Abstract
The adaptation of a population to a new environment is a result of selection operating on a suite of stochastically occurring mutations. This article presents an analytical approach to understanding the population dynamics during adaptation, specifically addressing a system in which periods of growth are separated by selection in bottlenecks. The analysis derives simple expressions for the average properties of the evolving population, including a quantitative description of progressive narrowing of the range of selection coefficients of the predominant mutant cells and of the proportion of mutant cells as a function of time. A complete statistical description of the bottlenecks is also presented, leading to a description of the stochastic behavior of the population in terms of effective mutation times. The effective mutation times are related to the actual mutation times by calculable probability distributions, similar to the selection coefficients being highly restricted in their probable values. This analytical approach is used to model recently published experimental data from a bacterial coculture experiment, and the results are compared to those of a numerical model published in conjunction with the data. Finally, experimental designs that may improve measurements of fitness distributions are suggested.
THE adaptation of asexual populations to a new environment is the result of selection operating on the suite of stochastically occurring mutations, each of which may confer a different selective advantage. The mutations occur throughout time, so that multiple clones of mutant cells are present simultaneously. Mathematical modeling of the population dynamics typically employs numerical simulation to calculate a particular instance of the system, sampling probability distributions to include stochastic effects. The calculations may involve detailed tracking of each mutant clone through the history of the population. Running the model many times allows determination of its characteristic behavior as a function of the parameters describing mutation and selection. Estimates of the values of these parameters in a living system are obtained by comparisons of the statistical properties of the model with those of experimental data.
By contrast, this article presents an analytical description of the population dynamics. The analysis begins by establishing the identity of the average behavior of sequential finite cultures separated by bottlenecks with the growth of an exponentially expanding effectively infinite culture. Consideration of the infinite system provides analytical expressions for characteristic properties of the finite populations. The results include quantitative descriptions of growth of the proportion of mutant cells with time and of the accompanying narrowing of the frequency distribution of their selection coefficients. Next, the stochastic behavior of finite systems is considered, resulting in a comprehensive and convenient description of the selection in the bottlenecks. The stochastic description is then used to develop a model of coculture experiments such as the one recently published by Hegreness and Shoresh (HS) (Hegreness et al. 2006). Application of the results obtained for the average behavior is used to simplify the model and facilitate its comparison with experimental data. Finally, the results are discussed and alternate experimental designs that may allow better measurement of the fitness distribution of the mutant cells are suggested. Significant additional information concerning the analysis is presented in the supplemental materials at http://www.genetics.org/supplemental/, including an Excel workbook for calculation of the statistical distributions that are developed in this article.
THE EXPERIMENTAL SYSTEM
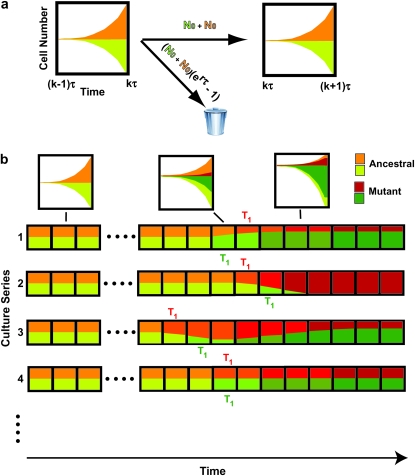
Consideration of the experimental system of HS (Figure 1a) provides concrete motivation for the analysis presented in the remainder of this article. Approximately 2 × 105 cells were seeded into a culture, one-half labeled with yellow fluorescent protein (YFP) and one-half with cyan fluorescent protein (CFP). Otherwise the ancestral cells were identical. Mutations with selection coefficients s occurred stochastically in both populations of ancestral cells with a probability distribution ρ(s) and an overall frequency μ per generation per cell. The cells grew exponentially for 24 hr, and the culture was then sampled to seed the next passage with ∼2 × 105 cells. After this bottleneck, exponential growth resumed, followed by sampling to seed the next daughter culture after another 24 hr, etc. The full experiment lasted ∼40 days or ∼450 doublings for the starting (ancestral) cells. Figure 1a illustrates the growth of the CFP and YFP populations just before and after the kth bottleneck for a time early in the series before the mutant populations have become significant. Each growth phase lasts a time τ = 11.7 population doublings and resuts in an erτ ∼ 3300-fold increase in cell number, where r = ln(2). At the bottleneck, a sampling of e−rτ ∼ 1/3300 of the mutant and ancestral cells proceeded to the subsequent daughter culture. The rest of the cells were discarded.
Figure 1.—
Schematic of the YFP/CFP coculture experiment. (a) The exponential expansion of the YFP and CFP populations (distinguished by green and red colors) during the growth periods before and after the kth bottleneck at a time early in the experiment when mutant cells are present at low abundance. The cultures begin with N0 of each type of cell and exponentially expand for time τ so that there are N0erτ of each. At the bottleneck N0 of each type of cell is removed and transferred to the next culture. The remainder is discarded. In the actual experiment, τ = 11.7 doublings, and the expansion factor is erτ ∼ 3300. (b) Four representative time courses for the behavior of the color ratios. Ancestral cells are indicated by light colors and mutants (of any selection coefficient) by dark colors. The colors are drawn separately for illustrative purposes, but in the actual system the cells are uniformly mixed. Schematic growth curves for the mutant and ancestral populations during three of the growth periods for the first series of cultures are shown in boxes above that series. The behavior of each culture series is described in the text. The red and green T1's indicate the times when mutant and ancestral cells are of equal abundance in the respective populations.
YFP/CFP fluorescence ratios in each of 72 series of such cultures were measured at each bottleneck, providing a measure of the relative numbers of the two cell types as a function of time. Because of the stochastic nature of the mutational process and the passage of mutants through the bottlenecks, one expects that under some conditions the ratio may change with time, differing in each series of cultures. Figure 1b shows examples of four of the possible time courses for the ratio, using green and red to distinguish the populations. The ancestral cells in both populations are represented as light colors, and the mutants (of any s) as dark colors. This distinction is made for illustrative purposes only since ancestral and mutant cells cannot be distinguished by their fluorescence. While the colors are illustrated as separated, in the actual experiment the two populations are thoroughly mixed.
In the first example, mutant cells are assumed to come to prominence initially in the “green” population. This time is indicated by a green T1, defined in our analysis as the time when the proportions of mutant and ancestral cells in the green population are equal. The overall growth rate of the green population detectably increases, and green cells begin to overgrow the “red” ones. Mutant cells with approximately the same s as those in the green population are assumed to come to prominence in the red population at a later time, indicated by the red T1. After this time, the green and red populations tend toward the same growth rate, and thus their ratio stabilizes. Mutant cells completely dominate both populations with time, as indicated by the darkening colors. This growth pattern corresponds to curves 2 and 3 in Figure 6, a and b. Graphs above series 1 schematically show the growth of the mutant and ancestral populations during three cultures of this series.
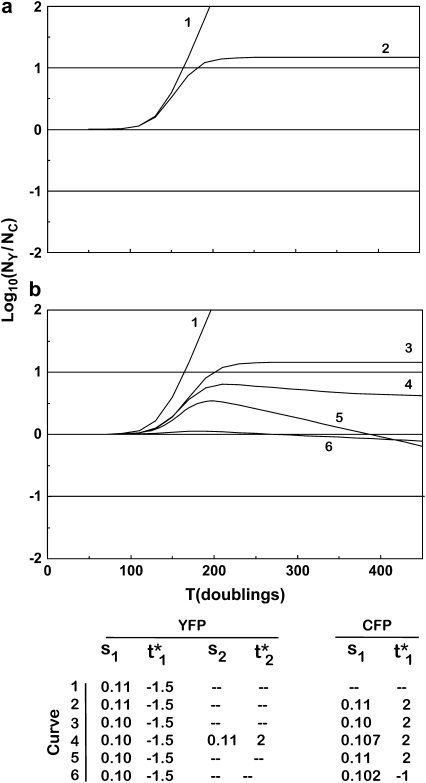
Figure 6.—
Characteristic behavior of Log10(NY/NC)as a function of time. (a) If all mutants have the same selection coefficient, ρ(s) equals a delta function, and two basic behaviors are possible for Log10(NY/NC). The ratio either departs from 0 and continues to increase (or decrease) until one population displaces the other (curve 1) or stabilizes at a constant value when mutants come to dominate both populations (curve 2). The effective mutation times 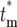 and the selection coefficients for the mutant clones for both a and b are shown at the bottom. (b) If the distribution of selection coefficients has finite width, then the behavior is more complex. The curves show the behavior of Equation 22 with either one or two (curve 4 only) mutant clones dominating
and the selection coefficients for the mutant clones for both a and b are shown at the bottom. (b) If the distribution of selection coefficients has finite width, then the behavior is more complex. The curves show the behavior of Equation 22 with either one or two (curve 4 only) mutant clones dominating 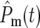 for each population. In curve 1, a mutant occurs first in the YFP population, and it completely dominates the culture. Curve 3 shows the behavior if mutants with the same selection coefficient occur in both populations but at different effective times. This is unlikely to be stable as time progresses unless both selection coefficients are at the maximum available value, since a clone with higher s will eventually become dominant. Curve 4 shows two significant mutant clones in the YFP population with slightly different values of s and effective mutation times and a single significant clone in the CFP population with a value of s between those in the YFP mutants. In the YFP population the mutant with larger s must occur at a later effective time or the lower-s mutant would never affect the behavior of the culture. Note that the curve departs from 0 in the positive direction and bends over to develop a negative slope, which slowly become less negative as the larger-s clone become more dominant in the YFP population. Curve 5 shows the behavior with single prominent mutant clones in both populations. Finally, curve 6 shows the effect of single mutants in each population with nearly identical selection coefficients and effective mutation times. The curve hardly departs from 0.
for each population. In curve 1, a mutant occurs first in the YFP population, and it completely dominates the culture. Curve 3 shows the behavior if mutants with the same selection coefficient occur in both populations but at different effective times. This is unlikely to be stable as time progresses unless both selection coefficients are at the maximum available value, since a clone with higher s will eventually become dominant. Curve 4 shows two significant mutant clones in the YFP population with slightly different values of s and effective mutation times and a single significant clone in the CFP population with a value of s between those in the YFP mutants. In the YFP population the mutant with larger s must occur at a later effective time or the lower-s mutant would never affect the behavior of the culture. Note that the curve departs from 0 in the positive direction and bends over to develop a negative slope, which slowly become less negative as the larger-s clone become more dominant in the YFP population. Curve 5 shows the behavior with single prominent mutant clones in both populations. Finally, curve 6 shows the effect of single mutants in each population with nearly identical selection coefficients and effective mutation times. The curve hardly departs from 0.
