Abstract
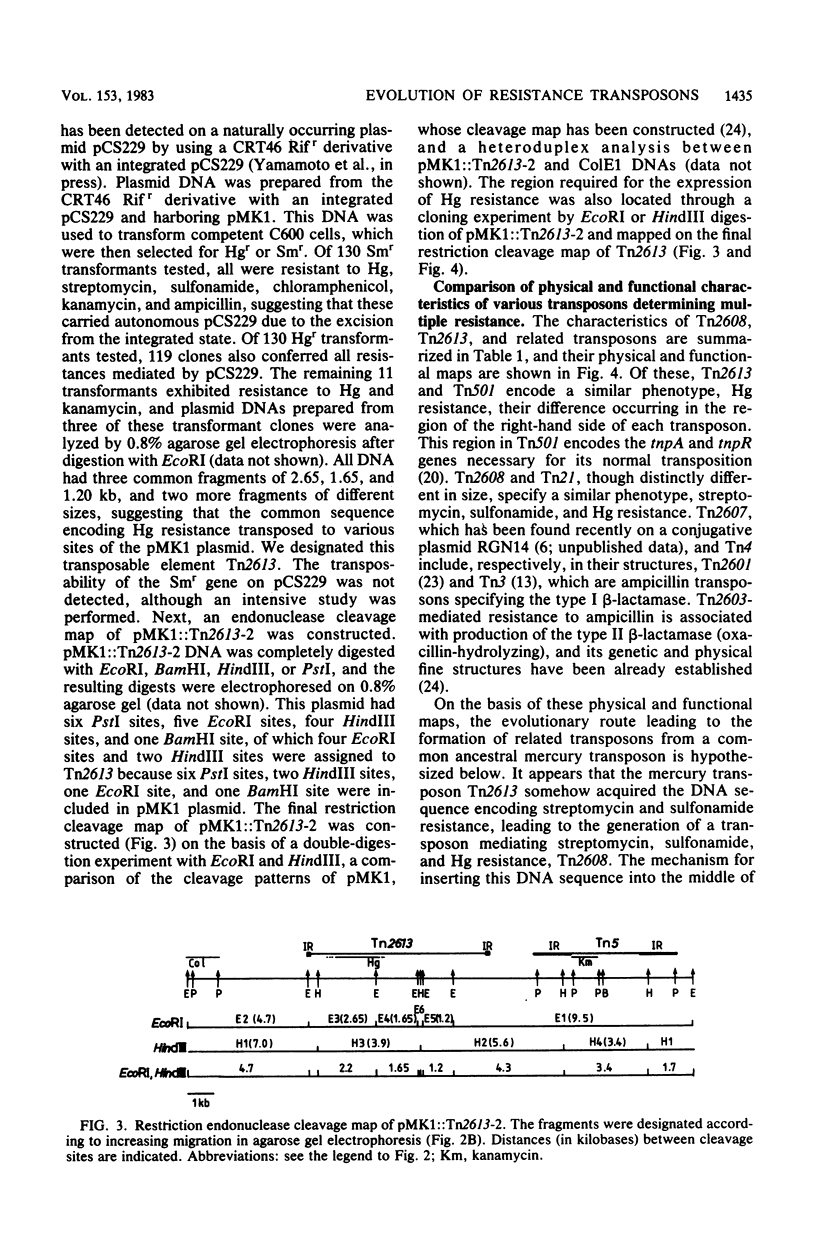
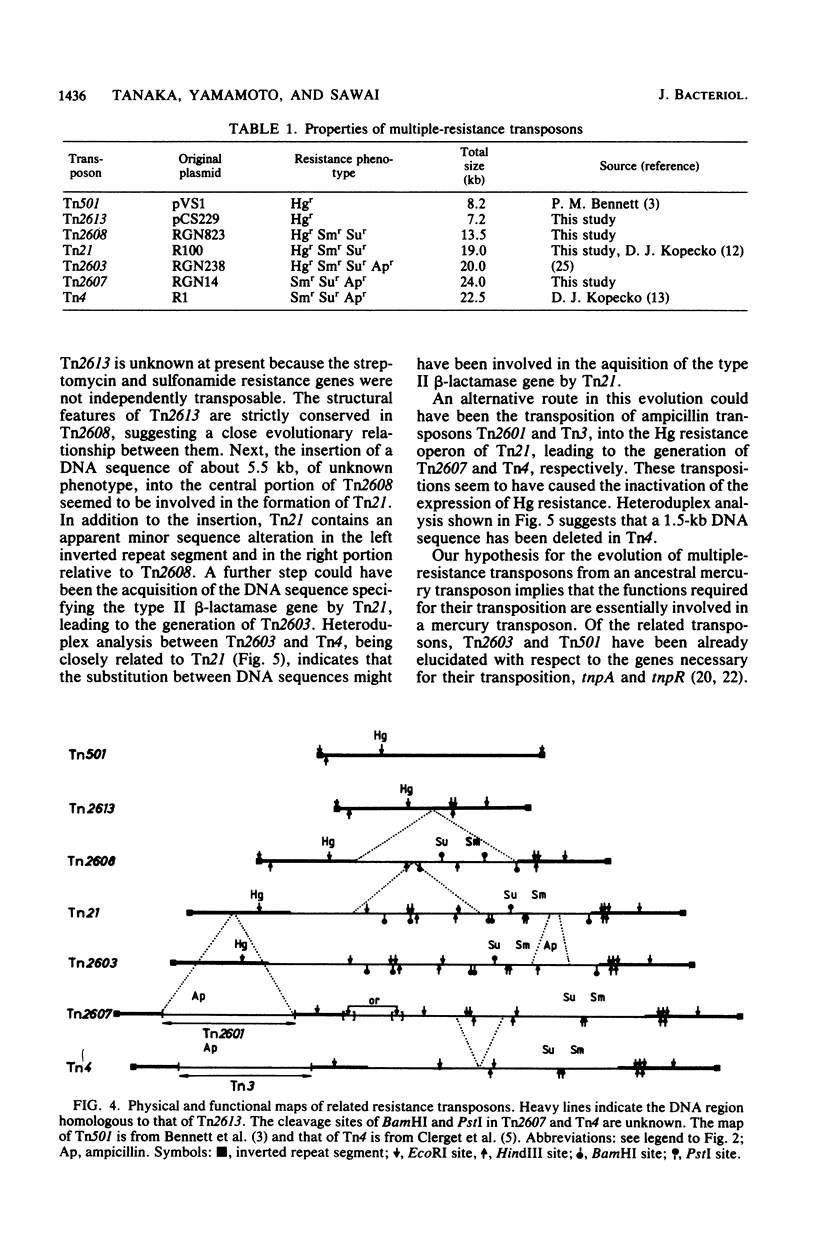
The molecular interrelationship of a transposon family which confers multiple antibiotic resistance and is assumed to have been generated from an ancestral mercury transposon was analyzed. Initially, the transposons Tn2613 (7.2 kilobases), encoding mercury resistance, and Tn2608 (13.5 kilobases), encoding mercury, streptomycin, and sulfonamide resistances, were isolated and their structures were analyzed. Next, the following transposons were compared with respect to their genetic and physical maps: Tn2613 and Tn501, encoding mercury resistance; Tn2608 and Tn21, encoding mercury, streptomycin, and sulfonamide resistance; Tn2607 and Tn4, encoding streptomycin, sulfonamide, and ampicillin resistance; and Tn2603, encoding mercury, streptomycin, sulfonamide, and ampicillin resistance. The results suggest that the transposons encoding multiple resistance were evolved from an ancestral mercury transposon.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Chou J., Lemaux P. G., Casadaban M. J., Cohen S. N. Transposition protein of Tn3: identification and characterisation of an essential repressor-controlled gene product. Nature. 1979 Dec 20;282(5741):801–806. doi: 10.1038/282801a0. [DOI] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. The structure of R1drd19: a revised physical map of the plasmid. Mol Gen Genet. 1981;181(2):183–191. doi: 10.1007/BF00268425. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Gill R. E., Heffron F., Falkow S. Identification of the protein encoded by the transposable element Tn3 which is required for its transposition. Nature. 1979 Dec 20;282(5741):797–801. doi: 10.1038/282797a0. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Terminal nucleotide sequences of Tn551, a transposon specifying erythromycin resistance in Staphylococcus aureus: homology with Tn3. Plasmid. 1980 Sep;4(2):148–154. doi: 10.1016/0147-619x(80)90004-9. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Caro L., Berg C. M., Hirota Y. Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. J Mol Biol. 1971 Feb 14;55(3):441–456. doi: 10.1016/0022-2836(71)90328-7. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Schwesinger M. D., Gruss A. D., Swanson E. C., Pattee P. A. Genetic translocation in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):400–404. doi: 10.1073/pnas.76.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka T., Hashimoto H., Mitsuhashi S. Comparison of penicillinases produced by R factors isolated from ampicillin-resistant gram-negative bacteria. Jpn J Microbiol. 1970 Mar;14(2):123–128. doi: 10.1111/j.1348-0421.1970.tb00499.x. [DOI] [PubMed] [Google Scholar]
