Abstract
Stresses leading to the accumulation of misfolded proteins in the endoplasmic reticulum (ER) elicit a highly conserved ER stress response in plants called the unfolded protein response (UPR). While the response itself is well documented in plants, the components of the signaling pathway are less well known. We have identified three membrane-associated basic domain/leucine zipper (bZIP) factors in Arabidopsis thaliana that are candidates for ER stress sensors/transducers. One of these factors, bZIP28, an ER-resident transcription factor, is activated in response to treatment by tunicamycin (TM), an agent that blocks N-linked protein glycosylation. Following TM treatment, bZIP28 is processed, releasing its N-terminal, cytoplasm-facing domain, which is translocated to the nucleus. Expression of a truncated form of bZIP28, containing only the cytoplasmic domain of the protein, upregulated the expression of ER stress response genes in the absence of stress conditions. Thus, bZIP28 serves as a sensor/transducer in Arabidopsis to mediate ER stress responses related to UPR.
INTRODUCTION
The ability to sense and respond to stress is a vital adaptive function in plants. The folding of proteins in the secretory pathway of plant cells is particularly sensitive to stress, and disturbances in protein folding in the endoplasmic reticulum (ER) induce an unfolded protein response (UPR). UPR is a quality control mechanism to bring the protein-folding capacity in the ER into line with the demands imposed by stress (Sitia and Braakman, 2003; Schroder and Kaufman, 2005a). UPR serves to mitigate the accumulation of unfolded or misfolded proteins in the ER by upregulating the expression of genes encoding chaperones and ER-folding proteins or by attenuating translation (Rutkowski and Kaufman, 2004).
UPR is conserved in eukaryotic organisms. In yeast, ER stress activates inositol requiring kinase 1, Ire1, an ER-localized type I transmembrane protein with a C-terminal RNase domain (Cox et al., 1993; Mori et al., 1993). Activated Ire1 splices a precursor mRNA encoding HAC1 (Cox and Walter, 1996; Sidrauski and Walter, 1997), a transcription factor that targets stress response genes possessing UPR promoter elements (Mori et al., 1992, 1996; Kohno et al., 1993).
UPR is more elaborate in mammals (Schroder and Kaufman, 2005b), where the accumulation of unfolded or misfolded proteins elicits signals by three types of ER stress sensor/transducer proteins: activating transcription factor 6 (ATF6), IRE1α or IRE1β, or dsRNA-activated protein kinase-like ER kinase (PERK) (Schroder and Kaufman, 2005a). ATF6 is a type II transmembrane protein normally retained in the ER by its association with the binding protein BiP/GRP78 (Chen et al., 2002; Shen et al., 2002). In response to stress, ATF6 dissociates from BiP/GRP78 and is transported to the Golgi, where it is subjected to proteolytic processing (Chen et al., 2002; Shen et al., 2002). Relocated ATF6 is cut on its lumen-facing side in the Golgi by a site-1 protease (S1P), and the N-terminal domain of the protein facing the cytosol is released by cleavage within the membrane by a site-2 protease (S2P) (Ye et al., 2000). The released N-terminal domain contains a basic domain/leucine zipper (bZIP) motif and is translocated to the nucleus (Yoshida et al., 1998), where it activates the transcription of genes with ER stress response elements (ERSE), such as BiP (Haze et al., 1999; Yoshida et al., 2000).
Mammalian cells also have a signaling system similar to the yeast Ire1-HAC1 pathway. ER stress activates Ire1α and Ire1β, which promote splicing of a pre-mRNA, giving rise to a mature mRNA encoding XBP1 (Yoshida et al., 2001; Calfon et al., 2002). XBP1 is a bZIP transcription factor related to ATF6 that also binds to ERSEs. In addition to its effects on transcription, UPR alleviates ER stress by attenuating bulk protein synthesis (Rutkowski and Kaufman, 2004). Protein synthesis is downregulated by phosphorylation of the eukaryotic translation initiation factor 2 α-subunit (eIF2) by PERK and by cleavage of 28S rRNA by Ire1β (Harding et al., 1999; Iwawaki et al., 2001).
Plants undergo ER stress responses, upregulating the expression of genes, such as BiP and Ca2+-dependent ER folding proteins to create a more optimal protein-folding environment (Jelitto-Van Dooren et al., 1999; Leborgne-Castel et al., 1999; Martinez and Chrispeels, 2003; Noh et al., 2003; Iwata and Koizumi, 2005a; Kamauchi et al., 2005). Homologs for the various components of the animal cell UPR have been identified in Arabidopsis thaliana, including three to four genes encoding bZIP transcription factors similar to ATF6 (Iwata and Koizumi, 2005b; Liu et al., 2007), two IRE1-related genes (Koizumi et al., 2001; Noh et al., 2002), and a number of genes related to PERK.
Iwata and Koizumi (2005a) described a membrane-associated bZIP transcription factor gene in Arabidopsis, bZIP60, which is upregulated in response to ER stress. They proposed that bZIP60 is an ER stress sensor/transducer but that it might operate in a different way than mammalian ATF6. The structure of bZIP60 differs from ATF6 in that bZIP60 does not have a canonical S1P cleavage site, and the lumen-facing domain of the protein, which would interact with chaperones, is much shorter than ATF6. Nonetheless, these authors reported that the truncated construct AtbZIP60ΔC was able to upregulate the expression of gene constructs bearing promoter elements with stress response motifs. If bZIP60 indeed plays a role in ER stresses, it may be activated by a proteolytic processing mechanism that is different from that acting on mammalian ATF6. With respect to the role of IRE1 in plants, Koizumi et al. (2001) examined whether the Arabidopsis IRE1 homologs functioned in the same way as its yeast and mammalian counterparts. They demonstrated that the N-terminal sensor domains of Arabidopsis IRE1 proteins functionally complemented a yeast Δire1 mutant.
