Abstract
Previous studies have suggested that MHC and non-MHC genes contribute to the development of autoimmune disease in F1 hybrids of New Zealand black (NZB) and white (NZW) mice. We conducted a genome-wide screen of 148 female (NZB × NZW)F1 × NZB backcross mice to map dominant NZW genetic loci linked with lupus disease traits. In this backcross analysis, inheritance of the NZW MHC (H2d/z vs. H2d/d) was strongly linked with the development of lupus nephritis (P ≈ 1 × 10−16), increasing the risk of disease by over 30-fold. H2d/z was also linked with elevated serum levels of IgG autoantibodies to single-stranded DNA, double-stranded DNA, histones, and chromatin but not with anti-gp70 autoantibodies, measured as circulating gp70–anti-gp70 immune complexes. Non-MHC contributions from NZW seemed weak in comparison to MHC, although NZW loci on chromosomes 7 and 16 were noted to be suggestively linked with autoantibody production. Strikingly, H2d/z (compared with H2d/d) enhanced antinuclear antibodies in a coordinate fashion but did not affect anti-gp70 production in the current backcross. However, the opposite influence was noted for H2d/z (compared with H2z/z) when (NZB × NZW)F1 × NZW backcross mice were analyzed. These results suggest that H2z and H2d haplotypes differentially regulate two different sets of nephritogenic autoantibody responses. This study confirms a critical role for H2z compared with other dominant NZW loci in (NZB × NZW)F1 mice and provides an explanation as to why H2d/z heterozygosity is required for full expression of disease in this model.
The F1 hybrid of the New Zealand black (NZB) and New Zealand white (NZW) mouse develops a systemic autoimmune syndrome that serves as a model for human systemic lupus erythematosus (1–3). (NZB × NZW)F1 mice produce autoantibodies to chromatin constituents and to an endogenous retroviral glycoprotein, gp70, and both of these autoantibody specificities seem to contribute to the development of a progressive lupus-like glomerulonephritis (2–10). The parental NZB and NZW strains have some autoimmune characteristics; however, neither of these two mouse strains develops a lupus nephritis with the frequency and severity of (NZB × NZW)F1 animals (1, 2, 10, 11). Thus, genes originating from both NZB and NZW mice must contribute to disease in the F1 mouse.
The genetic basis of disease in (NZB × NZW)F1 mice is complex (reviewed in ref. 12). However, data from backcross (9–14), intercross (15), and congenic experiments (16–19) have consistently shown linkage and association of the MHC from both NZB (H2d) and NZW (H2z) with lupus-like disease. Together, these studies have indicated that mice carrying H2d/z are more likely to develop disease than mice with either H2d/d or H2z/z haplotypes. At present, the mechanism for this effect and the identity of the underlying MHC gene (or genes) remain to be established. Recent studies with mice transgenic for Az or Ez molecules did not support the hypothesis that mixed haplotype class II MHC molecules underlie the enhancement of disease in H2d/z mice (20–22). In an attempt to characterize the contribution from the NZB MHC, we recently analyzed the autoantibodies affected by inheritance of H2d in (NZB × NZW)F1 × NZW backcross mice (10). This analysis indicated that H2d/z (compared with H2z/z) was linked preferentially with the production of autoantibodies to gp70, compared with antinuclear antibodies, and suggested that H2d contributes to disease by enhancing the anti-gp70 response (10).
Genetic analyses have also mapped non-MHC genetic contributions from both NZB and NZW mice (refs. 10, 14, 15, and 23–25; reviewed in ref. 12). In a genome-wide scan for susceptibility genes in New Zealand mixed (NZM; a mixture of NZB and NZW genes) mice, important effects have been isolated to regions derived from the NZW strain on chromosomes 1 and 7 (14, 26). Additional analyses of crosses with NZW mice have mapped regions on chromosome 14, 16, and 19 (10, 25).
In the present study, we undertook a complete genome scan of the (NZB × NZW)F1 × NZB backcross in an attempt to assess the effects of the NZW MHC in relation to dominant contributions from other NZW non-MHC genes. The results show the powerful disease-enhancing effect of H2d/z compared with H2d/d, although relatively weak influences from non-MHC loci on chromosomes 7 and 16 were also observed. Secondly, we performed an extensive serological analysis to determine what phenotypes show linkage with the NZW susceptibility genes. In marked contrast to the effects of H2d/z (vs. H2z/z) in (NZB × NZW)F1 × NZW backcross mice (10), H2d/z (vs. H2d/d) contributed to disease by preferentially promoting antinuclear rather than anti-gp70 autoantibodies. These studies provide evidence for an explanation as to why MHC alleles from both NZB and NZW are required for full expression of disease.
MATERIALS AND METHODS
Mice.
Parental NZB/BINJ and NZW/lacJ mice were obtained from The Jackson Laboratory and were maintained in the animal care facility at the National Jewish Medical and Research Center or University of Colorado Health Sciences Center. The backcross mice were bred in this facility, and all groups of mice used in these studies were housed in the same room and fed an identical diet. Only female animals were used, and backcross mice were studied until 12 months of age.
Evaluation of Nephritis and Collection of Serum.