In the second example, red mutants completely overtake the culture before green mutants of sufficiently large s come to prominence, although green mutations with low s will have occurred. This corresponds to curve 1 in Figure 6, a and b. In the third example, red mutants are assumed to come to prominence first and the fluorescence ratio begins to change in favor of red. At a later time, mutant cells with larger s come to prominence in the green population, and the ratio changes in favor of green. Still later, mutants that have a selection coefficient equivalent to the green population come to prominence in the red population, and the ratio stabilizes. The initial part of such behavior is shown by curve 5 in Figure 6b. Subsequent variations in the ratio may occur as mutants with ever larger s come to prominence in the two populations. But if ρ(s) has a sufficiently defined maximum s, then the ratio will finally stabilize when both populations are dominated by mutant cells with this maximal selection coefficient unless one population completely displaces the other as shown in example 2. In the final example, roughly equivalent mutants arise in both populations at about the same times, so that the ratio remains constant as both populations become dominated by mutant cells with selection coefficients tending toward the maximum possible. This is the behavior that always occurs if the size of the cultures is sufficiently large so that many mutations occur.
THE ANALYTICAL FRAMEWORK
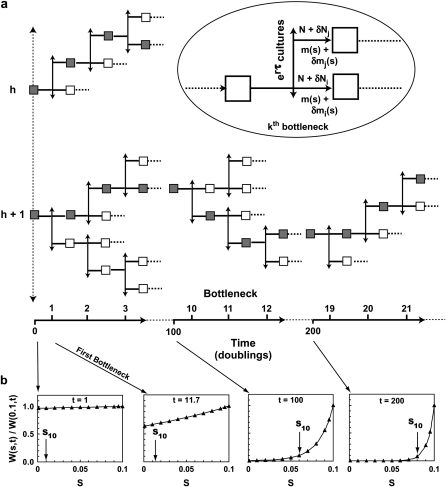
Imagine an arbitrarily large ensemble of initial cultures, each starting with N0 cells (Figure 2a). Only one cell population is shown for clarity. If additional populations are present as in the coculture experiment, their behaviors will be statistically independent. Each initial culture is sampled after growth time τ, but instead of seeding only one next passage culture as in the actual experiment, all of the material from each initial culture is used to seed erτ daughter cultures (3300 in the case of the specific experiment in question). The details of this exhaustive sampling are illustrated in the oval inset in Figure 2a for the kth bottleneck. Each of the (k +1)st cultures receive ∼N + δN ancestral and m(s) + δm(s) mutant cells, where the δ's indicate stochastic fluctuations that affect the individual cultures. The actual experiment is equivalent to selecting 72 of the initial cultures, and for each of these selecting a sequence of daughters, thereby producing 72 series with 40 sequential cultures in each. Two such series are indicated in Figure 2a by the shaded boxes. Examination of Figure 2a shows that the time dependence of the characteristics of the mutant cells averaged over a large number of series of daughters would be identical to the behavior of the total mutant population if all of the daughters were pooled. This pooled culture is just a population in unbounded exponential growth, which can be analyzed with straightforward approaches. Since the structure of this conceptual experiment preserves cell lineages, it also provides the basis for the subsequent calculation of the stochastic properties of the system.
Figure 2.—
The conceptual experiment. (a) An arbitrarily large number of initial cultures, each containing N0 cells, are started at t = 0. Two such cultures, h and h + 1, are shown. After time τ, the time of the first bottleneck, the cells in each initial culture are divided among erτ daughter cultures so that each receives N0 cells. Thus no cells are discarded. After each subsequent time interval τ, each daughter seeds erτ next-generation daughters. During each growth phase mutants received from the previous culture expand and de novo mutants randomly occur. At each bottleneck there is statistical variation in the number of mutant and ancestral cells received by the daughters. This process is indicated by the inset in the oval where m(s) and δm(s) indicate the average number and the fluctuation from the average of the mutants transmitted to the next daughter, and N and δN indicate the same for the ancestral cells. The time (in population doublings) and the number of the bottleneck is indicated along the bottom, assuming 11.7 doublings between bottlenecks. (b) The shape of W(s, t), normalized to its value at s = 0.1, is shown as a function of s for representative values of t. As t increases the weight of this function rapidly moves to larger s. The distribution of mutant cells in the pooled culture of a is given by ρ(s)W(s, t). For each of the possible paths through a series of daughters, the selection coefficients of the mutants will differ due to the stochastic variation, but the powerful shape of W(s, t) ensures that those that become prominent will be confined to a small range of s almost regardless of the shape of ρ(s). If ρ(s) is a constant out to some smax, then W(s, t) directly gives the expected distribution of the mutants. The values of s, termed s10, above which 90% of the mutant cells occur are indicated on the graphs for this case (see Equation 8).
This conceptual experiment is, of course, impossible to implement. Using the parameters of the actual experiment, by the 13th of the 40 days, the volume of culture medium required for the progeny of a single initial culture would fill a ball with a radius about 22 times that of our solar system out to Pluto, and the radius would be increasing faster than the speed of light. Coincidentally, this is about the characteristic time, 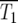 ∼ 150 doublings, that mutants became equal in abundance to the ancestral cells in the actual HS experiment (see below).
∼ 150 doublings, that mutants became equal in abundance to the ancestral cells in the actual HS experiment (see below).
DESCRIPTION OF THE AVERAGE CHARACTERISTICS
Basic relationships:
Consider one of the cell populations in the infinite pooled culture of Figure 2a. Its development is described by standard equations for exponential growth. Let N∞(t) and M∞(s, t) be the number of ancestral and mutant cells at time t. Then
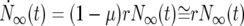 |
(1a) |
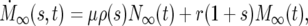 |
(1b) |
where the dots indicate the derivative with respect to time, μ is the overall mutation rate per generation for the ancestral cells, ρ(s) is the probability distribution for the selection coefficients s of the mutants, and r = ln(2). Equation 1a describes the increase of ancestral cells due to division and the decrease due to conversion to mutants. Since μ ≪ 1, it is neglected in what follows. The coefficient r scales time so that it is measured in units of the doubling time for the ancestral cells. Equation 1b describes the increase in the number of mutant cells with selection coefficient s through de novo mutation and the division of existing mutants with a rate 1 + s times that of the ancestral cells. In the real world, singly mutant cells are susceptible to additional mutations. The possibility of multiple mutations raises complex modeling issues that are discussed in section 2 of the supplemental materials at http://www.genetics.org/supplemental/. Multiple mutants are neglected in what follows.
This continuous growth model (with bottlenecks introduced below) does not include one important component of the actual experiment. In the real experiment the cultures enter a stationary phase where growth stops prior to seeding the daughter cultures. As mutants become prominent in the population and the overall growth rate increases somewhat, this stationary phase will be reached earlier during each passage. By contrast, the model allows expansion to continue during this stationary period. The error introduced by this simplification is small for the HS experiment. As is shown below, the maximum selection coefficient for the mutants in the experiment is ∼0.1. Thus, after cultures become dominated by mutants, in the time equivalent to 11.7 doublings of the ancestral cells the model allows about one additional doubling per passage (rsτ ∼ 1). Therefore, during the late phases of the experiment the timescale in the model may be somewhat accelerated compared to that of the actual cultures. However, inclusion of this small effect in the analysis is not warranted given the noise in the experimental data with which it will be compared. The presence of the stationary phase raises additional interpretive issues, since as indicated in the discussion the selective advantage of mutants may be expressed by changes in their behavior as they cease and resume proliferation.
The solution of Equation 1a is N∞(t) = N∞ert, where N∞ is the number of ancestral cells at t = 0. Inserting N∞(t) into Equation 1b allows solving for M∞(s, t). It is convenient for the subsequent discussion to calculate Rm(s, t), the ratio of mutant cells with selection coefficient s to the total number of ancestral cells, because this describes how significant the mutant population has become:
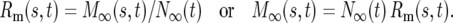 |
Inserting this into Equation 1b, one finds
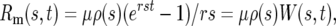 |
(2) |
where
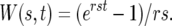 |
It is immediately apparent that the distribution of selection coefficients found in the mutant cells at time t is given by the product of ρ(s) with the weight factor W(s, t).
At small values of st, W(s, t) = t, and Rm(s, t) = μ tρ(s), indicating the buildup of de novo mutations linearly with time and with a distribution in s given by ρ(s). As s and/or t increase, W(s, t) increases dramatically due to the relative expansion of mutations that occurred early in the cultures. Figure 2b shows the shape of W(s, t) for 0 < s < 0.1 and times t = 1, 11.7 (the first bottleneck in the actual experiment), 100, and 200 population doublings. For plotting convenience these graphs have been normalized to the values of W(s, t) at s = 0.1. As time progresses, the weight of this function becomes concentrated at higher values of s. Thus for almost any shape of ρ(s) that one might choose, the width of the range of selection coefficients that are prominent in the mutant population will become narrower as time progresses. For most shapes of ρ(s) the “effective” selection coefficients will fall within a narrow range near the maximum s that is possible. An alternate derivation of Equation 2, which has the flexibility to address more complex systems, is given in section 1 of the supplemental materials at http://www.genetics.org/supplemental/.
Given Equation 2, the ratio of the number of mutant cells with selection coefficients between 0 and some value of s to the total number of ancestral cells at time t, denoted by Pm(s, t), can be calculated. Thus
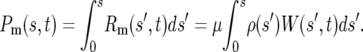 |
(3) |
Integrating over the whole range of s gives Pm(∞, t), the ratio of the total number of mutant cells to the ancestral cells at time t. The fraction of mutant cells with selection coefficients between any values s1 and s2 is then given by
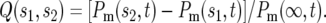 |
(4) |
These relationships allow determination of the average behavior of the system for various assumptions concerning μ and ρ(s). The range of effective selection coefficients and the characteristic times T1 and T100, for which the number of mutant cells are respectively equal to, or 100 times greater than, the ancestral population are now compared for two specific choices ρ(s).