In this study, we characterized the signaling pathway in Arabidopsis elicited by the ER stress agent, tunicamycin (TM) (Koizumi et al., 1999). We found that TM activates bZIP28, an ER resident stress sensor/transducer related to mammalian ATF6.
RESULTS
bZIP28 Mediates ER Stress Responses
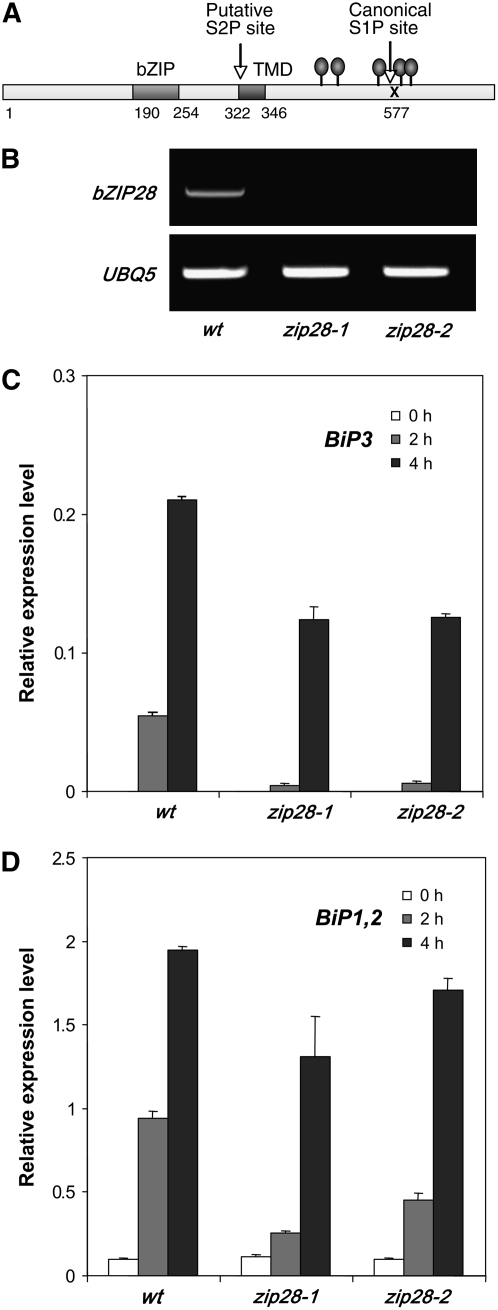
In a previous study (Liu et al., 2007), we searched the Arabidopsis genome for bZIP factors with predicted structures similar to ATF6 (i.e., proteins with a N-terminal bZIP domain, a transmembrane segment, and a canonical [RXXL or RXLX] S1P cleavage site on the C-terminal side of the transmembrane segment). Three candidate genes were identified: bZIP17 (At2g40950), -28 (At3g10800), and -49 (At3g56660) (Liu et al., 2007). We focused on bZIP28 (Figure 1A) because we found in another study that bZIP17 is associated with salt stress signaling. Also, bZIP28 was accessible because of the availability of T-DNA insertion mutations, zip28-1 (SALK_123659 with an insertion in the 5′ untranslated region) and zip28-2 (SALK_132285 with an insertion in the first exon of the gene). Both appeared to be full knockouts of bZIP28 (Figure 1B).
Figure 1.
bZIP28 Mediates Rapid ER Stress Responses.
(A) Map of bZIP28 showing the location of the bZIP DNA binding motif and the transmembrane domain (TMD). The N-terminal domain up to the TMD is predicted to be facing the cytoplasm, while the C-terminal domain is predicted to face the ER lumen. Position of a putative S2P site associated with the TMD and a canonical S1P site are indicated. Predicted N-glycosylation sites are indicated by gray ovals.
(B) RT-PCR demonstrates that the zip28-1 and zip28-2 mutants do not accumulate detectable bZIP28 transcripts.
(C) and (D) Quantitative RT-PCR was used to show that the zip28-1 and zip28-2 mutants reduce the expression of BiP3 (C) and BiP1 and -2 (D) at 2 and 4 h following treatment of seedlings with 5 μg/mL TM. BiP1, -2, and -3 expression was represented as the ratio of PCR efficiency in comparing the threshold cycles of the target cDNA to an actin gene reference. Primers did not distinguish BiP1 from BiP2, and so the expression of the two genes was measured together. Error bars indicate se (n = 3). Statistical analysis showed that differences between TM treatments and differences between the wild type and T-DNA knockout mutants are all significant (P = 0.05).
