Abstract
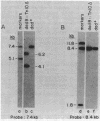
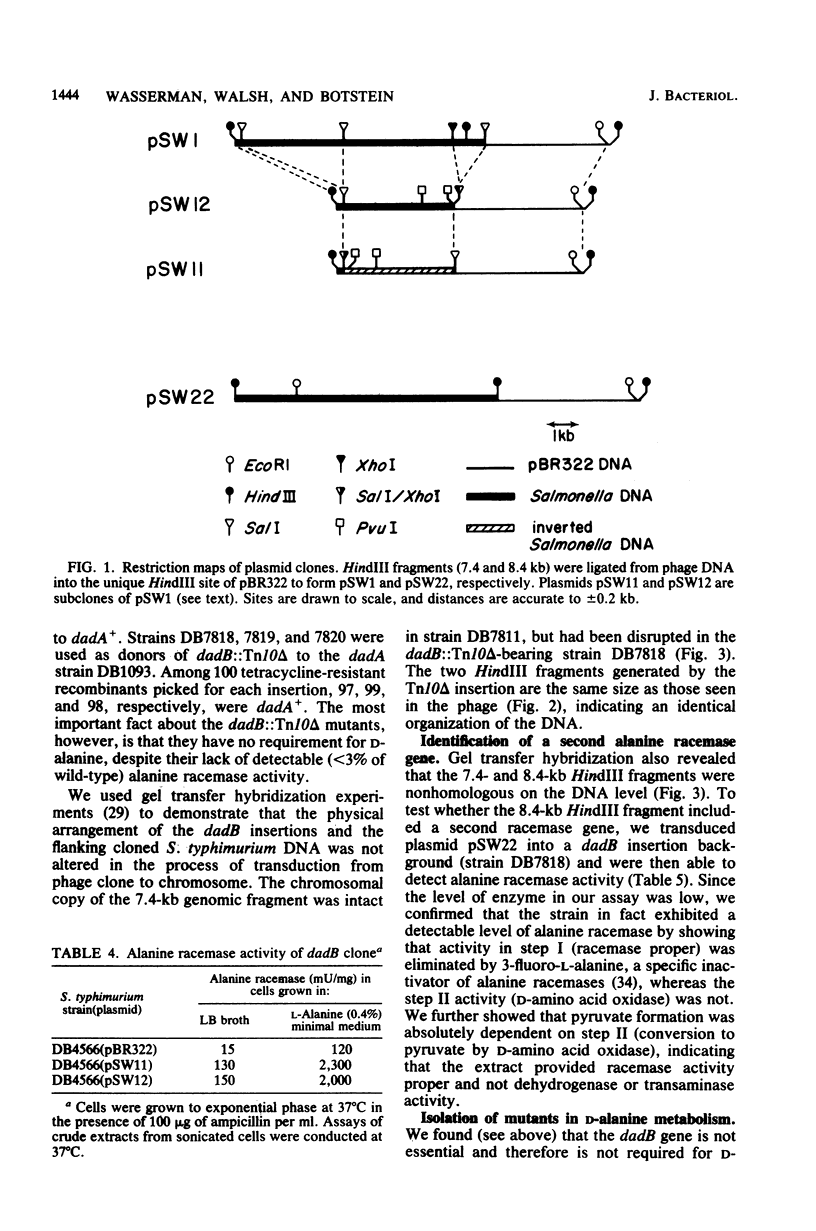
Mutations were isolated in a previously undescribed Salmonella typhimurium gene encoding an alanine racemase essential for utilization of L-alanine as a source of carbon, energy, and nitrogen. This new locus, designated dadB, lies within one kilobase of the D-alanine dehydrogenase locus (dadA), which is also required for alanine catabolism. The dadA and dadB genes are coregulated. Mutants (including insertions) lacking the dadB alanine racemase do not require D-alanine for growth unless a mutation is introduced at a second locus, designated dal. Two genes specifying alanine racemase activity were cloned from S. typhimurium. The two cloned DNA sequences do not cross-hybridize with each other; one was shown to contain the dadB gene.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atherton F. R., Hall M. J., Hassall C. H., Lambert R. W., Lloyd W. J., Ringrose P. S. Phosphonopeptides as antibacterial agents: mechanism of action of alaphosphin. Antimicrob Agents Chemother. 1979 May;15(5):696–705. doi: 10.1128/aac.15.5.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich R., Kaback M., Freese E. D-amino acids as inducers of L-alanine dehydrogenase in Bacillus subtilis. J Biol Chem. 1968 Mar 10;243(5):1006–1011. [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Bochner B. R., Savageau M. A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977 Feb;33(2):434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I., Taylor A. L. Mutants of Escherichia coli with a growth requirement for either lysine or pyridoxine. J Bacteriol. 1971 Mar;105(3):988–998. doi: 10.1128/jb.105.3.988-998.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREESE E., PARK S. W., CASHEL M. THE DEVELOPMENTAL SIGNIFICANCE OF ALANINE DEHYDROGENASE IN BACILLUS SUBTILIS. Proc Natl Acad Sci U S A. 1964 Jun;51:1164–1172. doi: 10.1073/pnas.51.6.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Venables W. A. Biochemical, genetic, and regulatory studies of alanine catabolism in Escherichia coli K12. Mol Gen Genet. 1976 Dec 8;149(2):229–237. doi: 10.1007/BF00332894. [DOI] [PubMed] [Google Scholar]
- Hilliker S., Botstein D. An early regulatory gene of Salmonella phage P22 analogous to gene N of coliphage lambda. Virology. 1975 Dec;68(2):510–524. doi: 10.1016/0042-6822(75)90291-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kollonitsch J., Barash L., Kahan F. M., Kropp H. Letter: New antibacterial agent via photofluorination of a bacterial cell wall constituent. Nature. 1973 Jun 8;243(5406):346–347. doi: 10.1038/243346a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lambert M. P., Neuhaus F. C. Factors affecting the level of alanine racemase in Escherichia coli. J Bacteriol. 1972 Mar;109(3):1156–1161. doi: 10.1128/jb.109.3.1156-1161.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M. P., Neuhaus F. C. Mechanism of D-cycloserine action: alanine racemase from Escherichia coli W. J Bacteriol. 1972 Jun;110(3):978–987. doi: 10.1128/jb.110.3.978-987.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg E. J., v Schijndel-van Dam A. Temperature-sensitive mutant of Escherichia coli K-12 with an impaired D-alanine:D-alanine ligase. J Bacteriol. 1973 Jan;113(1):96–104. doi: 10.1128/jb.113.1.96-104.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning J. M., Merrifield N. E., Jones W. M., Gotschlich E. C. Inhibition of bacterial growth by beta-chloro-D-alanine. Proc Natl Acad Sci U S A. 1974 Feb;71(2):417–421. doi: 10.1073/pnas.71.