Comparison of uniform and delta-function distributions for ρ(s):
Consider first a uniform distribution ρ(s) = 1/smax for 0 ≤ s ≤ smax and 0 otherwise. The product ρ(s)W(s, t) is W(s, t)/smax for 0 ≤ s ≤ smax and so has the shape of W(s, t) up to smax, whereupon it drops to 0. Figure 2b shows the shape of this function, indicating that as time increases the predominant mutants have selection coefficients progressively closer to smax. Applying Equations 3 and 4 allows calculation of the range of s of the effective mutations:
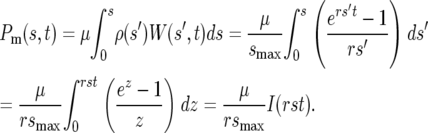 |
(5) |
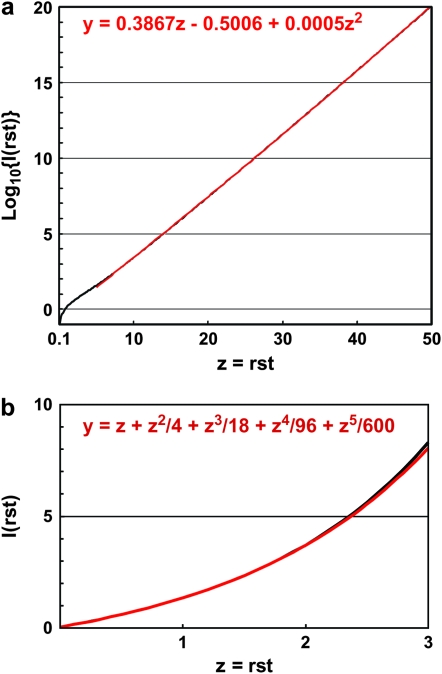
The integral I(rst) can be evaluated numerically in a straightforward manner. Figure 3a shows a graph of Log10[I(rst)] along with regression fit for the region 5 < rst < 50. Using this fit as an approximation gives:
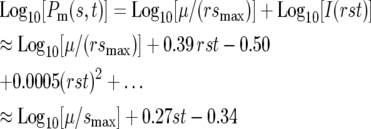 |
(6) |
or equivalently
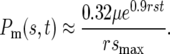 |
The use of the linear approximation for the fit to Log10[I(rst)] is particularly accurate for 7 < rst < 16, for which the deviation of the approximate value of Log10[Pm(s, t)] from the true value is <0.1. This corresponds to times on the order of ∼100 to ∼230 generations for s ∼ 0.1. Section 4 of the supplemental materials at http://www.genetics.org/supplemental/ and Figure 3b present an approximation to Equation 5 and I(rst) for low values of rst.
Figure 3.—
Approximate evaluation of the integral I(rst). (a) The black line shows the numerical evaluation of the integral in Equation 5 for 0.1 < rst < 50. The red line, coincident with the black line, shows a second-order fit to the values for 5 < rst < 50. The coefficients of the fit are sensitive to the exact range of the curve that is being fitted, but these are representative. (b) Evaluation of I(rst) for 0 < rst < 3 (black line). The result of integrating a Taylor series expansion of the integrand is shown in red. The formula for this curve is shown on the graph and used in section 4 of the supplemental materials at http://www.genetics.org/supplemental/.
The range of s containing 90% of the mutant cells is arbitrarily defined as the range of effective selection coefficients. Since Pm(s, t) is monotonically increasing, the effective range can be obtained by determining the selection coefficient, s10, for which 10% of mutants have lower s. Setting Q(0, s10) = 0.1 in Equation 4 yields s10. Since Pm(0, t) = 0, one finds
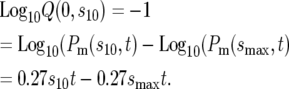 |
(7) |
Thus within the range of validity of the approximation for Log10[I(rst)],
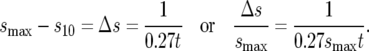 |
(8) |
Thus if smax = 0.1, at t = 100 doublings, 90% of the mutant cells have s between 0.063 and 0.1, while by t = 200 the same proportion will be between 0.083 and 0.1. The values of s10 are indicated on the graphs in Figure 2b.
The highly peaked distribution in s of the mutant population at long times even though ρ(s) was assumed to be flat suggests that some aspects of the behavior of cultures of these cells would be similar to a system in which one assumed a delta-function distribution of selection coefficients. Using ρ(s) = δ(s − χ) and a mutation rate of μδ in Equation 3, one finds analogous to quation 5,
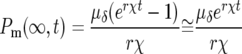 |
(9) |
or
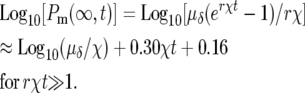 |
This result for the delta function is identical in form to the (approximate) result for the uniform distribution of ρ(s) found in Equation 6. Evaluating Equation 6 at s = smax so that the entire mutant population is represented, Equations 4 and 9 are quantitatively identical if one sets χ = 0.9smax and μδ = 0.29μ. Thus after the initial period where they are clearly distinct, the total number of mutant cells evolves with approximately the same time course for both of these forms for ρ(s). The delta-function approximation is appropriate for any ρ(s) that results in a highly peaked shape for ρ(s)W(s, t) at long times. The scaling of the overall mutation rates and the “equivalent” selection coefficients will depend on the details of the shape of ρ(s) and may also depend on which aspects of the cultures are being modeled. Some critical aspects of real systems cannot be modeled by the delta-function approximation, as shown in Figure 6 and the related text.
Characteristic times:
There are at least two times in the history of the culture that are interesting to calculate. The most relevant is T1, the time when the numbers of mutant and ancestral cells in the culture are equal. While this differs in each series of finite cultures as shown in Figure 1b, it has a well-defined value for the effectively infinite system of Figure 2a. This characteristic time, 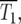 is the time when
is the time when 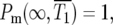 which can be determined from Equation 3 for any assumed ρ(s). For the specific case of the uniform fitness distribution,
which can be determined from Equation 3 for any assumed ρ(s). For the specific case of the uniform fitness distribution, 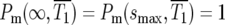 or
or 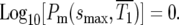 Similarly, for a delta-function distribution,
Similarly, for a delta-function distribution, 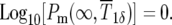 Using Equations 6 and 9 one finds
Using Equations 6 and 9 one finds
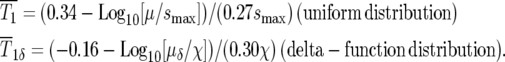 |
(10) |
The second relevant time is the time for “fixation” of the mutations, which following HS is defined as the time when mutants are 100 times the abundance of ancestral cells. At this time for the uniform distribution 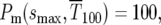 or
or 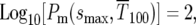 while similarly for the delta-function distribution
while similarly for the delta-function distribution 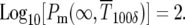 Thus from Equations 6 and 9,
Thus from Equations 6 and 9,
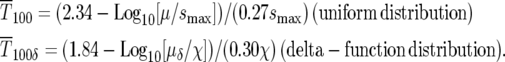 |
(11) |
The range of selection coefficients that are prominent in the population at these times for the uniform distribution can be calculated using Equation 8. Using μ = 10−5 and smax ∼ 0.12, values used by HS for their simulation of a uniform ρ(s) described in their Figure 1B, Equation 10 yields 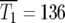 generations. At that time 90% of the mutant cells are in the range Δs/smax ∼ 0.23, or 0.089 < s < 0.12. Similarly, from Equation 11
generations. At that time 90% of the mutant cells are in the range Δs/smax ∼ 0.23, or 0.089 < s < 0.12. Similarly, from Equation 11 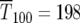 generations, and at that time Δs/smax = 0.16, or 0.10 < s < 0.12. Thus at these times 90% of the mutant cells have selection coefficients very near the maximum possible.
generations, and at that time Δs/smax = 0.16, or 0.10 < s < 0.12. Thus at these times 90% of the mutant cells have selection coefficients very near the maximum possible.
STOCHASTIC EFFECTS
Differences in behavior among different series of cultures are due to stochastic variation in the times 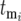 new mutants arise, the corresponding selection coefficients si of the mutants, and the effects of the selection bottlenecks. This section demonstrates that the stochastic effect of the multiple bottlenecks is equivalent to altering the actual mutation times
new mutants arise, the corresponding selection coefficients si of the mutants, and the effects of the selection bottlenecks. This section demonstrates that the stochastic effect of the multiple bottlenecks is equivalent to altering the actual mutation times 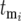 to effective times
to effective times 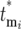 and derives probability distributions for
and derives probability distributions for 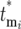 given
given 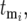 s, and the length of the growth periods, τ. This formalism, coupled with statistical sampling of the mutation times on the basis of the mutation rate and sampling of the selection coefficients on the basis of ρ(s), allows a complete description of the stochastic behavior of the system. The analysis proceeds by first calculating the statistics of the behavior of a single-mutant clone with a defined mutation time and selection coefficient and then combining multiple mutants to describe the system completely.
s, and the length of the growth periods, τ. This formalism, coupled with statistical sampling of the mutation times on the basis of the mutation rate and sampling of the selection coefficients on the basis of ρ(s), allows a complete description of the stochastic behavior of the system. The analysis proceeds by first calculating the statistics of the behavior of a single-mutant clone with a defined mutation time and selection coefficient and then combining multiple mutants to describe the system completely.