To monitor ER stress, we measured the levels of BiP3 (At1g09080), BiP1 (At5g28540), and BiP2 (At5g42020) expression by quantitative RT-PCR following treatment with TM. The zip28-1 and zip28-2 mutations reduced BiP1, -2, and -3 upregulation at 2 h following TM treatment; however, BiP1 and -2 expression, in particular, recovered somewhat by 4 h (Figures 1C and 1D). Later stages of the response (4 h and later) may involve alternative pathways, such as that mediated by At IRE1 (Koizumi et al., 2001; Noh et al., 2002). Similar observations have been made in mammalian cells in which IRE1 responses are delayed with respect to ATF6 responses (Yoshida et al., 2003). Nonetheless, bZIP28 appears to function in TM-induced ER stress signaling in Arabidopsis and appears to be more involved in the early phases of the response. To confirm that the T-DNA insertion in bZIP28 was in fact responsible for the zip28-2 mutation, we backcrossed zip28-2 mutants to the wild type and found by genotyping F2 progeny that the T-DNA insert segregated approximately in a 1:2:1 ratio (χ2 = 2.7, P = 0.253). The ratio was somewhat distorted by the underrecovery of homozygotes; nonetheless, several homozygous lines were grouped, tested, and found to be defective for TM-induced BiP3 expression (see Supplemental Figure 1 online). Unlike bZIP60, the expression of endogenous bZIP28 was not induced by TM treatment (see Supplemental Figure 2 online).
In Vivo Processing
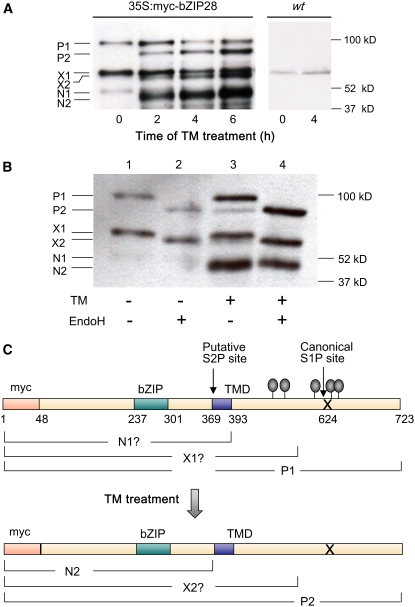
To determine whether bZIP28 is processed in response to ER stress, the protein was myc tagged at its N terminus, and 35S:myc-ZIP28 was expressed in transgenic seedlings. Prior to TM treatment, three bands reacting with the anti-myc antibody appeared in immunoblots of extracts: P1 (93 kD), X1 (60 kD), and N1 (48 kD) (Figure 2A, 0 time). We reasoned that P1 was a glycosylated form of myc-bZIP28 because P1 was converted to P2 (79 kD) when extracts were treated with β-N-acetylglucosaminidase H (EndoH) (Figure 2B, lanes 1 and 2). (For the location of predicted N-glycosylation sites, see Figure 2C.) P1 is about the predicted size for full-length myc-bZIP28 (Figure 2C). X1 appears to be largely made up of a truncated form of myc-bZIP28 and a minor amount of an endogenous protein that is present in the untransformed plant (Figure 2A, lanes marked wt) that cross-reacts with the anti-myc antibody. The component of X1 derived from myc-bZIP28 is glycosylated because it shifts, giving rise to X2 (58 kD) following EndoH treatment (Figure 2B, lanes 1 and 2). The nonglycosylated endogenous protein X1 component is fairly faint (Figure 2A, lanes marked wt) and cannot be detected in certain cases (Figure 2B, lanes 2 and 4).
Figure 2.
Processing of bZIP28 after TM-Induced ER Stress.
Transgenic lines expressing an N-terminal myc-tagged bZIP28 construct were analyzed for processing on protein gel blots using anti-myc antibodies as probe.
(A) Processing of myc-bZIP28 (50 μg total protein per lane) following treatment with TM at times indicated. Nontransgenic wild-type seedlings treated with TM for 0 and 4 h were included as controls. Interpretation of labeled bands is described in text and in protein maps below.
(B) EndoH treatment of samples from seedlings subjected to TM treatment for 0 and 2 h. The + indicates treatment, and − indicates lack of treatment. In each lane, 70 μg of total protein was analyzed.
(C) Map of myc-bZIP28 similar to the untagged form of bZIP28 shown in Figure 1A. Predicted locations of myc-tagged bZIP28 forms P1, P2, X1, X2, N1, and N2 are indicated. Note that map coordinates have been renumbered from those in Figure 1A to account for the myc tag.
Following TM treatment of 35S:myc-bZIP28 seedlings, three new bands appeared, P2 and X2, the nonglycosylated forms of myc-bZIP28 and truncated myc-bZIP28, respectively, and N2 (41 kD) (Figure 2A, 35S:myc-bZIP28, 2-6 h). N2 is not N-glycosylated (Figure 2B, compare lanes 3 and 4); therefore, we considered N2 to be the N-terminal component of the proteolytic processed form of P1 (and/or P2). N2 is about the size of a protein cleaved at the cytosolic face of the TMD at the S2P site (Figure 2C). We did not find a form that might represent S1P cleavage alone. Ye et al. (2000) made a similar observation on the processing of ATF6 in mammalian cells. They could only detect an S1P processing intermediate in a S2P mutant and from that they argued that S2P processing occurs so quickly following S1P cleavage that the intermediate is too short-lived to detect.