Mice were evaluated for proteinuria at monthly intervals by using tetrachlorophenol-tetrabromosulfophthalein paper (Chemstrip, Boehringer Mannheim) as described (10, 11, 19, 20). Urine samples were graded 0 to 3+, corresponding to approximate protein concentrations as follows: 0/trace = <0.3 g/liter; 1+ = ≈0.3 g/liter; 2+ = ≈1 g/liter; 3+ = >3 g/liter. Mice that scored 2+ or greater proteinuria on at least two consecutive occasions before 12 months of age were designated as positive for high-grade proteinuria and severe renal disease. The validity of using proteinuria to document severe renal disease in New Zealand hybrid mice has been shown (10, 11, 20) and more recently confirmed (T.J.V. and B.L.K., unpublished data). Serum samples were also obtained at monthly intervals as described (10, 11).
Genetic Mapping by Using Simple Sequence Length Polymorphisms.
Genome-wide mapping was conducted by using microsatellite markers polymorphic between NZB and NZW as well as PCR amplification with a PTC-100 thermal cycler (MJ Research, Watertown, MA) as described (10, 23). The animals were then scored as either BW (heterozygous for NZB and NZW alleles) or BB (homozygous for NZB alleles) for each marker. The positions of the simple sequence length polymorphism markers (and other genetic loci) with respect to the centromere are given in accordance with the Mouse Chromosome Committee Reports, obtainable from the Mouse Genome Database at www.informatics.jax.org.
Serological Assays.
Autoantibody levels were determined by ELISA. Antibodies to histone, chromatin, single-stranded DNA (ssDNA), and double-stranded DNA (dsDNA) were measured as described (10, 19). All assays were performed in duplicate and were quantified against a standard curve obtained with mAbs or sera. Values for the different antinuclear antibodies are reported as units/milliliter. The production of autoantibodies to gp70 was quantitated as serum levels of gp70–anti-gp70 immune complexes (gp70 IC), because the relative excess of gp70 in serum makes free anti-gp70 antibodies difficult to detect (27). These complexes were measured by ELISA after precipitation of the serum with polyethylene glycol (average molecular weight of 6,000) as described (27). The results are expressed as micrograms/milliliter gp70 complexed with anti-gp70 antibodies.
For certain comparisons and analyses examining the association of a particular locus with autoantibody levels, mice were separated into groups based on their serum levels of particular autoantibodies. The cutoffs used to group mice into negative/low, and positive/high groups in the current study are the same as those defined in studies of (NZB × NZW)F1 × NZW backcross mice (10). The cutoffs for anti-dsDNA, anti-chromatin, and gp70 IC autoantibodies correlated well with low levels produced in NZW and nonautoimmune strains and with high levels produced in (NZB × NZW)F1 mice. These cutoffs have also been used to analyze additional backcrosses (20). Cutoffs for low and high levels were as follows: for anti-dsDNA, <0.4 and >2.5 units/ml; for anti-chromatin, <1.0 and >4.6 units/ml; and for gp70 IC, <0.5 and >3.5 μg/ml, respectively.
Statistical Analysis.
The linkage of a particular locus with nephritis (categorized as positive or negative) was quantified by χ2 analysis, by using a standard (2 × 2) contingency matrix (28). Evidence that loci were linked with autoantibody levels was found by using the linkage program, mapmaker/qtl (29, 30), and the data are presented as maximal log10 likelihood of the odds (lod) scores (MLS). This program was used to map quantitative trait loci (QTL) in linkage with serum autoantibody levels. The autoantibody levels were log10 transformed before analysis with mapmaker/qtl, because this transformation tended to normalize their frequency distribution (10, 19), which improves the accuracy of mapmaker/qtl.
Because of the multiple-hypotheses testing that is inherent in a genome-wide search, a threshold for suggestive linkage was set at lod > 1.9 (P < 0.003; χ2 > 8.6; 1 df), based on the recommendation of Lander and Kruglyak (31). The threshold for linkage was set at lod > 3.3 (P < 1 × 10−4; χ2 > 15.1). Loci were considered to be corroborated if they were previously mapped at P < 0.01 and if a colocalizing chromosomal region was mapped in the current study at P < 0.01 (overall, P < 1 × 10−4; lod > 3.3; ref. 31).
RESULTS
Genome-Wide Scan for Loci Linked with Nephritis in (NZB × NZW)F1 × NZB Backcross Mice.
A previous analysis (11) of this (NZB × NZW)F1 × NZB backcross showed a strong association of H2z (i.e., H2d/z vs. H2d/d) with the development of severe lupus nephritis. The only non-MHC loci investigated in the initial report were the T cell receptor loci Tcra and Tcrb, which showed no linkage with nephritis or antibody production. In view of interim studies that have linked other NZW loci with lupus-like disease (refs. 10, 14, 15, and 25; reviewed in ref. 12), we extended our initial analysis to a genome-wide scan. The current screen involved 148 backcross mice, in which the development of high-grade proteinuria was a nearly perfect predictor for mortality before 12 months of age (11). We also assayed serial serum levels of IgG autoantibodies to chromatin, together with IgG1, IgG2a, and IgG3 anti-chromatin antibodies, IgG autoantibodies to dsDNA and histones, and IgG autoantibodies to gp70 in the form of gp70 IC. Linkage analysis included 75 polymorphic microsatellite markers (10, 23), which covered >90% of the genome and all genomic regions previously mapped to be linked with lupus-like disease in New Zealand mice (reviewed in ref. 12).