- Radford A. J., Oliver J., Kelly W. J., Reanney D. C. Translocatable resistance to mercuric and phenylmercuric ions in soil bacteria. J Bacteriol. 1981 Sep;147(3):1110–1112. doi: 10.1128/jb.147.3.1110-1112.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein S. J., Jorgensen R. A., Postle K., Reznikoff W. S. The inverted repeats of Tn5 are functionally different. Cell. 1980 Mar;19(3):795–805. doi: 10.1016/s0092-8674(80)80055-9. [DOI] [PubMed] [Google Scholar]
- Rubens C. E., McNeill W. F., Farrar W. E., Jr Evolution of multiple-antibiotic-resistance plasmids mediated by transposable plasmid deoxyribonucleic acid sequences. J Bacteriol. 1979 Nov;140(2):713–719. doi: 10.1128/jb.140.2.713-719.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Weiss R. B., Jacoby G. A. Transposition of mercury resistance from a transferable R plasmic of Escherichia coli. Plasmid. 1980 Jan;3(1):35–47. doi: 10.1016/s0147-619x(80)90032-3. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Harafuji H., Yamamoto T. A gene and its product required for transposition of resistance transposon Tn2603. J Bacteriol. 1982 Aug;151(2):723–728. doi: 10.1128/jb.151.2.723-728.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Katoh R., Shimazu A., Yamagishi S. Gene expression of ampicillin resistance transposons, Tn2601 and Tn2602. Microbiol Immunol. 1980;24(6):479–494. doi: 10.1111/j.1348-0421.1980.tb02852.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Baba R., Yamagishi S. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol Gen Genet. 1981;181(4):464–469. doi: 10.1007/BF00428737. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Nohara C., Fukunaga Y., Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981 Feb;145(2):808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Yamagata S., Horii K., Yamagishi S. Comparison of transcription of beta-lactamase genes specified by various ampicillin transposons. J Bacteriol. 1982 Apr;150(1):269–276. doi: 10.1128/jb.150.1.269-276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]