Abstract
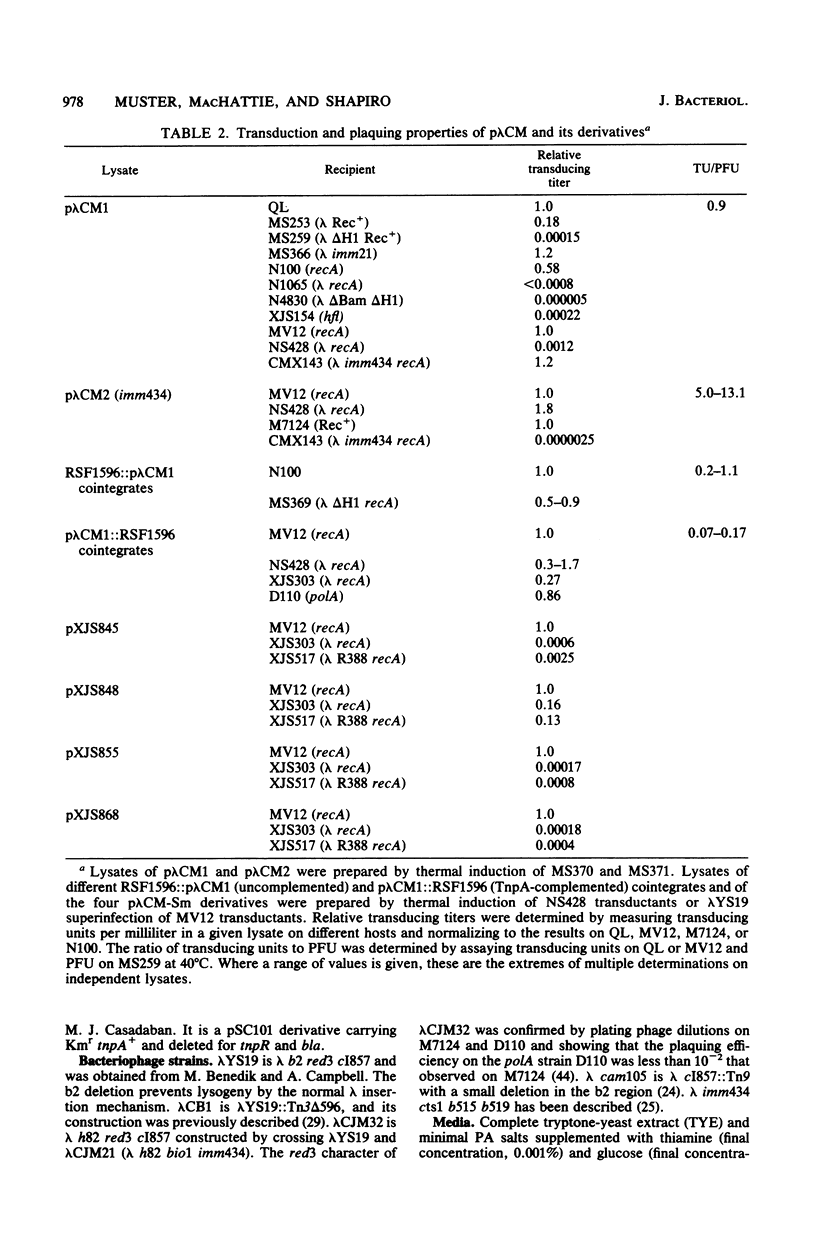
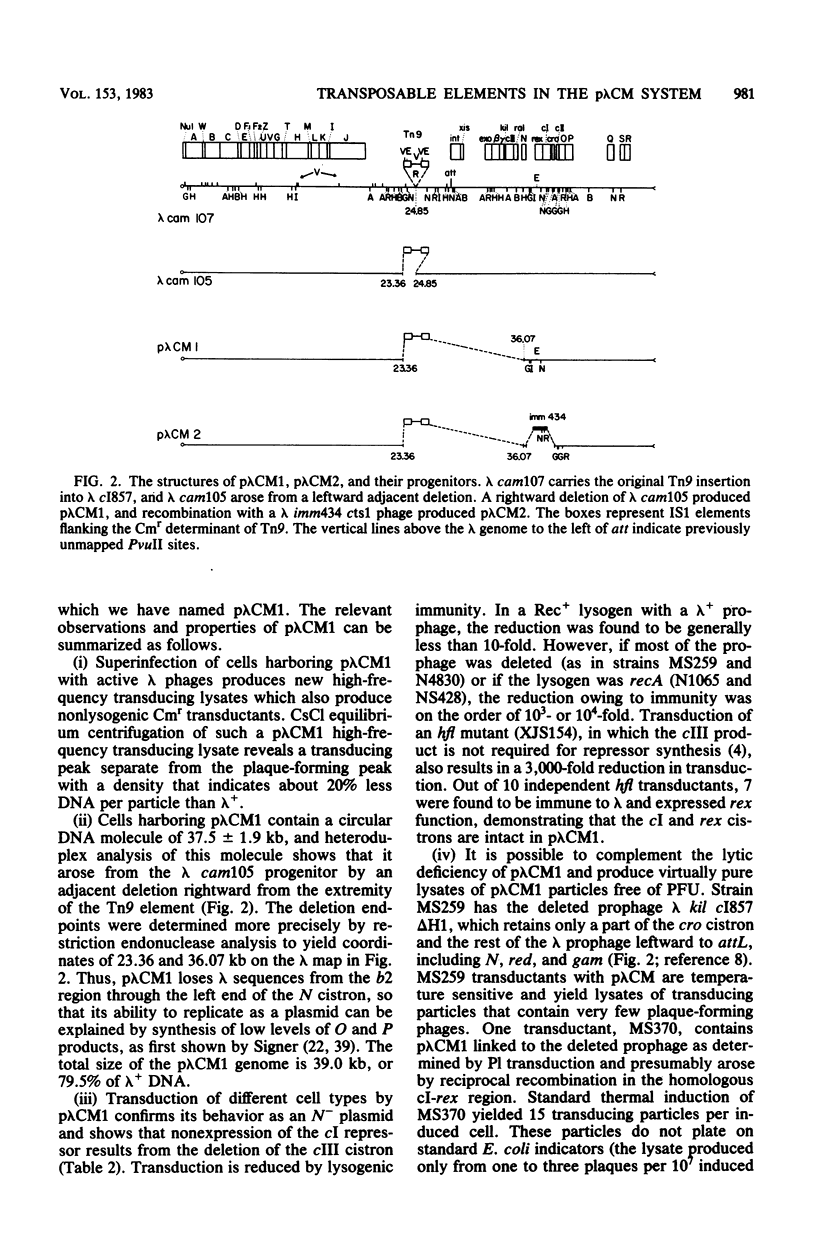
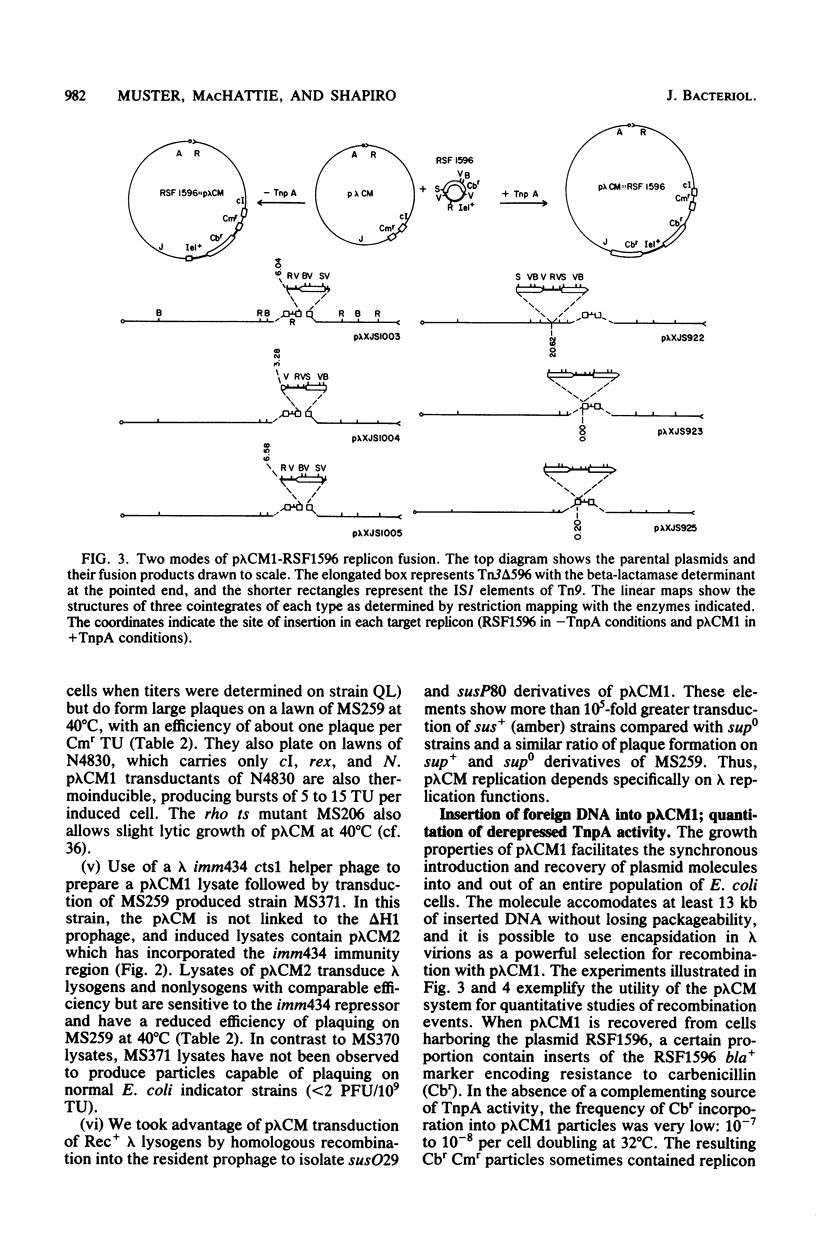
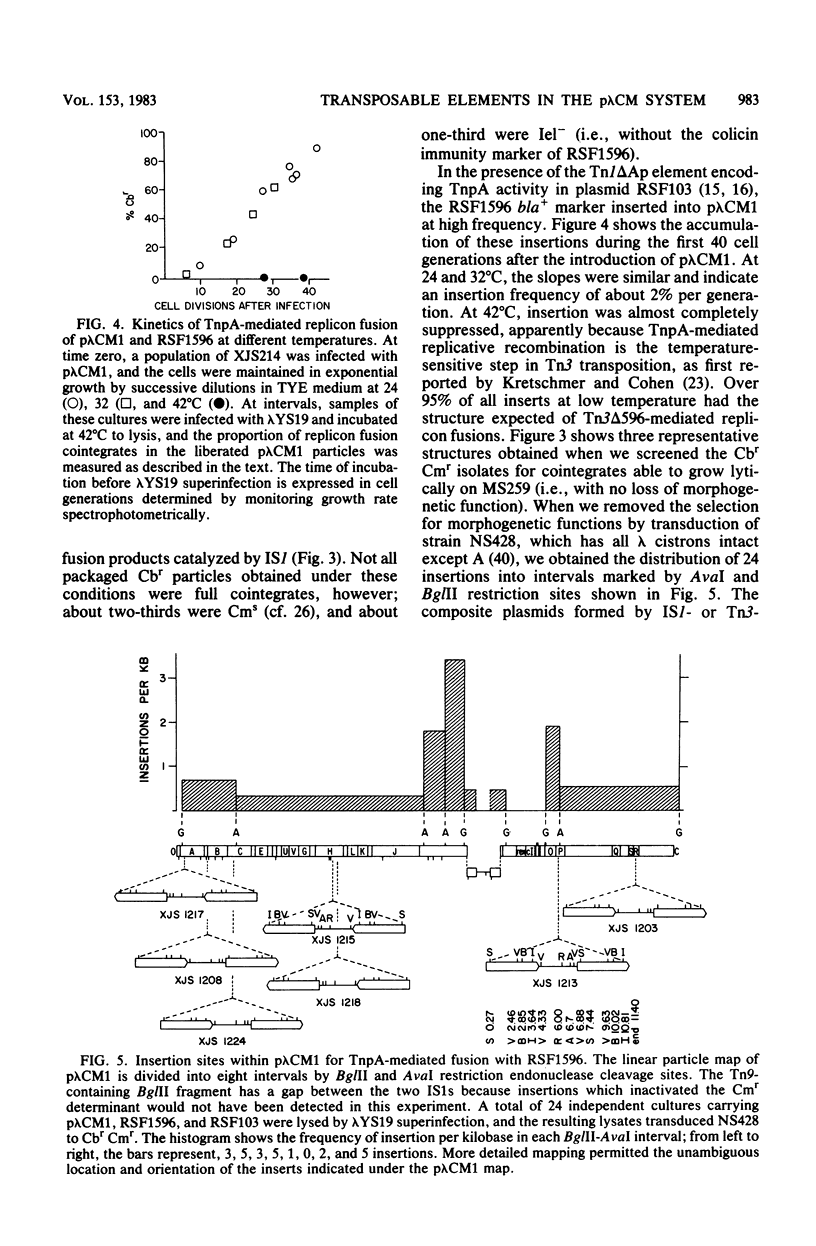
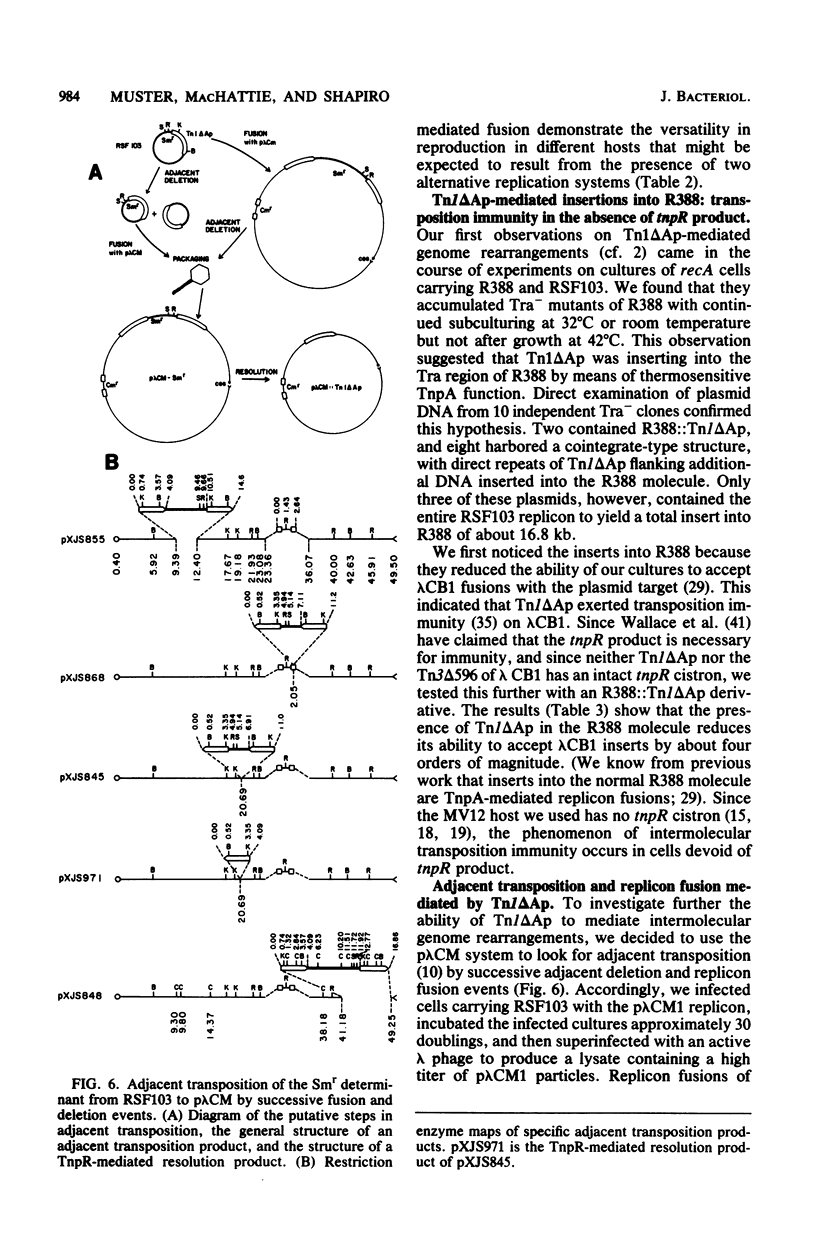
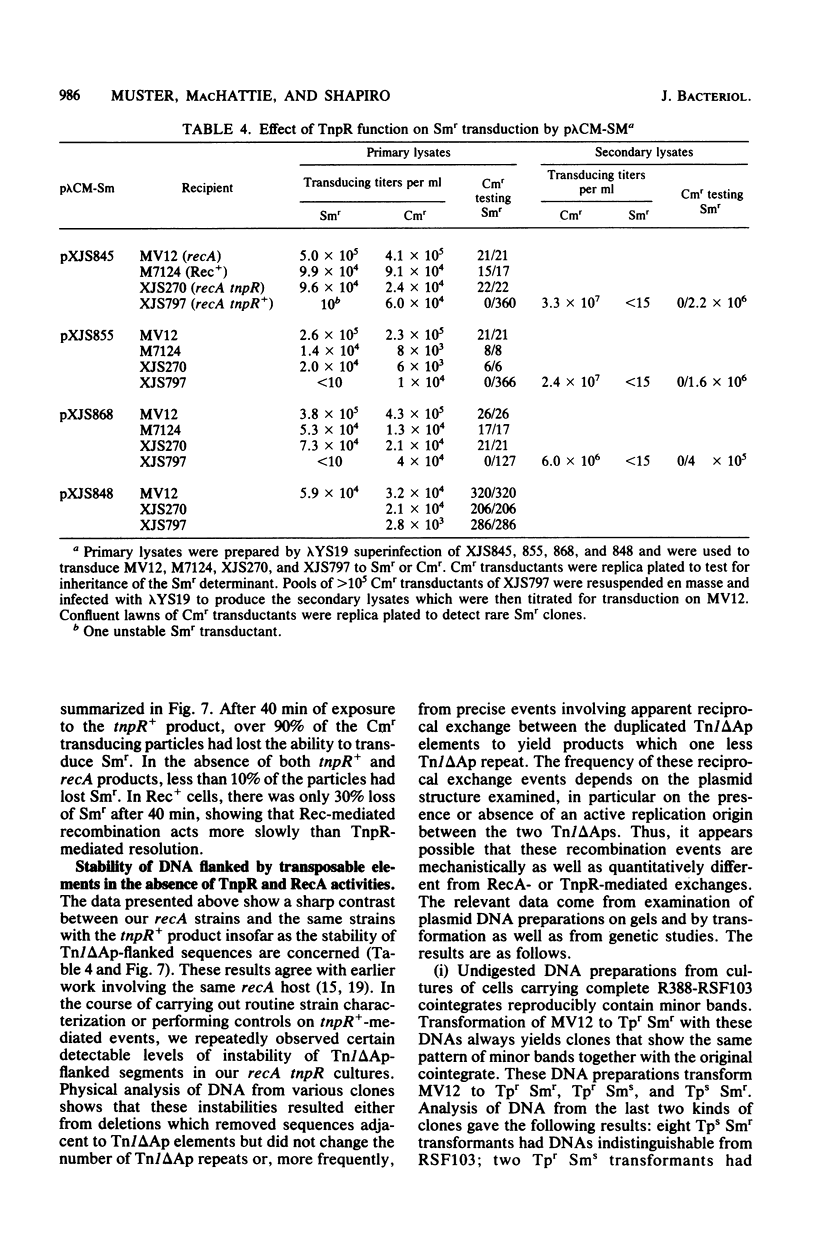
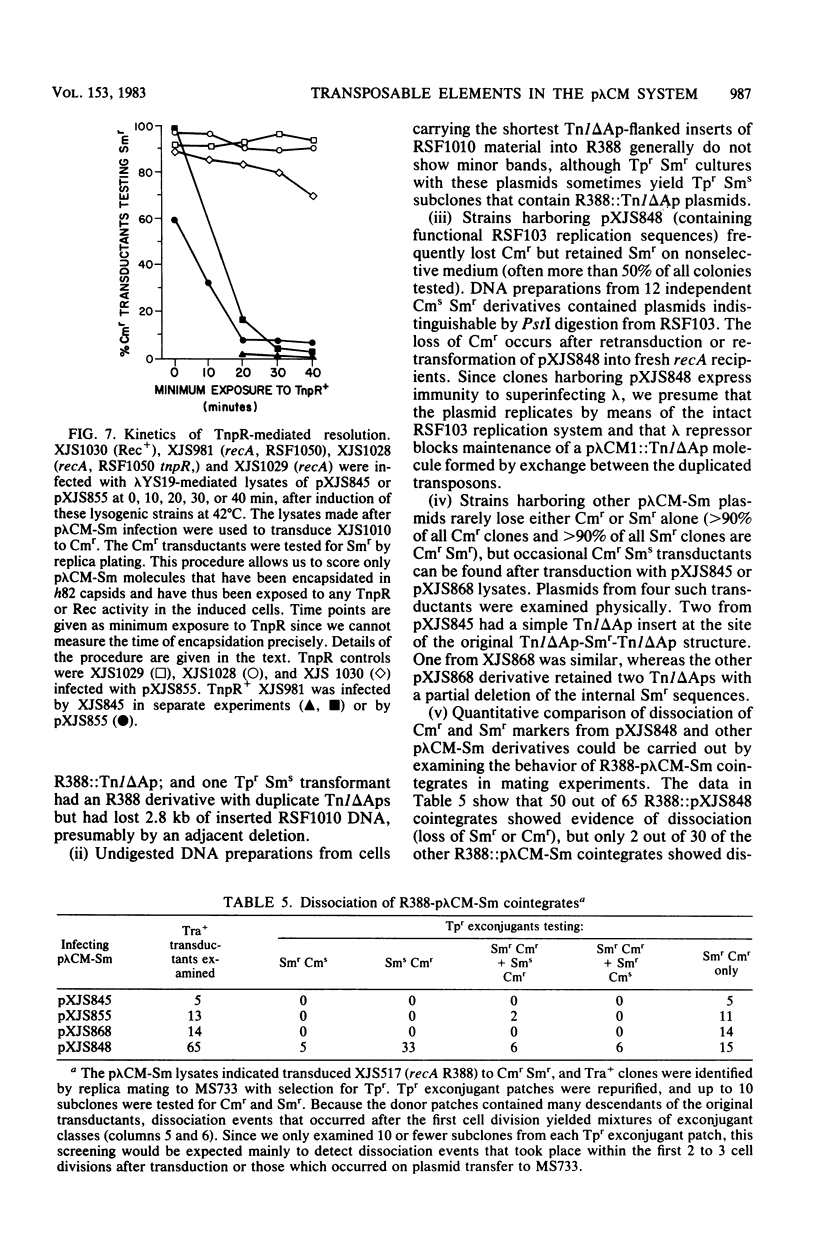
Transduction with phage derived from a 2-year-old lysate of λ cam105 (λ::Tn9) gave rise to chloramphenicol-resistant (Cmr) transductants harboring a plasmid (pλCM1) formed from λ cam105 by a Tn9-mediated adjacent deletion to position 36.07 kilobases in the N cistron of λ. The pλCM element can replicate as a plasmid, insert into the bacterial genome, or reproduce lytically as a phage on cells that provide N function. The feasibility of obtaining high titers in encapsidated form and the ease of synchronous introduction into and recovery from bacterial populations make pλCM very suitable for quantitative studies of recombination involving transposable elements. Replicon