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T., Matsuzawa H., Matsuhashi M., Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972 Nov;112(2):950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica T. Transduction by phage P1CM clr-100 in Salmonella typhimurium. Mol Gen Genet. 1975;138(2):113–126. doi: 10.1007/BF02428116. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Neuhaus F. C., Hammes W. P. Inhibition of cell wall biosynthesis by analogues and alanine. Pharmacol Ther. 1981;14(3):265–319. doi: 10.1016/0163-7258(81)90030-9. [DOI] [PubMed] [Google Scholar]
- Palva E. T., Liljeström P., Harayama S. Cosmid cloning and transposon mutagenesis in Salmonella typhimurium using phage lambda vehicles. Mol Gen Genet. 1981;181(2):153–157. doi: 10.1007/BF00268420. [DOI] [PubMed] [Google Scholar]
- Pioli D., Venables W. A., Franklin F. C. D-Alanine dehydrogenase. Its role in the utilisation of alanine isomers as growth substrates by Pseudomonas aeruginosa PA01. Arch Microbiol. 1976 Nov 2;110(23):287–293. doi: 10.1007/BF00690240. [DOI] [PubMed] [Google Scholar]
- Raunio R. P., Straus L. D., Jenkins W. T. D-alanine oxidase from Escherichia coli: participation in the oxidation of L-alanine. J Bacteriol. 1973 Aug;115(2):567–573. doi: 10.1128/jb.115.2.567-573.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso G., Takashima K., Adams E. Coenzyme content of purified alanine racemase from Pseudomonas. Biochem Biophys Res Commun. 1969 Jan 6;34(1):134–140. doi: 10.1016/0006-291x(69)90539-7. [DOI] [PubMed] [Google Scholar]
- Shortle D., Koshland D., Weinstock G. M., Botstein D. Segment-directed mutagenesis: construction in vitro of point mutations limited to a small predetermined region of a circular DNA molecule. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5375–5379. doi: 10.1073/pnas.77.9.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- UMBARGER H. E., BROWN B. Threonine deamination in Escherichia coli. II. Evidence for two L-threonine deaminases. J Bacteriol. 1957 Jan;73(1):105–112. doi: 10.1128/jb.73.1.105-112.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. L., Fox M. S. On the molecular basis of high negative interference. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1544–1548. doi: 10.1073/pnas.71.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman H. J. The characterization of an alanine racemase mutant of Escherichia coli. Genet Res. 1972 Dec;20(3):269–277. doi: 10.1017/s001667230001380x. [DOI] [PubMed] [Google Scholar]
- Wild J., Klopotowski T. D-Amino acid dehydrogenase of Escherichia coli K12: positive selection of mutants defective in enzyme activity and localization of the structural gene. Mol Gen Genet. 1981;181(3):373–378. doi: 10.1007/BF00425614. [DOI] [PubMed] [Google Scholar]
- Wild J., Kłopotowski T. Insensitivity of D-amino acid dehydrogenase synthesis to catabolic repression in dadR mutants of Salmonella typhimurium. Mol Gen Genet. 1975;136(1):63–73. doi: 10.1007/BF00275449. [DOI] [PubMed] [Google Scholar]
- Wild J., Walczak W., Krajewska-Grynkiewicz K., Klopotowski T. D-amino acid dehydrogenase: the enzyme of the first step of D-histidine and D-methionine racemization in Salmonella typhimurium. Mol Gen Genet. 1974;128(2):131–146. doi: 10.1007/BF02654486. [DOI] [PubMed] [Google Scholar]
- Winston F., Botstein D., Miller J. H. Characterization of amber and ochre suppressors in Salmonella typhimurium. J Bacteriol. 1979 Jan;137(1):433–439. doi: 10.1128/jb.137.1.433-439.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]