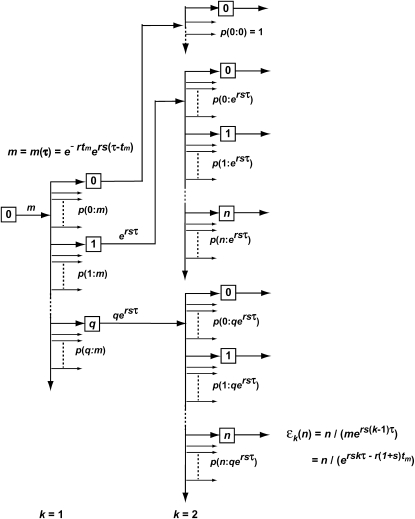
Stochastic behavior of a single-mutant clone:
Figure 4 shows the details of the behavior of the progeny of a mutant cell expanding through a series of daughter cultures such as described in the conceptual experiment Figure 2a. No cells are discarded. Assume that the mutation occurred at time 0 ≤ tm ≤ τ, where τ is the length of each of the culture periods. At the kth bottleneck, t = kτ, and the average number of mutants, m(kτ), transferred to each daughter is the total number of progeny from this mutation divided by the total number of daughter cultures:
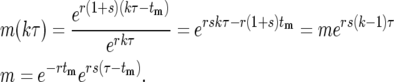 |
(12) |
m is the average number of mutant progeny that are transferred to each daughter at the first bottleneck.
Figure 4.—
Schematic of the stochastic transmission of the progeny of a single mutation through two bottlenecks. The boxes indicate individual cultures that are related to each other according to the exhaustive sampling scheme described in Figure 2a. A mutation is assumed to occur at some time during the initial culture period and on average m mutant progeny are passed to each of its erτ daughters. The daughters are arranged in order of the number of mutant cells they receive, which is indicated within the boxes. The small arrows indicate the additional daughter cultures that received the same number of mutant cells. The proportion of the first-generation daughters receiving an actual number, q, is given by a suitable probability distribution such as Poisson. Between the first and the second bottlenecks these mutants expand relative to the ancestral cells by a factor ersτ. These will again be probabilistically distributed to their daughters. From the second bottleneck onward, daughters with a particular number of cells, n, descend from multiple predecessor cultures. The probability of having n mutants after bottleneck k, 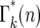 , is given by summing over these possibilities, as shown in Equation 13. The ratio of the actual number of mutant cells in an individual culture after the kth bottleneck to the average number for that time is ɛk (Equation 14).
, is given by summing over these possibilities, as shown in Equation 13. The ratio of the actual number of mutant cells in an individual culture after the kth bottleneck to the average number for that time is ɛk (Equation 14).
Due to statistical fluctuations in the bottlenecks, the actual number of cells each daughter receives is distributed around this average. As illustrated in Figure 4, after the second and subsequent bottlenecks, daughters with the same number of mutants can descend from different predecessors, so that the probability, 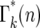 , of a daughter receiving n mutant cells after the kth bottleneck requires summing over these multiple possibilities. Suppose that q mutants were transferred into a daughter after the (k − 1)st bottleneck. At the next bottleneck, the kth, these have expanded so that on average qersτ mutant cells will be distributed to each of its erτ daughters. The statistical distribution in the number n received by these daughters is given by a probability distribution p(n:qersτ). But the probability of having a culture with q initial cells is given by
, of a daughter receiving n mutant cells after the kth bottleneck requires summing over these multiple possibilities. Suppose that q mutants were transferred into a daughter after the (k − 1)st bottleneck. At the next bottleneck, the kth, these have expanded so that on average qersτ mutant cells will be distributed to each of its erτ daughters. The statistical distribution in the number n received by these daughters is given by a probability distribution p(n:qersτ). But the probability of having a culture with q initial cells is given by 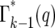 . Thus,
. Thus,
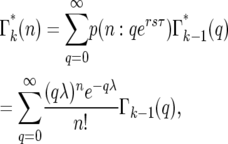 |
(13) |
where λ = ersτ, k ≥ 2, and the Poisson probability distribution has been used because it is appropriate, at least initially, when the number of mutant cells is substantially smaller than the total number of cells. As shown below, this condition is met for the entire time period during which stochastic variation is important. As can be seen from examination of Figure 4, 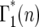 = p(n:m(τ)), where from quation 12, m(τ) = ersτ−r(1+s)tm = λe−r(1+s)tm. Evaluation of Equation 13 proceeds by using
= p(n:m(τ)), where from quation 12, m(τ) = ersτ−r(1+s)tm = λe−r(1+s)tm. Evaluation of Equation 13 proceeds by using 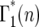 to calculate
to calculate 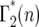 , etc. The supplemental materials at http://www.genetics.org/supplemental/ contains an Excel workbook that performs this calculation, as well as the calculations of the related statistical distributions discussed below. Note that
, etc. The supplemental materials at http://www.genetics.org/supplemental/ contains an Excel workbook that performs this calculation, as well as the calculations of the related statistical distributions discussed below. Note that 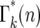 depends on τ, s, and tm through m and λ.
depends on τ, s, and tm through m and λ.
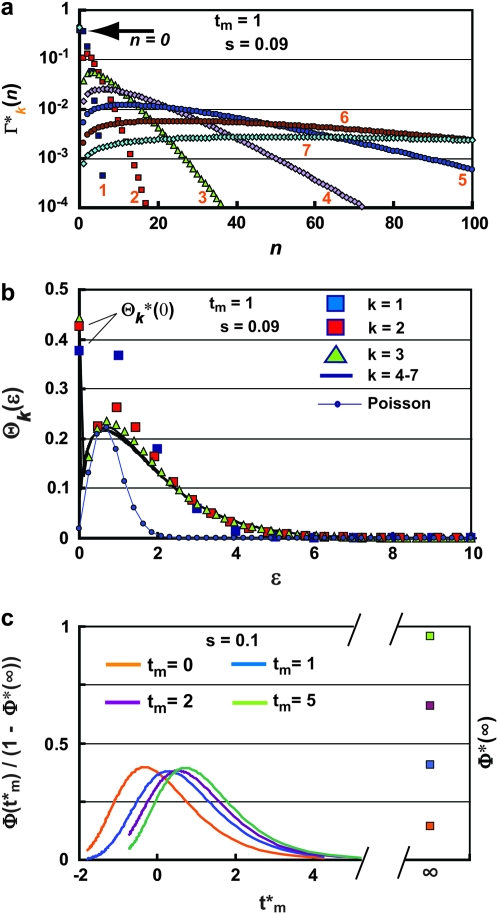
The basic behavior of Equation 13 is easily understood. Beginning with a Poisson distribution after the first bottleneck, it progressively broadens after subsequent bottlenecks. Figure 5a shows plots of 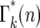 for k = 1–7 when tm = 1 and s = 0.09. Note that
for k = 1–7 when tm = 1 and s = 0.09. Note that 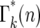 varies smoothly and progressively more slowly as k increases, except between n = 0 and 1 where there is a pronounced discontinuity that increases with increasing k. Initially the broadening of
varies smoothly and progressively more slowly as k increases, except between n = 0 and 1 where there is a pronounced discontinuity that increases with increasing k. Initially the broadening of 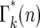 is due to the combination of stochastic events in the bottlenecks and the expansion of the mutant clone. As the number of mutant cells increases with time, stochastic variation in the number transferred from a particular culture to its immediate daughters eventually becomes insignificant compared to the mean number transferred since Δn/n ∼ n−1/2. Thus it is expected that the important stochastic variation induced by the bottlenecks occurs early in the experiment. This expectation can be quantitatively described in the following manner.
is due to the combination of stochastic events in the bottlenecks and the expansion of the mutant clone. As the number of mutant cells increases with time, stochastic variation in the number transferred from a particular culture to its immediate daughters eventually becomes insignificant compared to the mean number transferred since Δn/n ∼ n−1/2. Thus it is expected that the important stochastic variation induced by the bottlenecks occurs early in the experiment. This expectation can be quantitatively described in the following manner.
Figure 5.—
Statistical distributions for n, ɛ, and the effective mutation time 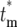 . (a) The probability
. (a) The probability 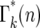 of having n progeny of a mutant after k bottlenecks for s = 0.09, τ = 11.7, and tm = 1. The value of k for each curve is shown by the orange numbers. Note that the distribution broadens to higher values of n with increasing k, and its maximum magnitude for n > 0 correspondingly decreases. For the chosen parameters, the probability that a culture will have 0 progeny of the mutant (indicated by the arrow) is basically established at the first bottleneck and subsequently increases only slightly. (b) Probability distribution for ɛk after each bottleneck. Distributions are shown for k = 1–7, for tm = 1, τ = 11.7, and s = 0.09. Symbols corresponding to each n are shown for the first three bottlenecks; for larger k they are so closely spaced that solid curves are shown. Note that differences become imperceptible for k > 4 and that the width is significantly broader than a Poisson distribution, which is shown in blue for comparison. The probability of a culture receiving 0 cells hardly increases after the first bottleneck for these parameters. These values are given by
of having n progeny of a mutant after k bottlenecks for s = 0.09, τ = 11.7, and tm = 1. The value of k for each curve is shown by the orange numbers. Note that the distribution broadens to higher values of n with increasing k, and its maximum magnitude for n > 0 correspondingly decreases. For the chosen parameters, the probability that a culture will have 0 progeny of the mutant (indicated by the arrow) is basically established at the first bottleneck and subsequently increases only slightly. (b) Probability distribution for ɛk after each bottleneck. Distributions are shown for k = 1–7, for tm = 1, τ = 11.7, and s = 0.09. Symbols corresponding to each n are shown for the first three bottlenecks; for larger k they are so closely spaced that solid curves are shown. Note that differences become imperceptible for k > 4 and that the width is significantly broader than a Poisson distribution, which is shown in blue for comparison. The probability of a culture receiving 0 cells hardly increases after the first bottleneck for these parameters. These values are given by 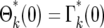 . (c) Dependence of the probability distribution Φ(
. (c) Dependence of the probability distribution Φ(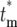 ) of the effective mutation times,
) of the effective mutation times, 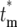 , on the actual mutation time, tm, for τ = 11.7 and s = 0.1. As a mutation occurs at later times, the probability that a daughter culture will receive 0 progeny increases, as shown by the boxes at
, on the actual mutation time, tm, for τ = 11.7 and s = 0.1. As a mutation occurs at later times, the probability that a daughter culture will receive 0 progeny increases, as shown by the boxes at 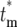 = ∞. The curves for finite
= ∞. The curves for finite 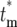 are plotted after normalization by 1 − Φ*(∞), where Φ*(∞) is the probability of having n = 0 progeny of the mutation. Note that the curves move to slightly higher values of
are plotted after normalization by 1 − Φ*(∞), where Φ*(∞) is the probability of having n = 0 progeny of the mutation. Note that the curves move to slightly higher values of 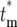 as the mutation time increases, but still overlap 0.
as the mutation time increases, but still overlap 0.
