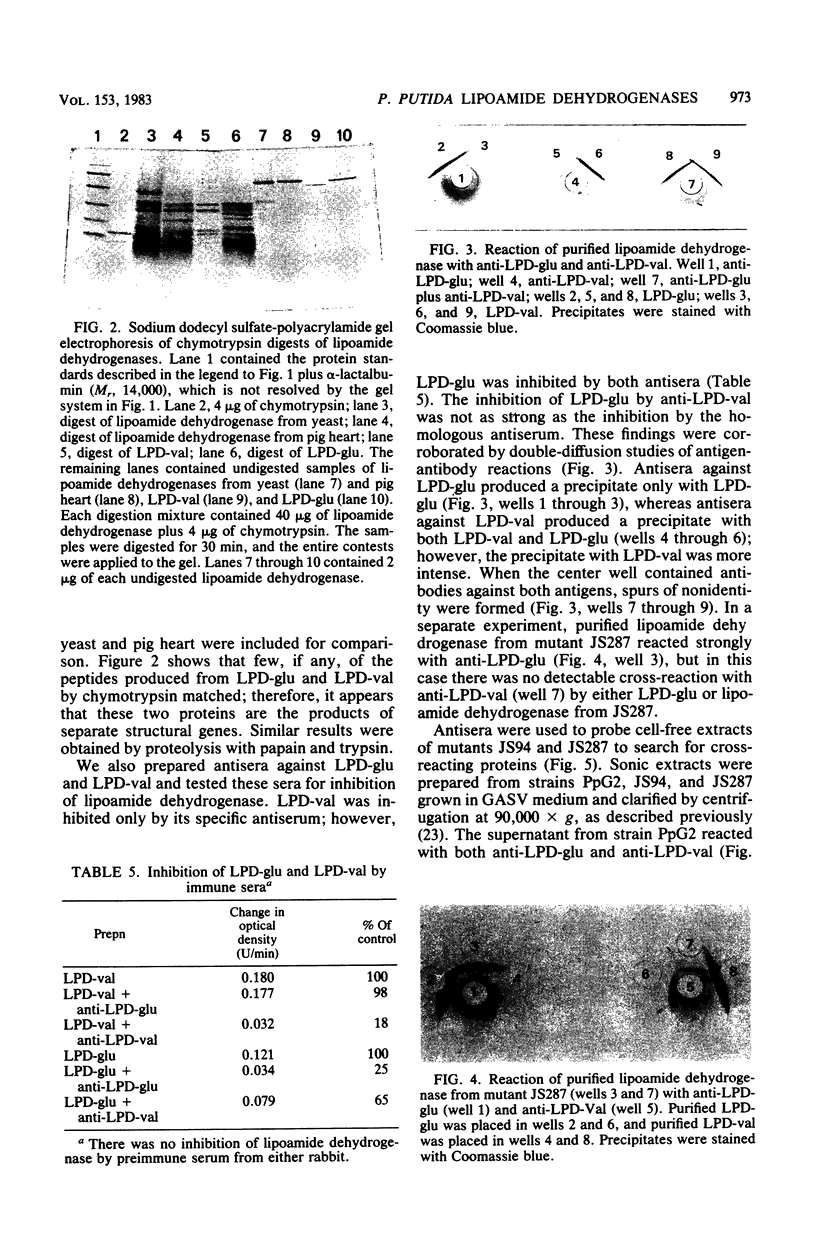
Pseudomonas putida grown on valine produces two lipoamide dehydrogenases, LPD-glu (Mr, 56,000 and LPD-val (Mr, 49,000). The 49,000-dalton protein is used by P. putida for branched-chain keto acid dehydrogenase, whereas the 56,000-dalton protein is presumably used for pyruvate and 2-ketoglutarate dehydrogenases. The objective of this study was to isolate and characterize mutants of P. putida with mutations affecting lipoamide dehydrogenases in order to study the relationship of these two proteins. Mutant JS287 lacked LPD-val, the lipoamide dehydrogenase which is induced by growth on valine and is specific for branched-chain keto acid dehydrogenase, and had normal amounts of LPD-glu, the lipoamide dehydrogenase which is formed during growth on glucose and which is probably used by both pyruvate and 2-ketoglutarate dehydrogenases. Mutant JS94 was a pleiotropic mutant with defects in 2-ketoglutarate, branched-chain, and lipoamide dehydrogenases. Proteolysis of LPD-glu and LPD-val produced completely different digestion products, suggesting that these two proteins are products of separate structural genes. Antisera prepared against LPD-glu reacted only with LPD-glu, whereas antisera prepared against LPD-val reacted with LPD-val and cross-reacted with LPD-glu. Although mutant JS94 did not produce active lipoamide dehydrogenase, cell-free extracts of this mutant contained a protein which cross-reacted with anti-LPD-val.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Duncan M. J., Fraenkel D. G. alpha-Ketoglutarate dehydrogenase mutant of Rhizobium meliloti. J Bacteriol. 1979 Jan;137(1):415–419. doi: 10.1128/jb.137.1.415-419.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L., Chang T. W. Regulation and significance of amino acid metabolism in skeletal muscle. Fed Proc. 1978 Jul;37(9):2301–2307. [PubMed] [Google Scholar]
- Guest J. R. Aspects of the molecular biology of lipoamide dehydrogenase. Adv Neurol. 1978;21:219–244. [PubMed] [Google Scholar]
- Guest J. R., Creaghan I. T. Gene-protein relationships of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: isolation and characterization of lipoamide dehydrogenase mutants. J Gen Microbiol. 1973 Mar;75(1):197–210. doi: 10.1099/00221287-75-1-197. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Gene-protein relationships of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: Chromosomal location of the lipoamide dehydrogenase gene. J Gen Microbiol. 1974 Feb;80(2):523–532. doi: 10.1099/00221287-80-2-523. [DOI] [PubMed] [Google Scholar]