fusions between pλCM1 and RSF1596 (pMB8::Tn3Δ596) occur by duplication of either IS1 (at low rate in the absence of TnpA activity) or Tn3Δ596 (in the presence of TnpA activity). At 24 or 32°C, the rate of increase of TnpA-mediated fusions per pλCM is about 2% per cell doubling. RSF103 contains the deleted Tn1ΔAp (which lacks intact beta-lactamase and TnpR resolvase coding sequences) adjacent to a streptomycin resistance (Smr) determinant. We observed that Tn1ΔAp mediates insertions of external RSF103 sequences into the R388 plasmid. R388::Tn1ΔAp plasmids show transposition immunity in cells lacking TnpR activity. Using the pλCM system, we isolated adjacent transpositions of the RSF103 Smr determinant. The resulting pλCM-Sm cosmids contain Smr genetic material flanked by direct repeats of Tn1ΔAp, and all are deleted for some RSF103 or pλCM sequences. The pλCM-Sm constructs will fuse into R388 by duplication of a single Tn1ΔAp element. In the presence of tnpR+ (but not tnpR) Tn1 or Tn3 elements, all Tn1ΔAp-mediated complex replicons break down completely and rapidly to simple Tn1ΔAp inserts. The equilibrium for resolution is at least 105:1, and resolution is more than 90% complete after 40 min of exposure to a tnpR+ cytoplasm. In the absence of TnpR, Rec, and Red activities, Tn1ΔAp-mediated complex replicons yield simple Tn1ΔAp inserts at a lower rate. The presence of intact RSF103 replication determinants between direct Tn1ΔAp repeats appears to accelerate this precise TnpR- and Rec-independent breakdown.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedik M., Fennewald M., Shapiro J. Transposition of a beta-lactamase locus from RP1 into Pseudomonas putida degradative plasmids. J Bacteriol. 1977 Feb;129(2):809–814. doi: 10.1128/jb.129.2.809-814.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Castellazzi M., Brachet P., Eisen H. Isolation and characterization of deletions in bacteriophage lambda residing as prophage in E. coli K 12. Mol Gen Genet. 1972;117(3):211–218. doi: 10.1007/BF00271648. [DOI] [PubMed] [Google Scholar]