One anomaly in these observations was the low levels of the P1 precursor relative to product in these experiments (Figure 2A). Some of that may be due to less efficient transfer of a larger protein from the gel to the protein gel blot; however, it is also possible that the precursor is less stable than the product and may turnover in the secretory pathway. We observed the same phenomenon in the processing of myc-bZIP17 following salt stress (Liu et al., 2007). Therefore, the possibility that the precursor turns over under nonstressed conditions is under examination.
We conclude that new forms of myc-bZIP28 appear in seedlings upon TM treatment, including a nonglycosylated precursor form (P2) that results from TM blocking glycosylation and a form (N2) representing the N-terminal or cytoplasm-facing domain of myc-bZIP28 that presumably arises by S1P cleavage and further processing at the S2P site.
Nuclear Relocation of bZIP28
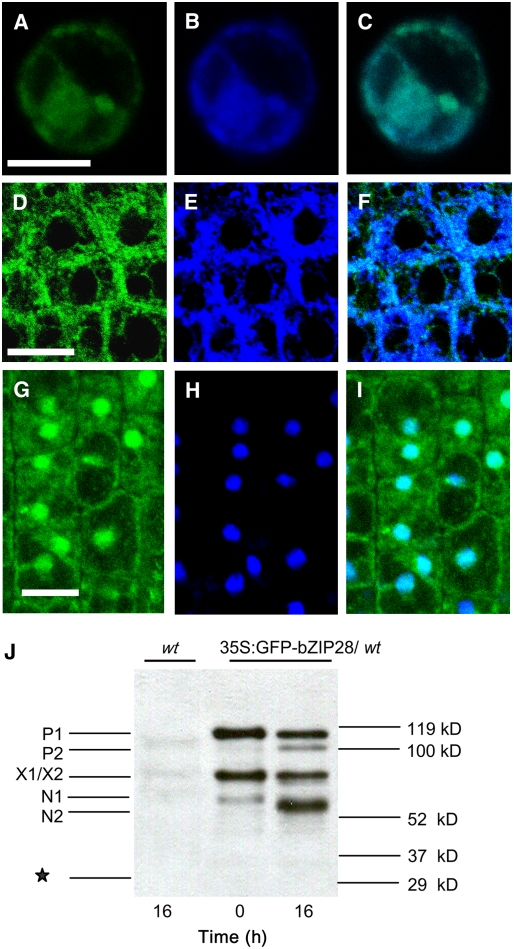
If the TM-induced stress signaling pathway in Arabidopsis operates at a cellular level similar to yeast or mammalian cells, then At bZIP28 should reside in the ER under unstressed conditions and the N-terminal component should relocate to the nucleus following stress. The subcellular localization of bZIP28 was determined using a construct in which modified green fluorescent protein (mGFP) was fused to the N terminus of the protein (35S:mGFP-bZIP28). Under normal conditions, most mGFP fluorescence in transfected Arabidopsis protoplasts was located in the cytoplasm and perinuclear region largely colocalizing with an ER marker (Figures 3A to 3C). Likewise, mGFP fluorescence colocalized with another ER marker, ER-tracker 2-aminoethyl sulfonamide (DPX), in intact roots of transgenic seedlings (Figures 3D to 3F). Under TM-induced stress conditions, mGFP-bZIP28 largely localized in nuclei, although much of the fluorescence remained in the cytoplasm (Figures 3G to 3I). We interpret this to mean that bZIP28 is processed under stress conditions and the N-terminal domain (cytoplasm-facing domain) relocates from the ER to nucleus. The fact that not all of the fluorescence relocates to the nucleus may be due to the fact that mGFP-bZIP28 is overexpressed in these cells. To demonstrate that correct processing of the mGFP-tagged bZIP28 occurs, root extracts from untreated and TM-treated plants were analyzed by protein gel blots using an anti-GFP antibody (Figure 3J). The results were comparable to the myc-tagged constructs in that the major cleavage product (N2, ∼62 kD) was the fusion between mGFP and the N terminus of bZIP28 cut at the putative S2P site. Since no free GFP was detected, we conclude that the likely form of fluorescence in the nucleus is the mGFP fusion and not free mGFP.
Figure 3.
bZIP28 Is Resident in the ER and Relocated to the Nucleus under ER Stress.
Confocal microscope images indicating the localization of bZIP28 in protoplasts from Arabidopsis suspension culture ([A] to [C]) or in roots of transgenic Arabidopsis seedlings ([D] to [I]). Bars = 10 μM.
(A) to (C) Colocalization of mGFP-bZIP28 (A) with an ER marker, SP-CFP-HDEL (B), and image overlay in protoplasts (C).
(D) to (F) Colocalization of mGFP-bZIP28 (D) with an ER marker, ER-tracker DPX (E), and image overlay in roots (F).
(G) to (I) Colocalization of mGFP-bZIP28 (G) with 4′,6-diamidino-2-phenylindole (DAPI)–stained nuclei (H) and image overlay in roots of seedlings subjected to 5 μg/mL TM treatment for 16 h (I).
(J) A protein gel blot showing the in vivo processing of mGFP-bZIP28 in roots following TM treatment for 0 and 16 h. The nontransgenic wild type treated for 16 h with TM was included as a control. Equal amounts of total protein (120 μg) were loaded in each lane. Interpretation of labeled bands is similar to Figure 2 and described in the text, except that the molecular sizes are different due to the mGFP tag. The asterisk indicates the predicted migration position of free mGFP.