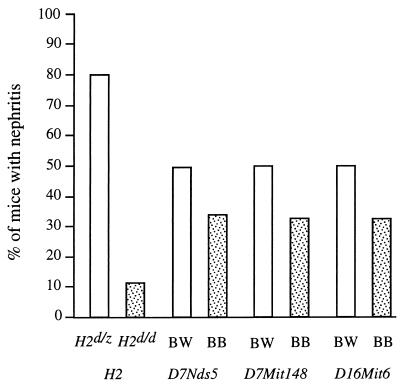
Table 1 shows the strong linkage of the NZW MHC (H2d/z vs. H2d/d) with the development of nephritis and early mortality. Inheritance of the NZW MHC provided a 31-fold increase in risk for the development of severe nephritis and seemed to overwhelm the contribution of any non-MHC loci. Of H2d/z mice, 80% developed severe proteinuria and succumbed to disease, compared with ≈10% of H2d/d mice (Fig. 1). Despite relatively low statistical support for linkage, loci on chromosomes 7 and 16 are also shown in Table 1 and Fig. 1 because of their contribution to IgG autoantibody production (see below). In addition, loci in similar chromosomal positions have been linked with nephritis in other genetic studies of NZW contributions to murine lupus (10, 25). In the current analysis, loci on chromosomes 7 and 16 showed only trends for linkage with nephritis (P < 0.05), and the marker with the strongest contribution to nephritis was D16Mit6 (odds ratio = 2.2).
Table 1.
Genome-wide scan for loci linked with severe nephritis in (NZB × NZW)F1 × NZB backcross mice*
| Locus | Position
|
Linkage
|
|||
|---|---|---|---|---|---|
| Chromosome no. | Distance from centromere, in centimorgans (cM) | χ2 | P value | Odds ratio† | |
| MHC (Tnf) | 17 | 19 | 69 | 8 × 10−17 | 31 |
| D7Nds5‡ | 7 | 23 | 4.4 | 0.036 | 2.1 |
| D7Mit148‡ | 7 | 46 | 4.7 | 0.030 | 2.1 |
| D16Mit6‡ | 16 | 60 | 4.9 | 0.027 | 2.2 |
| D19Mit11‡ | 19 | 41 | 4.6 | 0.032 | 2.1 |
| D1Nds1§ | 1 | 67 | 0.3 | >0.5 | ∼1.0 |
| D1Mit106§ | 1 | 85 | 0.01 | >0.5 | ∼1.0 |
| D4Mit71§ | 4 | 62 | 0.00 | >0.5 | ∼1.0 |
| D4Mit179§ | 4 | 71 | 0.03 | >0.5 | 1.1 |
Data were obtained from at least 145 backcross mice for each marker shown.
Odds ratio represents the increased risk for development of nephritis and is calculated as [(No. heterozygous diseased mice) × (No. NZB homozygous healthy mice)] ÷ [(No. NZB homozygous diseased mice) × (No. heterozygous healthy mice)].
Although these loci do not meet accepted thresholds (P < 0.0034) for suggestive linkage, their trends for linkage with nephritis are shown because they show suggestive linkage with various serological traits.
These loci have been reported to contribute strongly to nephritis in other analyses but failed to show even trends for linkage with nephritis or with serological traits in the current analysis.
Figure 1.
Association of nephritis with genotypes at MHC and particular non-MHC loci in 148 (NZB × NZW)F1 × NZB backcross mice. The results are shown as percentage of mice with severe nephritis, which correlated closely with early mortality (11), in relation to the genotypes shown.
Genome-Wide Scan for Loci Linked with Autoantibody Production in (NZB × NZW)F1 × NZB Backcross Mice.
Using a QTL method (mapmaker/qtl linkage program), we noted that all four of the antinuclear autoantibodies analyzed were linked with H2d/z (Table 2) with MLS between 2.2 for IgG anti-ssDNA and 4.4 for IgG anti-histone. Table 3 shows that the mean and median levels of anti-chromatin antibodies were markedly higher in H2d/z than in H2d/d mice (P = 6.2 × 10−4). There was also evidence of a differential linkage for H2 with the IgG subclasses of the anti-chromatin response (Table 2). Thus, H2d/z showed linkage with both IgG1 and IgG2a anti-chromatin antibody production (MLS = 2.8), whereas there was not even a trend for linkage of H2 with IgG3 anti-chromatin antibody levels.
Table 2.