- Chiang S. J., Clowes R. C. Intramolecular transposition and inversion in plasmid R6K. J Bacteriol. 1980 May;142(2):668–682. doi: 10.1128/jb.142.2.668-682.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deonier R. C., Hadley R. G. IS2-IS2 and IS3-IS3 relative recombination frequencies in F integration. Plasmid. 1980 Jan;3(1):48–64. doi: 10.1016/s0147-619x(80)90033-5. [DOI] [PubMed] [Google Scholar]
- Feiss M., Fisher R. A., Crayton M. A., Egner C. Packaging of the bacteriophage lambda chromosome: effect of chromosome length. Virology. 1977 Mar;77(1):281–293. doi: 10.1016/0042-6822(77)90425-1. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Gill R. E., Heffron F., Falkow S. Identification of the protein encoded by the transposable element Tn3 which is required for its transposition. Nature. 1979 Dec 20;282(5741):797–801. doi: 10.1038/282797a0. [DOI] [PubMed] [Google Scholar]
- Gill R., Heffron F., Dougan G., Falkow S. Analysis of sequences transposed by complementation of two classes of transposition-deficient mutants of Tn3. J Bacteriol. 1978 Nov;136(2):742–756. doi: 10.1128/jb.136.2.742-756.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Signer E. R. Genetic characterization of plasmid formation by N- mutants of bacteriophage lambda. Virology. 1977 Jun 1;79(1):160–173. doi: 10.1016/0042-6822(77)90342-7. [DOI] [PubMed] [Google Scholar]
- Kretschmer P. J., Cohen S. N. Effect of temperature on translocation frequency of the Tn3 element. J Bacteriol. 1979 Aug;139(2):515–519. doi: 10.1128/jb.139.2.515-519.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHattie L. A., Shapiro J. A. Chromosomal integration of phage lambda by means of a DNA insertion element. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1490–1494. doi: 10.1073/pnas.75.