Let ɛk(n) be the ratio of the actual number of mutant cells n in a particular culture to the average number (Equation 12) after the kth bottleneck. Then
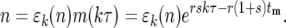 |
(14) |
The values of ɛk(n) for daughters receiving different numbers of mutants after the second bottleneck are illustrated in Figure 4. The average value of ɛk(n) = 1. Although n is rigorously an integer so that only specific values of ɛk(n) can occur, the slow variation of 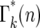 with n, Figure 5a, allows an accurate description of the system to be obtained by treating n as a continuous variable and defining Γk(n) as a continuous probability density that has the values
with n, Figure 5a, allows an accurate description of the system to be obtained by treating n as a continuous variable and defining Γk(n) as a continuous probability density that has the values 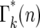 for integer values of n. For noninteger n, Γk(n) can be obtained by interpolation. This approximation is valid for n ≥ 1, but not for n = 0 due to the substantial discontinuity in
for integer values of n. For noninteger n, Γk(n) can be obtained by interpolation. This approximation is valid for n ≥ 1, but not for n = 0 due to the substantial discontinuity in 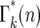 between n = 0 and 1. Thus in the calculations that follow, probabilities corresponding to n = 0 will be given by
between n = 0 and 1. Thus in the calculations that follow, probabilities corresponding to n = 0 will be given by 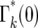 , while those corresponding to other values of n will be based on calculations that treat parameters as continuous. Using this approximation, which is increasingly accurate as k increases, the probability distribution, Θk(ɛk), for ɛk, after each bottleneck can be calculated using Equations 12–14,
, while those corresponding to other values of n will be based on calculations that treat parameters as continuous. Using this approximation, which is increasingly accurate as k increases, the probability distribution, Θk(ɛk), for ɛk, after each bottleneck can be calculated using Equations 12–14,
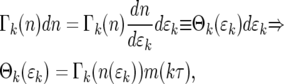 |
(15) |
where m(kτ) is given by Equation 12. Equation 15 holds for ɛk > 0. The probability of having ɛk = 0, which corresponds to n = 0, is 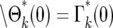 .
.
Figure 5b shows behavior of Θk(ɛk) and 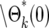 for k = 1–7, s = 0.09, τ = 11.7, and tm = 1. (The distributions for other parameter values can be calculated using the Excel sheet in the supplemental materials at http://www.genetics.org/supplemental/.) Note that the shape changes substantially for the first few bottlenecks, but for k ≥ 4 it becomes constant. This is the quantitative description of the previous statement that once the number of progeny of the mutated cell becomes large, no significant additional variation is introduced by the subsequent bottlenecks. Thus a single distribution, Θ(ɛ), calculable from first principles, can be used to describe the behavior of the mutants after sufficient time. The distribution for each mutant clone depends on the actual time the mutation occurred, the selection coefficient, and the length of the growth periods. In this example calculation, the stable distribution is reached by the analysis of cultures containing up to 100 mutant cells, 1000-fold less than the number of ancestral cells present in the actual experiment. Thus statistical stability is reached well before the time T1. For comparison, Figure 5b also shows a Poisson distribution scaled so that its maximum is located at the same position as the maximum of the limiting mutant distribution. The enhanced broadening due to the series of bottlenecks is evident.
for k = 1–7, s = 0.09, τ = 11.7, and tm = 1. (The distributions for other parameter values can be calculated using the Excel sheet in the supplemental materials at http://www.genetics.org/supplemental/.) Note that the shape changes substantially for the first few bottlenecks, but for k ≥ 4 it becomes constant. This is the quantitative description of the previous statement that once the number of progeny of the mutated cell becomes large, no significant additional variation is introduced by the subsequent bottlenecks. Thus a single distribution, Θ(ɛ), calculable from first principles, can be used to describe the behavior of the mutants after sufficient time. The distribution for each mutant clone depends on the actual time the mutation occurred, the selection coefficient, and the length of the growth periods. In this example calculation, the stable distribution is reached by the analysis of cultures containing up to 100 mutant cells, 1000-fold less than the number of ancestral cells present in the actual experiment. Thus statistical stability is reached well before the time T1. For comparison, Figure 5b also shows a Poisson distribution scaled so that its maximum is located at the same position as the maximum of the limiting mutant distribution. The enhanced broadening due to the series of bottlenecks is evident.
The fact that a stable, readily calculable probability distribution can be used to describe the progeny of each mutation after several bottlenecks allows a particularly simple general description of the stochastic behavior of the entire system. Consider the ratio,  of the average number of mutant cells to the number of ancestral cells per culture:
of the average number of mutant cells to the number of ancestral cells per culture:
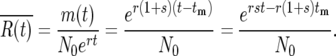 |
(16) |
The actual ratio in the daughter cultures, R(t), will differ from this ratio by the factor ɛ that has developed due to stochastic variation in the earlier bottlenecks. Therefore, for times long enough after the mutation for statistical stability to be established one has
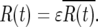 |
(17) |
Therefore,
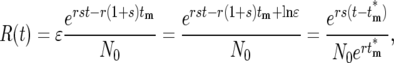 |
(18) |
where 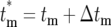 is the effective time the mutation occurred, adjusted from the actual tm by Δtm = −ln(ɛ)/r(1 + s) due to the stochastic effects in the bottlenecks, and
is the effective time the mutation occurred, adjusted from the actual tm by Δtm = −ln(ɛ)/r(1 + s) due to the stochastic effects in the bottlenecks, and 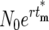 is the size of the ancestral population at the effective mutation time. Negative values of Δtm indicate a stochastic fluctuation that increases the abundance of a mutant clone relative to its average value; e.g., it appears that the mutation occurred earlier than it actually did, while positive values indicate a stochastic decrease relative to the average since the effective occurrence time was later than the actual time. If a mutant is lost from a series of cultures, Δtm = ∞. Note that Equation 18 assumes that 0 ≤ tm ≤ τ.
is the size of the ancestral population at the effective mutation time. Negative values of Δtm indicate a stochastic fluctuation that increases the abundance of a mutant clone relative to its average value; e.g., it appears that the mutation occurred earlier than it actually did, while positive values indicate a stochastic decrease relative to the average since the effective occurrence time was later than the actual time. If a mutant is lost from a series of cultures, Δtm = ∞. Note that Equation 18 assumes that 0 ≤ tm ≤ τ.
Calculation of the probability distribution, Φ(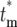 ), for
), for 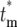 allows a statistical description of the behavior of the mutant cells. Using the continuous variable approximation discussed prior to Equation 15,
allows a statistical description of the behavior of the mutant cells. Using the continuous variable approximation discussed prior to Equation 15,
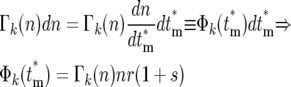 |
(19a) |
or
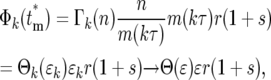 |
(19b) |
where the relationships among the various parameters required to evaluate Equation 19 are 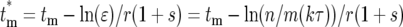 , and Equation 15 was used to relate Φk(
, and Equation 15 was used to relate Φk(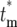 ) to
) to 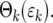 Equation 19b demonstrates that Φk(
Equation 19b demonstrates that Φk(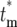 ) becomes independent of k after several bottlenecks since
) becomes independent of k after several bottlenecks since 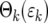 becomes independent of k. As a practical matter, Φk(
becomes independent of k. As a practical matter, Φk(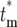 ) is calculated by choosing n and m(kτ) for any bottleneck after stabilization of the distributions, with m(kτ) given by Equation 12. Equation 19 is valid for finite
) is calculated by choosing n and m(kτ) for any bottleneck after stabilization of the distributions, with m(kτ) given by Equation 12. Equation 19 is valid for finite 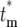 . For
. For 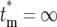 , which corresponds to having n = 0 progeny of the mutant, Φ*(∞) =
, which corresponds to having n = 0 progeny of the mutant, Φ*(∞) = 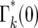 . The stabilization of the distribution for
. The stabilization of the distribution for 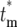 after several bottlenecks means that the
after several bottlenecks means that the 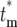 of a mutation in a particular daughter culture remains the same for all of its descendant cultures. Thus the effective mutation times provide a suitable basis for describing the stochastic character of the long-term evolution of the system.
of a mutation in a particular daughter culture remains the same for all of its descendant cultures. Thus the effective mutation times provide a suitable basis for describing the stochastic character of the long-term evolution of the system.
Figure 5c shows the behavior of Φ(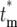 ) and Φ*(∞) for tm = 0, 1, 2, and 5, with s = 0.1 and τ = 11.7. Given the assumed parameters, if a mutation actually occurs at tm = 0, then on average ∼2.25 mutant cells from this clone will be transferred to each daughter at the first bottleneck, so that most will receive at least one cell. For larger tm, the proportion of daughter lineages that receive no progeny of the mutant (e.g., have
) and Φ*(∞) for tm = 0, 1, 2, and 5, with s = 0.1 and τ = 11.7. Given the assumed parameters, if a mutation actually occurs at tm = 0, then on average ∼2.25 mutant cells from this clone will be transferred to each daughter at the first bottleneck, so that most will receive at least one cell. For larger tm, the proportion of daughter lineages that receive no progeny of the mutant (e.g., have 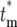 = ∞) increases, correspondingly reducing the magnitude of Φ(
= ∞) increases, correspondingly reducing the magnitude of Φ(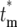 ) for finite
) for finite 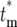 . Therefore, to conveniently visualize the behavior of Φ(
. Therefore, to conveniently visualize the behavior of Φ(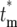 ) for different tm in one figure, Φ(
) for different tm in one figure, Φ(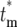 )/(1 − Φ*(∞)) has been plotted. Note that the distributions of effective mutation times for those daughter cultures that have at least one cell with this mutation include
)/(1 − Φ*(∞)) has been plotted. Note that the distributions of effective mutation times for those daughter cultures that have at least one cell with this mutation include 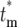 = 0 and have widths of several doubling times. Thus if the progeny of a mutation are not lost in a series of cultures, they behave as if their founding mutation occurred near the beginning of the culture period in which they originated. As tm increases the distributions move somewhat toward higher values
= 0 and have widths of several doubling times. Thus if the progeny of a mutation are not lost in a series of cultures, they behave as if their founding mutation occurred near the beginning of the culture period in which they originated. As tm increases the distributions move somewhat toward higher values 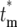 , but eventually stabilize except for a decrease in overall magnitude that has been normalized in this plot. The stabilization comes about since as tm increases eventually all daughter cultures receive either 0 or 1 cell at the first bottleneck, and those that receive a cell subsequently develop with similar statistical behavior.