Genome-wide scan for loci linked with autoantibody production in (NZB × NZW)F1 × NZB backcross mice
| Trait |
D7Nds5 to D7Mit145
|
D7Mit148 to D7Mit125
|
D16Mit5 to D16Mit6
|
H2z
|
||||
|---|---|---|---|---|---|---|---|---|
| MLS | P value* | MLS | P value* | MLS | P value* | MLS | P value* | |
| gp70 IC | 2.0 | 0.0024 | 2.1 | 0.0018 | 0.70 | — | 0.03 | — |
| IgG anti-dsDNA | 1.9 | 0.0031 | 2.1 | 0.0018 | 0.20 | — | 2.4 | 8.8 × 10−4 |
| IgG anti-histone | 1.3 | 0.014 | 1.4 | 0.011 | 0.10 | — | 4.4 | 6.7 × 10−6 |
| IgG anti-ssDNA | 1.9 | 0.0031 | 2.1 | 0.0018 | 0.21 | — | 2.2 | 0.0015 |
| IgG anti-chromatin | 0.65 | — | 0.34 | — | 1.7 | 0.0051 | 3.2 | 1.2 × 10−4 |
| IgG1 anti-chromatin | 0.11 | — | 0.05 | — | 1.6 | 0.0066 | 2.8 | 3.3 × 10−4 |
| IgG2a anti-chromatin | 0.19 | — | 0.32 | — | 2.1 | 0.0018 | 2.8 | 3.3 × 10−4 |
| IgG3 anti-chromatin | 1.2 | 0.019 | 0.50 | — | 0.89 | — | 0.21 | — |
, P values are shown for significant MLS at P < 0.02.
Table 3.
Comparison of levels of anti-chromatin antibodies and gp70 IC in F1 × NZB and F1 × NZW backcross mice
| Autoantibody trait | F1 × NZB
|
F1 × NZW
|
||
|---|---|---|---|---|
| H2d/z | H2d/d | H2d/z | H2z/z | |
| Anti-chromatin, units/ml | ||||
| Mean ± SEM | 2.8 ± 0.71 | 0.45 ± 0.06 | 5.3 ± 1.03 | 4.4 ± 1.23 |
| Median | 0.93 | 0.33 | 2.2 | 2.1 |
| 25th percentile | 0.53 | 0.18 | 1.2 | 0.55 |
| 75th percentile | 3.6 | 0.58 | 4.3 | 4.6 |
| gp70 IC μg/ml | ||||
| Mean ± SEM | 2.0 ± 0.43 | 1.8 ± 0.37 | 3.6 ± 0.57 | 1.2 ± 0.35 |
| Median | 0.50 | 0.55 | 2.0 | 0.25 |
| 25th percentile | 0.11 | 0.11 | 0.55 | 0.00 |
| 75th percentile | 2.5 | 2.0 | 5.0 | 1.5 |
Serum samples were obtained from 8-month-old animals, and a subset from each group was chosen to analyze for these autoantibodies. For the analysis of anti-chromatin antibodies, the data present samples from 69 F1 × NZB and 89 F1 × NZW backcross mice. For the analysis of gp70 IC, the data represent 131 F1 × NZB and 80 F1 × NZW backcross mice.
Previous studies have indicated that an important nephritogenic autoantibody in New Zealand hybrid mice is directed to the serum retroviral protein gp70 (7–10). It was therefore surprising that, despite the effect on nephritis and antinuclear antibody production, inheritance of H2z showed no linkage with levels of gp70 IC (Table 2). Consistent with this lack of linkage, H2z also had no effect on mean or median levels of this autoantibody (Table 3).
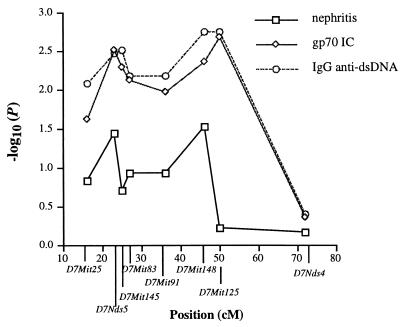
In the genome screen of backcross mice, NZW loci on chromosomes 7 and 16 showed evidence of suggestive linkage with various autoantibodies (Table 2). In regard to chromosome 7 loci, the results suggested that there may be at least two distinct positions linked with serum autoantibody levels. As shown in Fig. 2, the strongest linkage with both anti-dsDNA antibody and gp70 IC levels was mapped at ≈48 cM, between D7Mit148 (46.5 cM) and D7Mit125 (50 cM). Another peak of linkage seemed to be between D7Nds5 and D7Mit145 at ≈24 cM. For both loci, linkage with autoantibody levels was related to inheritance of the NZW allele vs. homozygosity for the NZB allele. The proximal NZW chromosome 7 interval was associated with a nearly 3-fold increase in the percentage of mice with elevated serum levels of gp70 IC. A similar but less dramatic trend was noted for the more distal chromosome 7 interval linked with autoantibody levels. As noted above, these associations were clearly greater than those seen for H2z, which had no effect on the percentage of mice with elevated gp70 IC.
Figure 2.
The pattern of linkage with nephritis (determined by χ2 analysis) as well as with IgG anti-dsDNA production and gp70 IC production (determined by mapmaker/qtl) in (NZB × NZW)F1 × NZB backcross mice is shown for loci on chromosome 7. There are two peaks of linkage: one at D7Nds5 (close to the position of Sle3; ref. 14) and a second at D7Mit148 (close to the position of Nba3; ref. 12). Data are derived from 148 mice followed for development of nephritis, of which 131 were studied for both anti-dsDNA and gp70 IC.