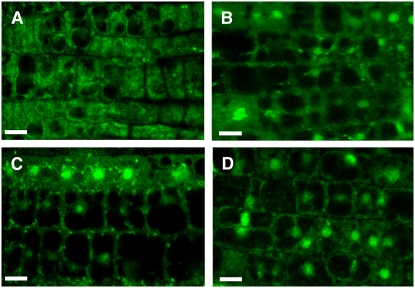
Since, upregulation of BiP genes occurs within a few hours of TM treatment, we examined the time course for the nuclear relocation of mGFP-bZIP28 (Figure 4). GFP fluorescence began to concentrate in some nuclei by 2 h (Figure 4B) and to more nuclei by 4 h (Figure 4C). In fact, about as much fluorescence had relocated to nuclei by 4 h as had during overnight (16 h) incubation (Figure 4D). Thus, the time course for nuclear translocation of the N-terminal component correlates closely with the kinetics of upregulation of BiP genes.
Figure 4.
The N-terminal Component of bZIP28 Is Rapidly Relocated to Nucleus after TM Treatment.
Transgenic seedlings expressing mGFP-bZIP28 were treated with 5 μg/mL TM, and root tips were imaged with a confocal microscope at 0 (A), 2 (B), 4 (C), and 16 h (D) after treatment.
It was difficult to determine from these experiments whether mGFP-bZIP28 transits through the Golgi prior to its relocation to the nucleus. We have seen some fluorescence in particulate structures at the 2 h time point; however, further studies are under way to determine the subcellular localization of that fluorescent signal.
Specificity of the Response
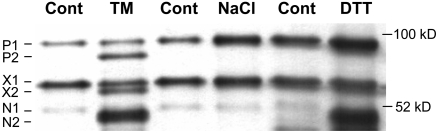
Since there are three Arabidopsis genes that encode bZIP proteins with features similar to ATF6, this begs the question whether responses to different stressors are mediated by different bZIP factors. We demonstrated in another study that bZIP17 functions in salt stress (Liu et al., 2007). In this study, we asked whether bZIP28 was processed in response to salt stress in the same way it was after TM treatment. As before, we observed that after TM treatment myc-bZIP28 was processed in vivo, yielding N2, an N-terminal fragment cut at the presumed S2P cleavage site (Figure 5, lane TM). We did not observe myc-bZIP28 processing and the appearance of N2 under salt stress (100 mM NaCl) (Figure 5, lane NaCl). However, treatment with another ER stress agent, DTT, resulted in myc-bZIP28 processing (Figure 5, lane DTT). DTT is thought to interfere with Cys bridge formation and the proper folding of ER proteins. We did not observe the appearance of P2 or X2 in response to salt or DTT stress because these two species are nonglycosylated proteins that only accumulate when N-linked glycosylation is inhibited by TM. Thus, we conclude from the above that bZIP17 and bZIP28 are involved in different ER stress responses and that bZIP28 is recruited in response to stress elicited by conventional UPR stress agents, TM or DTT.
Figure 5.
Processing of bZIP28 in Vivo after Treatment by Other Stressors.
Protein gel blot assay for processing myc-bZIP28 in vivo as described in Figure 2. Transgenic seedlings were subjected for 4 h to different stress agents, 5 μg/mL TM, 100 mM NaCl, 2 mM DTT, or control (Cont; dimethyl suloxide or water). Refer to text for description of labeled protein bands. Equal amounts of total protein (50 μg) were loaded in each lane.
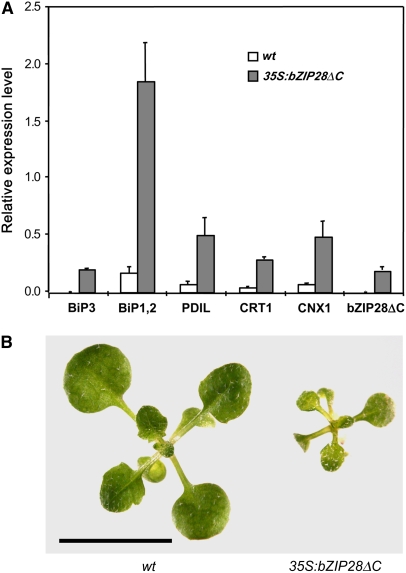
Other laboratories have profiled gene expression in Arabidopsis following treatment with ER stress agents. Martinez and Chrispeels (2003) showed that genes encoding chaperones, such as BiP, and Ca2+-dependent ER folding protein were prominent among the upregulated genes. A prediction from the hypothesis that bZIP28 processing mediates TM-induced ER stress responses is that expression of a truncated form of the protein without the transmembrane and C-terminal domain should constitutively activate the expression of ER stress response genes. The prediction was tested by expressing bZIP28ΔC in a wild-type background driven by the 35S promoter and comparing the gene expression patterns to the empty vector in the same background. Transgenic lines with modest overexpression levels were recovered. It was found that BiP1, -2, and -3 and several other ER folding enzyme genes were indeed upregulated in the bZIP28ΔC expressing lines without any ER stress inducer (Figure 6A). Thus, expression of the nonmembrane anchored form of bZIP28 is sufficient to upregulate ER stress response genes and mimics the condition of TM-induced ER stress with respect to the expression of these key marker genes. Constitutive expression of the truncated form of bZIP28 delayed growth of seedlings (Figure 6B) as might be expected for plants responding to stress conditions. However, mature transgenic and nontransgenic plants had similar form and stature.