, but eventually stabilize except for a decrease in overall magnitude that has been normalized in this plot. The stabilization comes about since as tm increases eventually all daughter cultures receive either 0 or 1 cell at the first bottleneck, and those that receive a cell subsequently develop with similar statistical behavior.
Description of multiple mutations:
Equations 18 and 19 allow a complete description of the multiple mutations that occur during growth in the cultures. The ratio of the total number of mutant to ancestral cells in daughter culture, 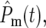 is obtained by summing over all mutations. Using Equation 18,
is obtained by summing over all mutations. Using Equation 18,
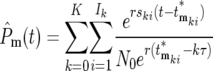 |
(20a) |
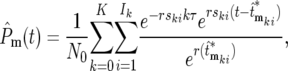 |
(20b) |
where the kith mutation occurs during the growth period after the kth bottleneck at time 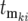 and with selection coefficient ski and effective mutation time of
and with selection coefficient ski and effective mutation time of 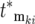 , and the times in the denominator of Equation 20a are adjusted to account for the fact that after each bottleneck the culture returns to having N0 ancestral cells. To simplify interpretation of Equation 20a and calculation of the effective mutation times, let
, and the times in the denominator of Equation 20a are adjusted to account for the fact that after each bottleneck the culture returns to having N0 ancestral cells. To simplify interpretation of Equation 20a and calculation of the effective mutation times, let 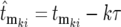 and correspondingly
and correspondingly 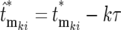 , where
, where 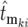 is the actual mutation time and
is the actual mutation time and 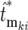 is the effective mutation time measured relative to the time of the kth bottleneck. Thus the distributions for the
is the effective mutation time measured relative to the time of the kth bottleneck. Thus the distributions for the 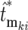 can be calculated using Equation 19, employing the adjusted mutation times
can be calculated using Equation 19, employing the adjusted mutation times 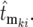
The evaluation of Equation 20 formally requires random sampling from the mutation rate distribution times the instantaneous population of ancestral cells to obtain the mutation times, assigning a selection coefficient to each mutation by sampling from the distribution ρ(s), and finally assigning the corresponding effective mutation times by sampling from the appropriate Φ(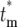 ) calculated from Equation 19. However, many aspects of its general behavior can be understood much more simply, as discussed in section 3 of the supplemental materials at http://www.genetics.org/supplemental/. Note that
) calculated from Equation 19. However, many aspects of its general behavior can be understood much more simply, as discussed in section 3 of the supplemental materials at http://www.genetics.org/supplemental/. Note that 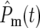 is the finite-system analog of the previously derived Pm(∞, t) from Equation 3. Pm(∞, t) is the average value of Equation 20 over multiple finite cultures.
is the finite-system analog of the previously derived Pm(∞, t) from Equation 3. Pm(∞, t) is the average value of Equation 20 over multiple finite cultures.
ANALYSIS OF THE YFP/CFP COMPETITION EXPERIMENT
Calculation of the fluorescence ratio:
Equation 20 allows a full description of the stochastic behavior of the YFP/CFP cocultures. In the coculture the differentially labeled populations grow independently and are independently subject to the statistics of mutation formation and bottlenecks. Thus, if NY and NC are the total numbers of YFP and CFP cells, respectively, and NYA and NCA are the corresponding numbers of ancestral cells, then
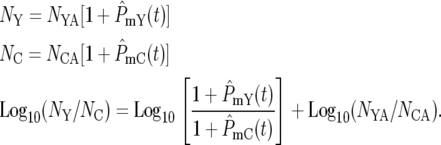 |
(21) |
At the beginning of an experiment Log10(NY/NC) remains constant (equal to 0 if the initial population sizes are equal) until a sufficient number of mutant cells arise such that 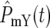 and/or
and/or 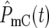 ∼ 1. Around this time, whose characteristic value is
∼ 1. Around this time, whose characteristic value is 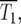 variations in Log10(NY/NC) may develop. The behavior depends on the experimental design and mutational properties of the YFP and CFP populations, which are incorporated into evaluation of Equation 20.
variations in Log10(NY/NC) may develop. The behavior depends on the experimental design and mutational properties of the YFP and CFP populations, which are incorporated into evaluation of Equation 20.
While Equation 20 appears complex, it can be substantially simplified in a manner that preserves its quantitative accuracy and facilitates understanding of the essential factors that affect the behavior of the cultures. The simplification results from recognizing that only a small subset of the terms is significant and that key parameters are restricted in their values. First, most of the terms in Equation 20 are equal to 0 because the effective mutation time for most mutants is infinite, as indicated by the approach of Φ*(∞) to 1 as 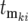 increases during a culture period (Figure 5c) (e.g., most are lost due to “drift” in the bottlenecks). Additionally, the nonzero terms have effective mutation times clustered near 0, as also indicated by Figure 5c. Moreover, Equation 2 shows that as time progresses the cells that constitute an appreciable proportion of the mutant population will have selection coefficients in the range where ρ(s)W(s, t) has significant magnitude, which is typically very narrow. Finally, a mutant clone originating at an effective time
increases during a culture period (Figure 5c) (e.g., most are lost due to “drift” in the bottlenecks). Additionally, the nonzero terms have effective mutation times clustered near 0, as also indicated by Figure 5c. Moreover, Equation 2 shows that as time progresses the cells that constitute an appreciable proportion of the mutant population will have selection coefficients in the range where ρ(s)W(s, t) has significant magnitude, which is typically very narrow. Finally, a mutant clone originating at an effective time 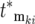 contributes substantially to Equation 20 only if its selection coefficient is larger than any of the selection coefficients of mutations with smaller
contributes substantially to Equation 20 only if its selection coefficient is larger than any of the selection coefficients of mutations with smaller 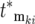 . Therefore the most significant contributors to the mutant population come from the ordered subset of mutations [
. Therefore the most significant contributors to the mutant population come from the ordered subset of mutations [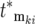 , ski], where both monotonically increase and the ski are restricted to a narrow range just below the maximum available selection coefficient, while the
, ski], where both monotonically increase and the ski are restricted to a narrow range just below the maximum available selection coefficient, while the 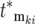 are near 0.
are near 0.
Thus as time passes clonal succession occurs, with the population being dominated by mutants with selection coefficients tending toward the largest available. Section 4 of the supplemental materials at http://www.genetics.org/supplemental/ calculates the average number of newly arising mutant cells transferred from a culture to its daughters, elucidating the buildup of the total mutant population.
Consider the YFP/CFP coculture at the time period around 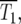 the time when
the time when 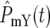 and
and 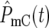 ∼ 1 on average. The behavior of the population ratio depends on the stochastically generated differences in
∼ 1 on average. The behavior of the population ratio depends on the stochastically generated differences in 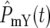 and
and 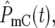 which are related to how densely the ranges of effective mutation times and selections coefficients are sampled in the two mutant populations. Imagine that N0 for both the YFP and the CFP populations is very large, so that at
which are related to how densely the ranges of effective mutation times and selections coefficients are sampled in the two mutant populations. Imagine that N0 for both the YFP and the CFP populations is very large, so that at 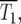 which is independent of N0, many terms are required to make
which is independent of N0, many terms are required to make 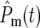 ∼ 1. Note that
∼ 1. Note that 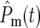 contains the factor 1/N0 (Equation 20) so that increasingly more terms are required as N0 increases. Biologically the increase in the number of terms comes from the proportional increase in the number of mutations that occur due to the larger population size. But if there are many significant terms, then the intervals of both the effective mutation times and selection coefficients between sequential terms in the ordered subset [
contains the factor 1/N0 (Equation 20) so that increasingly more terms are required as N0 increases. Biologically the increase in the number of terms comes from the proportional increase in the number of mutations that occur due to the larger population size. But if there are many significant terms, then the intervals of both the effective mutation times and selection coefficients between sequential terms in the ordered subset [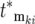 , ski] must be small due to their limited ranges of the effective values. Although the specific values in [
, ski] must be small due to their limited ranges of the effective values. Although the specific values in [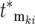 , ski] will differ for the YFP and CFP populations due to the stochastic effects, the behavior of the
, ski] will differ for the YFP and CFP populations due to the stochastic effects, the behavior of the 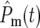 's is insensitive to these differences and for both populations approaches that of Pm(∞, t) given by Equation 3. Thus the YFP and CFP populations evolve indistinguishably in terms of cell numbers, and the population ratio remains constant. All cocultures will appear to behave the same, and no indication of the internal stochastic differences will be measurable.
's is insensitive to these differences and for both populations approaches that of Pm(∞, t) given by Equation 3. Thus the YFP and CFP populations evolve indistinguishably in terms of cell numbers, and the population ratio remains constant. All cocultures will appear to behave the same, and no indication of the internal stochastic differences will be measurable.