A locus on chromosome 16 showed suggestive linkage with IgG anti-chromatin autoantibody levels, particularly of the IgG2a subclass (Table 2). Inheritance of the NZW allele at this locus was associated with an increase from 18% to 34% of the mice positive for anti-chromatin antibody levels.
Selective Effects of NZW vs. NZB MHC Haplotypes on Autoantibody Production.
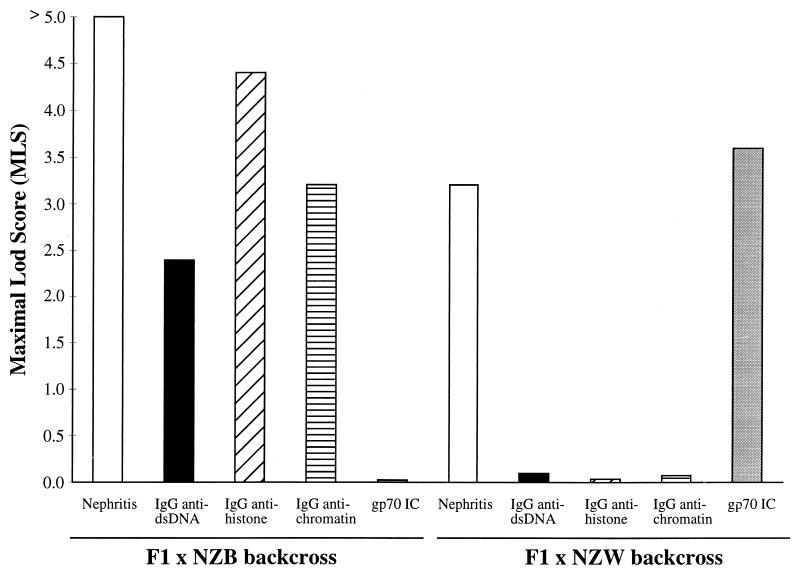
Fig. 3 compares a linkage analysis of H2z in the current F1 × NZB backcross with a similar analysis done for H2d in a separate F1 × NZW backcross (10). In the current backcross, H2d/z mice are compared with H2d/d mice, whereas H2d/z mice are compared with H2z/z mice in the former backcross. Although each MHC in the respective backcrosses showed strong linkage with the development of nephritis, the effects of the two different MHC haplotypes on autoantibody levels were remarkably different. Thus, H2z seemed to enhance the production of IgG antinuclear antibodies, such as anti-dsDNA, anti-histone, and anti-chromatin, and was not linked with the production of gp70 IC. In contrast, H2d was linked with elevated levels of gp70 IC and had little apparent effect on the antinuclear antibodies measured in F1 × NZW backcross mice.
Figure 3.
Selective effects of H2z in F1 × NZB backcross mice vs. H2d in F1 × NZW backcross mice. The data are presented as linkage values (MLS) for H2d/z vs. H2d/d in F1 × NZB backcross mice and for H2d/z vs. H2z/z in F1 × NZW backcross mice. MLS > 1.4, P < 0.01; MLS > 2.35, P < 1 × 10−3; MLS > 3.3, P <1 × 10−4; MLS > 4.23, P < 1 × 10−5. Data are derived from 148 F1 × NZB mice followed for nephritis, 131 F1 × NZB mice studied serologically, 108 F1 × NZW mice followed for nephritis, and 92 F1 × NZW mice studied serologically.
The above linkage results were based on a comparison of H2d/z with H2d/d mice or H2d/z with H2z/z mice in the F1 × NZB or F1 × NZW backcrosses, respectively. Within each backcross, H2d/z (vs. H2d/d or H2z/z) was also associated with significantly higher mean levels of anti-chromatin autoantibodies or gp70 IC (Table 3). Table 3 also allows a comparison of absolute levels of anti-chromatin and gp70 IC between the two different backcrosses. Overall, the F1 × NZW backcross mice had higher levels of both autoantibodies, which is probably best shown by comparing MHC-matched H2d/z mice from each backcross. Mean levels of anti-chromatin and gp70 IC were almost 2-fold greater in the H2d/z mice from the F1 × NZW compared with those from the F1 × NZB backcross (P = 0.01). This comparison shows the importance of non-MHC genes for autoantibody production in general and possibly implicates recessive NZW genes not mapped in the current analysis. In contrast, a comparison of H2d/d mice with H2z/z mice in the two different backcrosses shows important selective and opposite effects on the two autoantibodies measured. Although F1 × NZW mice had higher levels of gp70 IC compared with F1 × NZB mice, H2z/z mice from the F1 × NZW backcross had lower levels of this autoantibody compared with H2d/d mice from the F1 × NZB backcross (P = 0.04). In contrast, H2z/z mice had 10-fold higher mean levels of IgG anti-chromatin antibodies (P = 1 × 10−4).