- HAGER L. P., KORNBERG H. L. On the mechanism of alpha-oxoglutarate oxidation in Escherichia coli. Biochem J. 1961 Jan;78:194–198. doi: 10.1042/bj0780194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Biochemical and genetic studies with lysine+methionine mutants of Escherichia coli: lipoic acid and alpha-ketoglutarate dehydrogenase-less mutants. J Gen Microbiol. 1968 Oct;53(3):363–381. doi: 10.1099/00221287-53-3-363. [DOI] [PubMed] [Google Scholar]
- Jacobson L. A., Bartholomaus R. C., Gunsalus I. C. Repression of malic enzyme by acetate in Pseudomonas. Biochem Biophys Res Commun. 1966 Sep 22;24(6):955–960. doi: 10.1016/0006-291x(66)90343-3. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Marshall V. D., Sokatch J. R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972 Jun;110(3):1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. R., Marshall V. D., Sokatch J. R., Unger L. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J Bacteriol. 1973 Jul;115(1):198–204. doi: 10.1128/jb.115.1.198-204.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey L. K., Sokatch J. R., Conrad R. S. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976 Mar;40(1):42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. IV. Regulation. J Biol Chem. 1966 Aug 25;241(16):3800–3810. [PubMed] [Google Scholar]
- Pettit F. H., Reed L. J. Alpha-keto acid dehydrogenase complexes. 8. Comparison of dihydrolipoyl dehydrogenases from pyruvate and alpha-ketoglutarate dehydrogenase complexes of Escherichia coli. Proc Natl Acad Sci U S A. 1967 Sep;58(3):1126–1130. doi: 10.1073/pnas.58.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit F. H., Yeaman S. J., Reed L. J. Purification and characterization of branched chain alpha-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REED L. J., KOIKE M., LEVITCH M. E., LEACH F. R. Studies on the nature and reactions of protein-bound lipoic acid. J Biol Chem. 1958 May;232(1):143–158. [PubMed] [Google Scholar]
- Roberts C. M., Sokatch J. R. Branched chain amino acids as activators of branched chain ketoacid dehydrogenase. Biochem Biophys Res Commun. 1978 Jun 14;82(3):828–833. doi: 10.1016/0006-291x(78)90857-4. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R. Isolation of two lipoamide dehydrogenases from Pseudomonas putida grown on valine. Biochem Biophys Res Commun. 1981 Jun 16;100(3):1071–1076. doi: 10.1016/0006-291x(81)91932-x. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Gebrosky J., Sokatch D. J. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):639–646. doi: 10.1128/jb.148.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Roberts C. M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahren J., Felig P., Cerasi E., Luft R. Splanchnic and peripheral glucose and amino acid metabolism in diabetes mellitus. J Clin Invest. 1972 Jul;51(7):1870–1878. doi: 10.1172/JCI106989. [DOI] [PMC free article] [PubMed] [Google Scholar]