Figure 6.
Truncated bZIP28 Upregulates the Expression of ER Stress Genes without TM Induction.
bZIP28ΔC, a truncated form of bZIP28 without the TMD and lumen-facing domain, was expressed in transgenic seedlings.
(A) Several genes, known to be upregulated by TM treatment, were assessed by quantitative RT-PCR for expression promoted by bZIP28ΔC without TM treatment. PDIL (protein disulfide isomerase-like protein; At1g21750), CRT1 (calreticulin-1; At1g56340), and CNX1 (calnexin-1; At5g61790). Relative expression is represented as the ratio of PCR efficiency in comparing the threshold cycles of the target cDNA to an actin gene reference. Error bars indicate se (n = 3). Statistical analysis showed that differences between empty vector control (wt) and 35S:bZIP28ΔC transgenic seedlings were significant (P = 0.05).
(B) Comparison in growth of 10-d-old wild-type and bZIP28ΔC transgenic seedlings. Bar = 10 mm.
DISCUSSION
The image that emerges from this study is a plant cell poised to respond to stress. Transcription factor precursors lying in the ER membranes represent a state of preparedness to environmental threats. Among the battery of membrane-associated transcription factor precursors in Arabidopsis (Jakoby et al., 2002) are three in the bZIP family that are candidates for sensors/transducers of ER stress signals (Jakoby et al., 2002). These bZIP factors are all type II membrane proteins with structures similar to mammalian ATF6. We have shown in this study that one of the factors, bZIP28, transduces ER stress generated by TM. In transducing the ER stress signal, bZIP28 is subjected to proteolytic processing and is relocated from the ER to the nucleus.
In a previous study, we found that another membrane-associated bZIP factor, bZIP17, is activated by salt stress in Arabidopsis (Liu et al., 2007). We have shown here that bZIP28 is activated by both ER stress-inducing agents, TM and DTT, but not by salt stress, and upregulates genes that are distinct from those induced by salt stress. Therefore, bZIP17 and -28 are able to distinguish between these two types of stresses. It is assumed that bZIP28 senses misfolded proteins that accumulate in the presence of stress agents that interfere with N-linked glycosylation or disulfide bridge formation. It is not clear how bZIP17 senses salt stress. It is possible that both sense misfolded proteins but that the populations of misfolded proteins differ as, perhaps, do the chaperones that interact with these proteins.
The problem in sorting out stress responses is probably much larger than what we see here. Recent reports suggest that there are transcription factors in other gene families predicted to be membrane associated and may respond to different stresses (Kim et al., 2006). In particular, 13 members of the NAC transcription factor family, one of the largest transcription factor families in Arabidopsis, have membrane-spanning domains in their C-terminal tails (Kim et al., 2006). It is not clear whether any or all of these membrane-associated NAC transcription factors are activated by membrane release; however, Kim et al. (2006) showed that transgenic plants overexpressing a truncated construct for one of the factors (NTL6) showed a strong phenotype similar to abiotic stress. Surprisingly, a number of the NAC transcription factor genes were also regulated at the transcript accumulation level by abiotic stresses, adding an additional level of complexity to the responses (Kim et al., 2006). Recently, Kim et al. (2007) reported that one of these NAC transcription factor genes, NTM1, appears to mediate the role of cytokinin in cell cycle regulation. They suggest that NTM1 is released from membranes by a calpain protease and that cytokinins stabilize the protein.
We have shown that transgenic expression of a constitutively active form of bZIP28 (bZIP28ΔC) leads to the upregulation of BiP genes and genes encoding other ER protein folding factors. These genes were shown by Martinez and Chrispeels (2003) and Kamauchi et al. (2005) to be upregulated by ER stress agents used to induce UPR. We have not yet demonstrated that bZIP28 directly targets and activates these genes, and we do not know its DNA binding specificity. Oh et al. (2003) have identified a 24-bp cis-element ATTGGTCCACGTCATC involved in plant UPR. The element (P-UPRE) contains two overlapping sequences (ERSE-II and Xbp1 binding sequences) responsible for the UPR in animals, and either of the two sequences is sufficient for the plant UPR. ERSE-like and Xbp1 binding-like cis-elements were also found in promoter regions of other chaperone genes induced upon the UPR (Noh et al., 2003).
A fascinating aspect of UPR is its cell biology. The bZIP28 sensor/transducer is located in the ER, and upon activation, the protein is translocated to the nucleus. In another study (Liu et al., 2007), we showed that bZIP17 processing requires S1P and that this protease is located in the Golgi. Presuming that this is also the case for bZIP28, then activation would involve relocation to the Golgi apparatus for processing. We have not yet been able to track mGFP-bZIP28 through the Golgi, perhaps because its residency in that organelle may be brief. We hope to catch bZIP28 en route by observing the response in an S1P mutant.
METHODS
Plant Materials and Growth Conditions
T-DNA insertion lines for bZIP28 were obtained from the ABRC, and homozygous plants were screened by PCR using a left border T-DNA primer and gene-specific primers listed in Supplemental Table 1 online. T2 and/or T3 generations of transgenics were studied, and where indicated, homozygous lines were selected. Seeds were germinated on agar plates containing half-strength Murashige and Skoog (MS) salts, 1% sucrose, and 0.05% MES, pH 5.7, after stratification at 4°C for at least 2 d. One-week-old seedlings grown in an illuminated growth chamber at 23°C were harvested for further experiments, except in some experiments, where only roots were used for protein gel blots with an anti-GFP antibody and confocal imaging. For RT-PCR, in vivo processing, or confocal analysis, seedlings were transferred to liquid MS medium plus TM (5 μg/mL), DTT (2 mM), or NaCl (100 mM) for various periods as indicated.