DISCUSSION
(NZB × NZW)F1 mice are primarily a model of lupus-like autoantibody production in which nephritogenic IgG autoantibodies accumulate in glomeruli as immune complexes and cause a diffuse and progressive lupus nephritis (1–3). Genes from each parent contribute to the full expression of disease (reviewed in ref. 12). The results of the current genome-wide screen clearly show that the most important dominant NZW contribution to disease is the NZW MHC. The statistical support for linkage of H2z with nephritis was striking, and inheritance of H2z in backcross mice was associated with a 31-fold increased risk for severe proteinuria. Because proteinuria correlated closely with mortality in this backcross (11), H2z also was the major genetic determinant of death from disease in this study. These results are consistent with a large body of other studies showing that H2d/z heterozygosity, compared with two copies of H2d or two copies of H2z, is associated with an increased frequency of lupus nephritis in this model (9–20, 22). In various studies, both H2z and H2d seemed to be important for this codominant effect.
The genes within H2 and the mechanism by which H2 heterozygosity enhances nephritis in this model are unknown. Some investigators have suggested that mixed haplotype class II molecules such as AαdAβz or EαdEβz are important for autoantigen presentation and T cell help for autoantibody production (17, 32, 33). Evidence supports the cell surface expression and presentation by these mixed molecules (33). However, recent studies with mice transgenic for Az (21, 22) or Ez (20) in crosses that allowed for mixed-molecule formation did not show any effect of the transgenes on autoantibody production or nephritis.
The present work provides support for a different model in which H2z and H2d separately contribute to nephritis by enhancing two different types of nephritogenic autoantibodies. Thus, within the F1 × NZB backcross, H2d/z (vs. H2d/d) was linked with antibodies directed to chromatin constituents such as dsDNA and histones but was not linked to gp70 IC. Selective effects on IgG autoantibody production in the opposite direction were linked with H2d/z (vs. H2z/z) in the other backcross. Consistent with the current results, in a previous analysis of (B6.H2z × NZB)F1 × NZB backcross mice (19), linkage of H2z with elevated IgG anti-chromatin antibodies (lod = 3.1–6.1; P = 2 × 10−4 to 1 × 10−7) was much stronger statistically than the linkage observed for gp70 IC (lod = 0.8–1.4; P = 0.05–0.01).
The opposite effects of H2z vs. H2d on autoantibody production were also observed when absolute levels were compared between the two backcrosses. This comparison was made somewhat difficult by the higher levels of all antibodies measured in the F1 × NZW backcross. However, despite this overall difference, H2d/d mice of the F1 × NZB backcross still had higher levels of gp70 IC compared with H2z/z mice of the F1 × NZW backcross. These data support the contention that H2z selectively enhances anti-chromatin autoantibodies (vs. anti-gp70 autoantibodies). Perhaps the best study design to confirm the selective effects of H2d vs. H2z in the same set of mice would be a (NZB × NZW)F2 intercross. The basis for the overall difference in autoantibody levels between the two crosses is not known and may relate to the fact that they were bred at different times in our animal facility. The differences more likely relate to the non-MHC genetic backgrounds of the two backcrosses, especially the presence of homozygous NZW alleles in the F1 × NZW backcross but not in the F1 × NZB backcross. We emphasize that certain recessive NZW contributions were unlikely to be mapped in the F1 × NZB backcross, but these contributions have been demonstrated when NZW or NZM mice were crossed with nonautoimmune mice (14, 25).
The combined and selective effect of different MHC haplotypes on autoantibody production is consistent with the ability of certain MHC genes to mediate antigen-selective processing and presentation. In past studies, non-MHC susceptibility loci have not shown linkage for one type of autoantibody without showing at least trends for linkage with all other autoantibodies. (10, 19, 20). The one exception to this rule was a susceptibility locus on proximal chromosome 14 (10), corresponding to the position of the Tcra complex, which is another set of genes that theoretically could mediate selective immune recognition. It would seem reasonable to hypothesize that class II H2z genes underlie the selective effect of MHC on autoantibody production. However, as noted above, we recently completed two studies with mice transgenic for Az or transgenic for Ez molecules (20, 22). In neither study could the class II transgenes be linked with IgG autoantibody production or disease. It may be that both Az and Ez molecules are required for this effect, and future studies are required to test this idea. Previous studies also suggest that the effect of H2z on autoantibody production and especially nephritis may be complex and involve different genes within the MHC such as Tnfa (34, 35).
Compared with the MHC, non-MHC contributions from NZW seemed weak in this backcross analysis. Two loci on chromosome 7 showed suggestive linkage with autoantibody production and trends for linkage with nephritis. Although statistical support for linkage is low in the present study, our mapping data are likely to reflect disease-susceptibility gene(s), as they confirm previous findings in (NZB × NZW)F1 × NZW backcross mice (10). In that backcross, an NZB chromosome 7 locus, subsequently named Nba3, was mapped to ≈50 cM and was linked with nephritis, anti-dsDNA production, and gp70 IC production. Interestingly, in that study, a heterozygous (BW) genotype [vs. a homozygous (WW) genotype] was linked with the lupus phenotype. In the present study, the BW (vs. BB) genotype was disease-enhancing. Thus, in both reciprocal backcrosses, a locus, Nba3, was linked with nephritis and autoantibody production with BW alleles conferring greater risk than either WW or BB alleles. It is unclear whether the explanation for this finding is the interaction of the NZB and NZW alleles at a single gene, or whether there is a separate contribution from two closely linked NZB and NZW disease genes on mid chromosome 7. Another NZW locus more proximal on chromosome 7 was also mapped. It is likely that this locus is separate and distinct from the more distal Nba3, because a locus at ≈25 cM has been described by other groups (14, 15). Kono et al. (15) mapped a locus in (NZB × NZW)F2 mice, Lbw5 at Ngfg (23 cM), that was linked with mortality. Morel et al. (14) studied (B6 × NZM)F1 × NZM backcross mice and mapped a locus, Sle3, that was linked with nephritis at pink eye, p (28 cM). Sle3 originates in NZW mice and has been shown to promote autoantibody production in congenic mice (26). It may therefore be identical to the more proximal chromosome 7 locus mapped in the current study. Chromosome 7 evidently carries a large number of genes linked with murine lupus. An NZW locus, Sle5, has been mapped (36) at an even more proximal position (≈5 cM), and locus Lrdm1 has been mapped at a similar position in MRL–lpr/lpr lupus-prone mice (37). These loci are likely to be distinct from the proximal locus mapped in this study. We also noted a locus on chromosome 16 with weak effects on autoantibody production and nephritis. These results seem to confirm the existence of an NZW locus mapped on chromosome 16 in (NZB × NZW)F1 × NZW backcross mice (10), previously linked with nephritis and autoantibody production.