As N0 is decreased, fewer terms are required in Equation 20 to result in 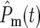 ∼ 1 at t ∼
∼ 1 at t ∼ 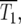 so the intervals between sequential terms in [
so the intervals between sequential terms in [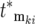 , ski] increase. The stochastic differences between the exact terms included in [
, ski] increase. The stochastic differences between the exact terms included in [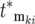 , ski] for the two populations lead to increasing possibilities for differential behavior between
, ski] for the two populations lead to increasing possibilities for differential behavior between 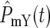 and
and 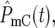 so that greater variation in the population ratio will occur among a group of “identical” cocultures. The easily measurable effects will include increased variability in the T1's, increased magnitude of the ratio variations that develop, and a larger proportion of cultures in which the ratio variations are so large that one population effectively displaces the other. If the initial YFP and CFP populations differ substantially in size, for example, one is “small” and the other is “large,” then their stochastic behaviors may be quite different in character. Section 5 of the supplemental materials at http://www.genetics.org/supplemental/ presents a rough method of estimating the number of significant terms in Equation 20 or, conversely, estimating the population size above which ratio variations are not expected.
so that greater variation in the population ratio will occur among a group of “identical” cocultures. The easily measurable effects will include increased variability in the T1's, increased magnitude of the ratio variations that develop, and a larger proportion of cultures in which the ratio variations are so large that one population effectively displaces the other. If the initial YFP and CFP populations differ substantially in size, for example, one is “small” and the other is “large,” then their stochastic behaviors may be quite different in character. Section 5 of the supplemental materials at http://www.genetics.org/supplemental/ presents a rough method of estimating the number of significant terms in Equation 20 or, conversely, estimating the population size above which ratio variations are not expected.
The effect of population size on the stochastic variations in the population ratio can be appreciated by examination of the experimental data of HS (reproduced in section 6 of the supplemental materials at http://www.genetics.org/supplemental/). If their initial population size were increased by a factor of 10, the expected ratio behavior could be estimated by averaging randomly selected sets of 10 of their experimental curves, after first properly transforming the data prior to averaging. Clearly the ratio deviations would begin at approximately the same time, indicating the constancy of 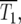 but would be of substantially reduced magnitude.
but would be of substantially reduced magnitude.
Equation 22 shows Equation 21 with the first few nonzero terms explicitly displayed,
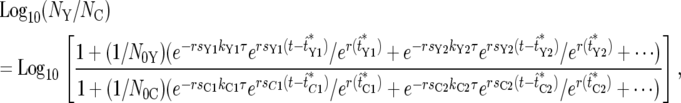 |
(22) |
where the offset term due to unequal populations has been neglected. As just discussed, in the regime where substantial population variations occur only a few terms are important. Figure 6a shows the behavior of Equation 22 under the assumptions that the initial population sizes are equal, ρ(s) is a delta function, and only one mutant clone is contributing significant progeny in each population. Since all the selection coefficients are equal, if ratio fluctuations occur due to differences in the effective mutation times, the ratio will stabilize at some constant value after mutant cells dominate both populations. Mathematically the function always reaches a stable value, curve 2. However, this value may be so extreme that it represents the extinction of one of the populations, as illustrated by curve 1. The parameters for the curves are shown at the bottom of Figure 6.
The appropriateness of Equation 22 for describing real experiments can be assessed by comparison of its behavior to the experimental data of HS. The overall shapes of the curves in the time period where they begin to depart from Log10(NY/NC) = 0 and the rough magnitudes of the ranges of variation of the curves for the model and the experimental data are qualitatively similar. Thus a model containing single significant mutant clones in the two populations, and employing effective mutation times consistent with the range defined by Figure 5c, reasonably describes these aspects of the data. However, Figure 6a does not properly describe the long-term behavior of the ratio.
The experimental data clearly show that as mutant cells become dominant in both populations, the slope of Log10(NY/NC) becomes small, but it is not typically 0. This is a clear indication of differences in the selection coefficients of the YFP and CFP mutants that are most prevalent in that time period. This behavior is modeled very well by employing differences in both the selection coefficients and the effective mutation times in Equation 22, as shown in Figure 6b. All but one of the curves in Figure 6b use one exponential term (one mutant clone) to describe the mutants in each population. The slopes of the curves after the mutants have overgrown the ancestral cells are directly related to selection coefficients of the dominant mutant clones. Taking the derivative of Equation 22 and assuming only one exponential term in the numerator and the denominator, one finds
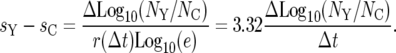 |
(23) |
If more than one clone has significant prevalence in these populations, then the effective selection coefficients are weighted averages of the several clones. However, if more than one clone is prominent, then one expects the slope to progressively change with time as the one with the highest selection coefficient increases in prevalence relative to the others. This is illustrated by curve 4 in Figure 6b. Note that on the timescale of the actual experiment, the change in slope due to clonal succession is very subtle, since the selection coefficients are necessarily closely spaced.
In summary, if the population sizes are sufficiently large, multiple mutations will contribute significant proportions of mutant cells at times on the order of  This corresponds to having many significant terms in Equation 20, and ratio variations will be of low amplitude. If the population sizes are sufficiently small, then Equation 22 with only one exponential term in the numerator and the denominator describes the experimental system, and one expects substantial ratio variations. Moreover, the smaller the population sizes are the larger the proportion of culture series that will be entirely overtaken by either the YFP or the CFP cells, e.g., curve 1 in Figure 6, a and b.
This corresponds to having many significant terms in Equation 20, and ratio variations will be of low amplitude. If the population sizes are sufficiently small, then Equation 22 with only one exponential term in the numerator and the denominator describes the experimental system, and one expects substantial ratio variations. Moreover, the smaller the population sizes are the larger the proportion of culture series that will be entirely overtaken by either the YFP or the CFP cells, e.g., curve 1 in Figure 6, a and b.
Measurement of bacterial characteristics from the experimental data:
As indicated in the discussion following Equation 22, the experimental data from HS are qualitatively described by a very simple model containing only one prominent mutant clone in the YFP and CFP populations. Quantitative determination of biological parameters of the bacteria based on the model is straightforward in this case. Once these parameters are obtained, they can be checked to determine if they are quantitatively consistent with the single-mutant clone description.
Estimation of  :
:
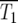 can be determined by examining the timing of the initial departures of Log10(NY/NC) from 0. T1 is the time at which
can be determined by examining the timing of the initial departures of Log10(NY/NC) from 0. T1 is the time at which 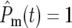 for a particular population. If this happens in the YFP population significantly prior to the CFP population, then T1 is just the time when Log10(NY/NC) = 0.3 (or −0.3 if the reverse occurs). More generally, both
for a particular population. If this happens in the YFP population significantly prior to the CFP population, then T1 is just the time when Log10(NY/NC) = 0.3 (or −0.3 if the reverse occurs). More generally, both 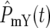 and
and 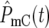 have some significant value, so that |Log10(NY/NC)| will be <0.3 at t ∼
have some significant value, so that |Log10(NY/NC)| will be <0.3 at t ∼ 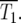 Thus a reasonable estimate for
Thus a reasonable estimate for 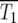 is the time when many of the experimentally measured traces of Log10(NY/NC) from a collection of culture series have significant departures from 0, but do not fully reach ±0.3. For the experimental data of HS,
is the time when many of the experimentally measured traces of Log10(NY/NC) from a collection of culture series have significant departures from 0, but do not fully reach ±0.3. For the experimental data of HS, 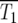 ∼ 150 generations. A more complex statistical fitting of Equation 21 to the data would produce a better estimate for
∼ 150 generations. A more complex statistical fitting of Equation 21 to the data would produce a better estimate for 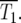 The value of such a procedure depends on the noise level of the data.
The value of such a procedure depends on the noise level of the data.
Estimation of the maximum effective selection coefficient for the mutations:
The maximum value of s can be estimated by examining the population-ratio curves for all of the experimental cultures to find the maximum slope of Log10(NY/NC) after its initial departure from 0. This typically occurs in a culture series where one population completely and rapidly overtakes the other, presumably because a mutation with a near-maximum selection coefficient had a very early effective occurrence time (or perhaps was preexisting in the population). This approach leads to an estimate of smax ∼ 0.11, using Equation 23 with one selection coefficient set equal to 0. To assist with the analysis, lines with various slopes have been added to the data figures of HS that are reproduced in the supplemental materials at http://www.genetics.org/supplemental/.
The analysis assuming a single-mutant clone is self consistent. Using the procedure in section 5 of the supplemental materials at http://www.genetics.org/supplemental/, Equation S8 estimates that there are typically 1.1 significant mutant clones in both populations. Of course, some cultures will by chance have more than one significant mutant clone in one or both populations, but as mentioned before, these cultures will typically have relatively low population-ratio deviations, and initial slopes of the curves will be relatively small compared to the others.
Estimation of the width of the effective selection coefficients:
Examination of the slopes of Log10(NY/NC) for the experimental data at times after mutants dominate both populations finds ∼32 cultures series with 0 < |sY − sC| < 0.02 and only 5 with 0.02 < |sY − sC| < 0.04. (Given the large number of curves contained in the HS figures, and the measurement noise, it is difficult to be precise in these estimates.) Therefore most of the mutants must have nearly identical selection coefficients, and these must be very close to smax given the large slope of the curves during their initial departure from 0. Thus one would estimate that most of the selection coefficients of the mutants that are significant in these experiments fall into the range of 0.09–0.11. This is just the behavior expected for a system with an effective mutation distribution described by ρ(s)W(s, t) from Equation 2. Given the strong dependence of W(s, t) on s, and the noise in the data, it is very difficult to determine much about the actual shape of ρ(s) for the bacteria. Alternate experimental designs that may reveal more details of ρ(s) are proposed in the discussion.
Estimation of the mutation rate:
Finally, assuming a uniform distribution for the selection coefficients for new mutations, Equation 10a finds that μ ∼ 10−5 if 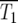 ∼ 150 generations. The numerical value obtained for μ depends on the assumed form for ρ(s). The inferred mutation rate would be much lower if a larger proportion of the weight of ρ(s) were at higher selection coefficients and would be much higher if ρ(s) were preferentially weighted toward 0. For comparison, if ρ(s) were a delta function, the estimate from Equation 10b for the mutation rate would be ∼8.5 × 10−7. Since all shapes of ρ(s) lead to a very narrow distribution of effective selection coefficients, the value of the mutation rate for the delta-function distribution is an estimate of an effective mutation rate for the system—the other mutations that may occur will have limited discernible effect on the cultures. Therefore it represents a “lower bound” estimate of the actual mutation rate for these cells.