Transcript Level Analysis
Total RNA from plant tissues pooled from three separate agar plates was isolated using an RNeasy kit, treated with RNase-free DNase I according to the manufacturer's instructions (Qiagen), and quantified by 260/280-nm UV light absorption. One microgram of total RNA was reverse transcribed using the Supertranscript III RT kit (Invitrogen). For quantitative RT-PCR, 6.8 μL of 10 times dilution of cDNA was used with a total reaction volume of 20 μL. The final primer concentration was 0.2 μM, and all primers are listed in Supplemental Table 1 online. Quantitative RT-PCR was performed with the Stratagene Mx4000 multiplex quantitative PCR system with SYBR Green PCR master mix (Applied Biosystems). The efficiency of amplification of various cDNAs was assessed relative to amplification of transcripts from two actin genes (actin 2, At3g18780; actin 8, At1g49240). Each RNA sample was assayed in triplicate. Expression levels were calculated relative to actin using a comparative threshold cycle method with ΔΔCt = ΔCtreference − ΔCtsample, where ΔCtsample was the Ct value for the assay sample normalized to actin, and ΔCtreference is the Ct value for calibration, also normalized to actin. Statistical two-way analysis of variance was performed using the SAS system (SAS Institute). Tukey's studentized range (honestly significant difference) test was used to determine significant differences among genotypes or treatment time points. An α level of 0.05 was used for statistical significance.
Plasmid Construction
The open reading frame of bZIP28 (At3g10800) was amplified from 1-week-old seedlings by RT-PCR (primer OE800) and cloned into pSKM36 at the AscI and SpeI sites, resulting in pSK800. For confocal analysis, mGFP was amplified (primer GFP-AscI) from an m-GFP-ER vector and inserted into pSK800 at the AscI site to generate an N-terminal mGFP-tagged bZIP28. The ER marker (SP-CFP-HDEL) was made by amplifying CFP from pSKC36 with a forward primer containing a chitinase signal peptide and reverse primer with ER retention signal HDEL and inserting into pCHF1 at SmaI and BamHI sites. For in vivo processing analyses, a 4× epitope myc tag (EQKLISEEDLRN) was amplified from pSKM36 using the primer MYC-AscI (includes ATG before the myc sequence) and inserted into pSK800 at an AscI site to generate an N-terminal myc-tagged, full-length bZIP28. To generate the truncated bZIP28 construct (pSKM800ΔC), in which the transmembrane domain and C-terminal lumen-facing sequence were eliminated, the first 966 nucleotides of the bZIP28 coding sequence were amplified using primer OE800ΔC and inserted into pSKM36 at AscI and SpeI sites. All the primers used in this study are listed in Supplemental Table 1 online, and all the clones were confirmed by sequencing the whole inserts.
Confocal Microscopy
For subcellular localization of mGFP-bZIP28, roots were stained with DAPI (5 μg/mL; Invitrogen) or the ER marker ER-tracker Blue-White DPX (1 μM; Invitrogen) and observed under a laser confocal microscope (Olympus Fluo View FV1000). A sequential scanning mode was used when DAPI or ER marker staining was combined with GFP to minimize the crosstalk between the two partially overlapping emission spectra. An Arabidopsis thaliana Columbia-0 suspension cell culture was obtained from D.C. Bassham (Iowa State University) and maintained by subculturing weekly in MS Minimal Organics medium (Gibco BRL), 2% (w/v) sucrose, 1 mg/mL naphthalene acetic acid (Sigma-Aldrich), and 1 mg/mL kinetin (Sigma-Aldrich). Cultures were grown in Erlenmeyer flasks at room temperature, under ambient light, with constant shaking (115-rpm rotation). Protoplasts isolated from Arabidopsis suspension culture were cotransfected with mGFP-bZIP28 and a CFP ER marker using a polyethylene glycol (PEG 4000) method. Protoplasts were examined under a confocal microscope after overnight incubation in the dark at room temperature.
In Vivo bZIP28 Processing
In vivo processing of myc-bZIP28 and mGFP-bZIP28 was examined using protein gel blotting according to Liu et al. (2007). SDS was omitted from the extraction buffer, and the Bradford method was used for protein quantification. The 6% protogels (National Diagnostic) were used for detection of mGFP-bZIP28 processing; otherwise, 10% protogels were used. For EndoH treatment, an equal amount (70 μg) of total protein was diluted with reaction buffer and denaturation solution and heated at 100°C for 5 min, cooled on ice, 4 μL of EndoH (Sigma-Aldrich), or water as a control were added, and the mixture was incubated at 37°C overnight. After digestion, protein loading buffer was added and loaded into the gels after denaturing at 65°C for 10 min. The anti-c-myc antibody (9E10) was obtained from Santa Cruz Biotechnology, and anti-GFP antibody was purchased from Medical and Biological Laboratories.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At3g10800 (bZIP28), At2g40950 (bZIP17), At3g56660 (bZIP49), At5g28540 (BiP1), At5g42020 (BiP2), At1g09080 (BiP3), At1g21750 (PDIL), At1g56340 (CRT1), At5g61790 (CNX1), At3g18780 (ACT2), and At1g49240 (ACT8).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Inability to Upregulate BiP3 Expression by TM Segregates with T-DNA Insertion in bZIP28.