The present genome-wide scan for lupus-susceptibility loci is also notable for certain chromosomal regions not found to be linked with nephritis or autoantibody production. For example, loci on distal chromosome 1, encompassing an interval that includes the NZW locus, Sle1/Lbw7 (14, 15), or the NZB locus, Nba2 (18–20, 24), showed no trends for linkage with nephritis. In previous studies, although Nba2 was one of the few NZB non-MHC loci that showed consistent linkage in the context of different nonautoimmune backgrounds (18, 19, 24), it has not shown linkage in crosses that involved NZB and NZW backgrounds as in the current study (10, 23). It is currently unclear whether the NZB and NZW contributions on distal chromosome 1 involve alleles of the same gene (discussed in ref. 12). In (NZB × NZW)F1 × NZW backcross mice, an NZB locus on distal chromosome 4, Nba1, was the non-MHC locus with the strongest linkage with nephritis (10, 23). Markers on chromosome 4 showed no influence on disease in the current cross, which compares NZB homozygosity with NZB/NZW heterozygosity. This absence of linkage is consistent with its previously determined dominant mode of influence and suggests that this NZB susceptibility gene is almost completely dominant with respect to nephritis.
We have substantially extended the genetic analysis of (NZB × NZW)F1 × NZB backcross mice. The results indicate that H2 is by far the most influential dominant NZW locus with respect to nephritis. In addition, this MHC haplotype seems to enhance selectively antinuclear autoantibody production rather than autoantibodies to gp70, a complete reverse of the contribution of H2d in (NZB × NZW)F1 × NZW mice. These results suggest that enhancement of disease by H2d/z heterozygosity in (NZB × NZW)F1 mice is related to separate contributions from H2z and H2d and production of separate nephritogenic autoantibodies.
Acknowledgments
This work was supported by National Institutes of Health Grant AR 37070 and a grant from the Swiss National Foundation for Scientific Research.
ABBREVIATIONS
- NZB
New Zealand black
- NZW
New Zealand white
- NZM
New Zealand mixed
- BW
black/white (heterozygous for NZB and NZW alleles)
- BB
black/black (homozygous for NZB alleles)
- ssDNA and dsDNA
single- and double-stranded DNA
- gp70 IC
gp70–anti-gp70 immune complexes
- lod
log10 likelihood of the odds in favor of linkage
- MLS
maximal lod score
- QTL
quantitative trait locus
- cM
centimorgan
References
- 1.Howie J B, Helyer B J. Adv Immunol. 1968;9:215–266. doi: 10.1016/s0065-2776(08)60444-7. [DOI] [PubMed] [Google Scholar]
- 2.Theofilopoulos A N. In: Systemic Lupus Erythematosus. Lahita R G, editor. New York: Churchill Livingstone; 1992. pp. 121–194. [Google Scholar]
- 3.Kotzin B L. Cell. 1996;85:303–306. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- 4.Madaio M P, Carlson J, Cataldo J, Ucci A, Migliorini P, Pankewycz O. J Immunol. 1987;138:2883–2889. [PubMed] [Google Scholar]
- 5.Raz E, Brezis M, Rosenmann E, Eilat D. J Immunol. 1989;142:3076–3082. [PubMed] [Google Scholar]
- 6.Tsao B P, Ohnishi K, Cheroutre H, Mitchell B, Teitell M, Mixter P, Kronenberg M, Hahn B H. J Immunol. 1992;149:350–358. [PubMed] [Google Scholar]
- 7.Izui S, McConahey P J, Theofilopoulos A N, Dixon F J. J Exp Med. 1979;149:1099–1116. doi: 10.1084/jem.149.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izui S, Elder J H, McConahey P J, Dixon F J. J Exp Med. 1981;153:1151–1160. doi: 10.1084/jem.153.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruyama N, Furukawa F, Nakai Y, Sasaki Y, Ohta K, Ozaki S, Hirose S, Shirai T. J Immunol. 1983;130:740–746. [PubMed] [Google Scholar]