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo H., Ohtsubo E. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc Natl Acad Sci U S A. 1982 Jan;79(2):277–281. doi: 10.1073/pnas.79.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Laux R., Boch G., Hinds M., Bayly R., Shapiro J. A. Broad-host-range IncP-4 plasmid R1162: effects of deletions and insertions on plasmid maintenance and host range. J Bacteriol. 1982 Oct;152(1):140–150. doi: 10.1128/jb.152.1.140-150.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster C. J., Shapiro J. A. Recombination involving transposable elements: on replicon fusion. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):239–242. doi: 10.1101/sqb.1981.045.01.036. [DOI] [PubMed] [Google Scholar]
- Nieder M., Shapiro J. Physiological function of the Pseudomonas putida PpG6 (Pseudomonas oleovorans) alkane hydroxylase: monoterminal oxidation of alkanes and fatty acids. J Bacteriol. 1975 Apr;122(1):93–98. doi: 10.1128/jb.122.1.93-98.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., McCormick M., Machida C., Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. doi: 10.1101/sqb.1981.045.01.041. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Reif H. J., Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137(1):17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- Robinson M. K., Bennett P. M., Richmond M. H. Inhibition of TnA translocation by TnA. J Bacteriol. 1977 Jan;129(1):407–414. doi: 10.1128/jb.129.1.407-414.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., MacHattie L. A. Integration and excision of prophage lambda mediated by the IS 1 element. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1135–1142. doi: 10.1101/sqb.1979.043.01.127. [DOI] [PubMed] [Google Scholar]
- Signer E. R. Plasmid formation: a new mode of lysogeny by phase lambda. Nature. 1969 Jul 12;223(5202):158–160. doi: 10.1038/223158a0. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Tiemeier D., Enquist L. In vitro packaging of a lambda Dam vector containing EcoRI DNA fragments of Escherichia coli and phage P1. Gene. 1977 May;1(3-4):255–280. doi: 10.1016/0378-1119(77)90049-x. [DOI] [PubMed] [Google Scholar]
- Wallace L. J., Ward J. M., Richmond M. H. The tnpR gene product of TnA is required for transposition immunity. Mol Gen Genet. 1981;184(1):87–91. doi: 10.1007/BF00271200. [DOI] [PubMed] [Google Scholar]
- Ward J. M., Grinsted J. Mapping of functions in the R-plasmid R388 by examination of deletion mutants generated in vitro. Gene. 1978 Apr;3(2):87–95. doi: 10.1016/0378-1119(78)90053-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- de Wet J. R., Daniels D. L., Schroeder J. L., Williams B. G., Denniston-Thompson K., Moore D. D., Blattner F. R. Restriction maps for twenty-one Charon vector phages. J Virol. 1980 Jan;33(1):401–410. doi: 10.1128/jvi.33.1.401-410.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]