Supplemental Figure 2. bZIP28 Expression Is Not Upregulated by TM Treatment.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
This work supported by the National Science Foundation 2010 program (IBN0420015).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stephen H. Howell (shh@iastate.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Calfon, M., Zeng, H., Urano, F., Till, J.H., Hubbard, S.R., Harding, H.P., Clark, S.G., and Ron, D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415 92–96. [DOI] [PubMed] [Google Scholar]
- Chen, X., Shen, J., and Prywes, R. (2002). The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 277 13045–13052. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., Shamu, C.E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73 1197–1206. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., and Walter, P. (1996). A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87 391–404. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397 271–274. [DOI] [PubMed] [Google Scholar]
- Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, Y., and Koizumi, N. (2005. a). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 102 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, Y., and Koizumi, N. (2005. b). Unfolded protein response followed by induction of cell death in cultured tobacco cells treated with tunicamycin. Planta 220 804–807. [DOI] [PubMed] [Google Scholar]
- Iwawaki, T., Hosoda, A., Okuda, T., Kamigori, Y., Nomura-Furuwatari, C., Kimata, Y., Tsuru, A., and Kohno, K. (2001). Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 3 158–164. [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. [DOI] [PubMed] [Google Scholar]
- Jelitto-Van Dooren, E.P., Vidal, S., and Denecke, J. (1999). Anticipating endoplasmic reticulum stress. A novel early response before pathogenesis-related gene induction. Plant Cell 11 1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi, S., Nakatani, H., Nakano, C., and Urade, R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272 3461–3476. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., Kim, S.G., Kim, Y.S., Seo, P.J., Bae, M., Yoon, H.K., and Park, C.M. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: Implications for membrane biology in genome regulation. Nucleic Acids Res. 35 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.S., Kim, S.G., Park, J.E., Park, H.Y., Lim, M.H., Chua, N.H., and Park, C.M. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, K., Normington, K., Sambrook, J., Gething, M.J., and Mori, K. (1993). The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, N., Martinez, I.M., Kimata, Y., Kohno, K., Sano, H., and Chrispeels, M.J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127 949–962. [PMC free article] [PubMed] [Google Scholar]
- Koizumi, N., Ujino, T., Sano, H., and Chrispeels, M.J. (1999). Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 121 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leborgne-Castel, N., Jelitto-Van Dooren, E.P., Crofts, A.J., and Denecke, J. (1999). Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell 11 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.X., Srivastava, R., Che, P., and Howell, S.H. (2007). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to ER stress signaling. Plant J. 51 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, K., Kawahara, T., Yoshida, H., Yanagi, H., and Yura, T. (1996). Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1 803–817. [DOI] [PubMed] [Google Scholar]
- Mori, K., Ma, W., Gething, M.J., and Sambrook, J. (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74 743–756. [DOI] [PubMed] [Google Scholar]
- Mori, K., Sant, A., Kohno, K., Normington, K., Gething, M.J., and Sambrook, J.F. (1992). A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 11 2583–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, S.J., Kwon, C.S., and Chung, W.I. (2002). Characterization of two homologs of Ire1p, a kinase/endoribonuclease in yeast, in Arabidopsis thaliana. Biochim. Biophys. Acta 1575 130–134. [DOI] [PubMed] [Google Scholar]
- Noh, S.J., Kwon, C.S., Oh, D.H., Moon, J.S., and Chung, W.I. (2003). Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311 81–91. [DOI] [PubMed] [Google Scholar]
- Oh, D.H., Kwon, C.S., Sano, H., Chung, W.I., and Koizumi, N. (2003). Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem. Biophys. Res. Commun. 301 225–230. [DOI] [PubMed] [Google Scholar]
- Rutkowski, D.T., and Kaufman, R.J. (2004). A trip to the ER: Coping with stress. Trends Cell Biol. 14 20–28. [DOI] [PubMed] [Google Scholar]
- Schroder, M., and Kaufman, R.J. (2005. a). The mammalian unfolded protein response. Annu. Rev. Biochem. 74 739–789. [DOI] [PubMed] [Google Scholar]
- Schroder, M., and Kaufman, R.J. (2005. b). ER stress and the unfolded protein response. Mutat. Res. 569 29–63. [DOI] [PubMed] [Google Scholar]
- Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3 99–111. [DOI] [PubMed] [Google Scholar]
- Sidrauski, C., and Walter, P. (1997). The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90 1031–1039. [DOI] [PubMed] [Google Scholar]
- Sitia, R., and Braakman, I. (2003). Quality control in the endoplasmic reticulum protein factory. Nature 426 891–894. [DOI] [PubMed] [Google Scholar]
- Ye, J., Rawson, R.B., Komuro, R., Chen, X., Dave, U.P., Prywes, R., Brown, M.S., and Goldstein, J.L. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6 1355–1364. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Haze, K., Yanagi, H., Yura, T., and Mori, K. (1998). Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273 33741–33749. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Hosokawa, N., Kaufman, R.J., Nagata, K., and Mori, K. (2003). A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 4 265–271. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107 881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2000). ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20 6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.