- 10.Vyse T J, Drake C G, Rozzo S J, Roper E, Izui S, Kotzin B L. J Clin Invest. 1996;98:1762–1772. doi: 10.1172/JCI118975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotzin B L, Palmer E. J Exp Med. 1987;165:1237–1251. doi: 10.1084/jem.165.5.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vyse T J, Kotzin B L. Annu Rev Immunol. 1998;16:261–292. doi: 10.1146/annurev.immunol.16.1.261. [DOI] [PubMed] [Google Scholar]
- 13.Knight J G, Adams D D. J Exp Med. 1978;147:1653–1660. doi: 10.1084/jem.147.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morel L, Rudofsky U H, Longmate J A, Schiffenbauer J, Wakeland E K. Immunity. 1994;1:219–229. [PubMed] [Google Scholar]
- 15.Kono D H, Burlingame R W, Owens D G, Kuramochi A, Balderas R S, Balomenos D, Theofilopoulos A N. Proc Natl Acad Sci USA. 1994;91:10168–10172. doi: 10.1073/pnas.91.21.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose S, Ueda G, Noguchi K, Okada T, Sekigawa I, Sato H, Shirai T. Eur J Immunol. 1986;16:1631–1633. doi: 10.1002/eji.1830161226. [DOI] [PubMed] [Google Scholar]
- 17.Hirose S, Kinoshita K, Nozawa S, Nishimura H, Shirai T. Int Immunol. 1990;2:1091–1095. doi: 10.1093/intimm/2.11.1091. [DOI] [PubMed] [Google Scholar]
- 18.Rozzo S J, Vyse T J, Drake C G, Kotzin B L. Proc Natl Acad Sci USA. 1996;93:15164–15168. doi: 10.1073/pnas.93.26.15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vyse T J, Rozzo S J, Drake C G, Izui S, Kotzin B L. J Immunol. 1997;158:5566–5574. [PubMed] [Google Scholar]
- 20.Vyse T J, Rozzo S J, Drake C G, Appel V B, Lemeur M, Izui S, Palmer E, Kotzin B L. J Immunol. 1998;160:2757–2766. [PubMed] [Google Scholar]
- 21.Nishimura H, Ishikawa S, Nozawa S, Awaji M, Saito J, Abe M, Gotoh Y, Tokushima M, Kimoto M, Akakura S, et al. Int Immunol. 1996;8:967–976. doi: 10.1093/intimm/8.6.967. [DOI] [PubMed] [Google Scholar]
- 22.Rozzo S J, Vyse T J, David C S, Palmer E, Izui S, Kotzin B L. J Immunol. 1999;162:2623–2630. [PubMed] [Google Scholar]
- 23.Drake C G, Babcock S K, Palmer E, Kotzin B L. Proc Natl Acad Sci USA. 1994;91:4062–4066. doi: 10.1073/pnas.91.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drake C G, Rozzo S J, Hirschfeld H F, Smarnworawong N P, Palmer E, Kotzin B L. J Immunol. 1995;154:2441–2447. [PubMed] [Google Scholar]
- 25.Vyse T J, Morel L, Tanner F J, Wakeland E K, Kotzin B L. J Immunol. 1996;157:2719–2727. [PubMed] [Google Scholar]
- 26.Morel L, Mohan C, Yu Y, Croker B P, Tian N, Deng A, Wakeland E K. J Immunol. 1997;158:6019–6028. [PubMed] [Google Scholar]
- 27.Izui S, Lange G. Clin Exp Immunol. 1983;71:45–49. [PMC free article] [PubMed] [Google Scholar]
- 28.Rosner B. Fundamentals of Biostatistics. Boston: PWS Kent; 1990. pp. 318–396. [Google Scholar]
- 29.Lincoln S E, Daly M, Lander E S. Whitehead Institute Technical Report. 2nd Ed. Cambridge, MA: Whitehead Inst.; 1992. [Google Scholar]
- 30.Lander E S, Botstein D. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lander E S, Kruglyak L. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 32.Nygard N R, McCarthy D M, Schiffenbauer J, Schwartz B D. Immunol Today. 1993;14:53–56. doi: 10.1016/0167-5699(93)90058-s. [DOI] [PubMed] [Google Scholar]
- 33.Gotoh Y, Takashima H, Noguchi K, Nishimura H, Tokushima M, Shirai T, Kimoto M. J Immunol. 1993;150:4777–4787. [PubMed] [Google Scholar]
- 34.Jongeneel C V, Acha-Orbea H, Blankenstein T. J Exp Med. 1990;171:2141–2146. doi: 10.1084/jem.171.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob C O, Fronek Z, Lewis G D, Koo M, Hansen J A, McDevitt H O. Proc Natl Acad Sci USA. 1990;87:1233–1237. doi: 10.1073/pnas.87.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wakeland E K, Morel L, Mohan C, Yui M. J Clin Immunol. 1997;17:272–281. doi: 10.1023/a:1027370514198. [DOI] [PubMed] [Google Scholar]
- 37.Vidal S, Kono D H, Theofilopoulos A N. J Clin Invest. 1998;101:696–702. doi: 10.1172/JCI1817. [DOI] [PMC free article] [PubMed] [Google Scholar]