∼ 150 generations. The numerical value obtained for μ depends on the assumed form for ρ(s). The inferred mutation rate would be much lower if a larger proportion of the weight of ρ(s) were at higher selection coefficients and would be much higher if ρ(s) were preferentially weighted toward 0. For comparison, if ρ(s) were a delta function, the estimate from Equation 10b for the mutation rate would be ∼8.5 × 10−7. Since all shapes of ρ(s) lead to a very narrow distribution of effective selection coefficients, the value of the mutation rate for the delta-function distribution is an estimate of an effective mutation rate for the system—the other mutations that may occur will have limited discernible effect on the cultures. Therefore it represents a “lower bound” estimate of the actual mutation rate for these cells.
DISCUSSION
This article has presented an analytical description of competing asexual populations that highlights the essential processes underlying their behavior. Beginning with a qualitative consideration of the competing populations (Figure 1), it introduced a hypothetical construction that established the relationship of a single large culture in unbounded exponential growth to a set of embedded sequential finite cultures that are separated by bottlenecks (Figure 2). The finite cultures are subject to stochastic variation due to the mutational processes and the random assortment of mutants at the bottlenecks, but their average behavior is identical to the “infinite” culture. Since the infinite culture is described by the standard equations of exponential growth, average characteristics of the stochastic processes could be readily calculated.
Consideration of the infinite, continuously expanding system immediately yielded the result that the distribution of selection coefficients in the mutant cells is given by ρ(s)W(s, t) (Equation 2). As time increases, W(s, t) increases more strongly with increasing s (Figure 2b) so that the distribution of effective selection coefficients becomes progressively more confined to a narrow range near the maximum s permitted by ρ(s). Since this must also be true on average for the embedded finite cultures, Equation 2 is the analytical description of the “equivalence principle” that HS found through extensive numerical simulation.
The qualitative description of the behavior of a series of finite cultures (Figure 1b) highlighted the importance of the times T1, when the mutant and ancestral populations are of equal magnitude, for determining when discernible variations in the population ratios may begin to occur in competition experiments. Analysis of the infinite culture also allowed determination of the characteristic value for this time, 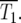 Thus the distribution of selection coefficients that are prominent in the mutant populations, and the characteristic times at which ratio variations might occur in competition experiments, could be calculated without consideration of the details of the stochastic processes.
Thus the distribution of selection coefficients that are prominent in the mutant populations, and the characteristic times at which ratio variations might occur in competition experiments, could be calculated without consideration of the details of the stochastic processes.
The stochastic aspects of the cultures were addressed by analyzing the fate of the progeny of a single mutation, again considering a hypothetical experiment in which all of the cells in a culture are transferred to daughter cultures (Figure 4). This analysis demonstrated that after a sufficient number of bottlenecks, the progeny of a mutated cell expand in a lineage of daughter cultures as a simple exponential, but with an apparent mutation time that is shifted from the actual mutation time according to readily calculable statistical distributions (Equations 15 and 19 and Figure 5). Summing over multiple mutants provided a complete description of the system (Equation 20).
Full evaluation of Equation 20 requires substantial effort. However, the results of the average value calculations, specifically those for 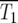 and the demonstration of the narrow range of effective selection coefficients, and the restricted range of effective mutation times derived from the statistical analysis, permitted a qualitative understanding of the basic behavior of Equation 20. These considerations lead to a dramatically simplified single-clone (or several-clone) description of a system of competing populations (Equation 22), which is applicable to the experimental regime where variations in the population ratios are prominent. Thus the interplay between a detailed statistical analysis and simple calculations of average values of various parameters for infinite populations leads to a tractable analytical description of competition experiments.
and the demonstration of the narrow range of effective selection coefficients, and the restricted range of effective mutation times derived from the statistical analysis, permitted a qualitative understanding of the basic behavior of Equation 20. These considerations lead to a dramatically simplified single-clone (or several-clone) description of a system of competing populations (Equation 22), which is applicable to the experimental regime where variations in the population ratios are prominent. Thus the interplay between a detailed statistical analysis and simple calculations of average values of various parameters for infinite populations leads to a tractable analytical description of competition experiments.
The analysis of experimental data to extract quantitative estimates of parameters of the mutational process is straightforward if the population sizes are small enough so that only a single exponential term is required in Equation 22 to represent the mutant cells. The maximum slope that the population ratio achieves after its initial departure from horizontal gives an estimate of the maximum value of the selection coefficient available to the mutant cells, and the average time that nonzero slopes appear allows an estimate of 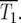 If the populations sizes are intermediate, so that ratio changes are observable but progeny of multiple mutations contribute, then the maximum deviations in the ratios are reduced since mutant clones become significant in both populations at about the same time and with about the same values of selection coefficients. In this case extraction of the biological parameters from the data would require a more complex statistical fitting process.
If the populations sizes are intermediate, so that ratio changes are observable but progeny of multiple mutations contribute, then the maximum deviations in the ratios are reduced since mutant clones become significant in both populations at about the same time and with about the same values of selection coefficients. In this case extraction of the biological parameters from the data would require a more complex statistical fitting process.
Application of the single-mutant clone version of Equation 22 to the experimental data of HS estimates that the maximum selection coefficient of the mutations is s ∼ 0.11 and that the mutation rate is μ = 10−5 or 8.5 × 10−7, depending on whether one assumes that ρ(s) is uniform or a delta function. By contrast, HS found s ∼ 0.054 and μ ∼ 10−6.7 = 2 × 10−7 when using their procedures to fit a delta-function model to their data (their Figure 3B). The reasons for the considerable differences in the estimates produced by the two modeling approaches require elucidation.
One of the benefits of the analytical approach taken here is that it reveals general relationships that may be obscured in more complex computational analyses. For example, exponential fitness distributions, ρ(s) = (1/k)e−ks, have been postulated as being relevant to real biological systems (Orr 2003). However, if the expected behavior of a culture with such a distribution is calculated using Equation 3, one finds that at times t > k/r the product ρ(s)W(s, t) is unbounded with increasing s. Thus the fraction of mutant cells becomes infinite at finite times. This is a physically unreasonable result and indicates that in real systems the fitness distribution must go to 0 more rapidly than exponentially. An exponential truncated at some maximum s has the desired properties, and indeed truncation is assumed in the distributions proposed by Orr (2003).
The information concerning the shape of ρ(s) that can be obtained from the HS measurements is limited because the experimental behavior is so strongly dominated by the limited range of selection coefficients defined by ρ(s)W(s, t). Altering aspects of the experimental design may allow one to obtain more information. For example, one could substantially reduce the population sizes in the experiment and correspondingly increase the number of culture series. Such a change would not affect the characteristic time,  at which the mutant populations will become significant, but would dramatically increase the variation in the T1's for the YFP and CFP populations in each culture series. This would increase the fraction of culture series in which one population completely displaces the other, e.g., the curves labeled “1” in Figure 6, a and b. These displacements are predominantly due to single-mutant clones, and determination of the frequency with which different slopes are obtained during the displacement process provides a direct measure of the shape of ρ(s). This design should allow measurement of selection coefficients that are significantly below the maximum since the small population size would give these mutants a greater chance of becoming dominant prior to appearance of another mutant clone with larger s. The smaller the population size is, the lower the values of s that could be addressed in such an experiment, but the larger the number of series of cultures that would be needed.
at which the mutant populations will become significant, but would dramatically increase the variation in the T1's for the YFP and CFP populations in each culture series. This would increase the fraction of culture series in which one population completely displaces the other, e.g., the curves labeled “1” in Figure 6, a and b. These displacements are predominantly due to single-mutant clones, and determination of the frequency with which different slopes are obtained during the displacement process provides a direct measure of the shape of ρ(s). This design should allow measurement of selection coefficients that are significantly below the maximum since the small population size would give these mutants a greater chance of becoming dominant prior to appearance of another mutant clone with larger s. The smaller the population size is, the lower the values of s that could be addressed in such an experiment, but the larger the number of series of cultures that would be needed.
Another possible approach would be to perform single-cell comparative growth experiments on a massive scale. This could be done by first growing several series of large cultures with all bacteria containing the same label, say YFP, for a time on the order of  Approximately half of the cells in these mass cultures would be mutant, with the mutant population distributed in s according ρ(s)W(s, t). Growing the cultures for different times would provide different proportions of mutant and ancestral cells and somewhat different frequency distributions given the change in W(s, t) with time. Shorter growth times would yield a broader distribution of selection coefficients for the mutants but a greater proportion of the cells would be normal, while longer times would produce a greater proportion of mutant cells but a narrower frequency distribution for s. Selection of a multitude of single YFP cells from the mass cultures and individually comparing their growth rates to normal CFP-labeled cells would allow measurement of the shape of ρ(s)W(s, Tm), where Tm is the length of the initial mass culture. Division by W(s, Tm) would provide a determination of ρ(s). The range of s for which a reasonable estimate could be obtained would depend on the number of mutants that are measured and the accuracy of the measurements.
Approximately half of the cells in these mass cultures would be mutant, with the mutant population distributed in s according ρ(s)W(s, t). Growing the cultures for different times would provide different proportions of mutant and ancestral cells and somewhat different frequency distributions given the change in W(s, t) with time. Shorter growth times would yield a broader distribution of selection coefficients for the mutants but a greater proportion of the cells would be normal, while longer times would produce a greater proportion of mutant cells but a narrower frequency distribution for s. Selection of a multitude of single YFP cells from the mass cultures and individually comparing their growth rates to normal CFP-labeled cells would allow measurement of the shape of ρ(s)W(s, Tm), where Tm is the length of the initial mass culture. Division by W(s, Tm) would provide a determination of ρ(s). The range of s for which a reasonable estimate could be obtained would depend on the number of mutants that are measured and the accuracy of the measurements.
The better the experimental means of measuring the two cell populations is, the shorter the culture time required for the measurement and the more accurate the result. Thus high-sensitivity fluorescence monitoring of growth in microtiter plates, or single-cell analytical procedures such as flow cytometry or fluorescence microscopy, or perhaps highly parallel microfluidic measuring systems, would be beneficial. Ideally one might add one YPF cell to one ancestral CFP cell and determine the relative numbers at several time points during a single growth phase without the introduction of any bottlenecks. However, it is important to note that such an approach would measure the selection coefficients under conditions different from those of the original selection. Specifically, if a mutation provided its advantage during the initial mass culture by increasing the ability of cells to divide as the culture enters and/or leaves stationary phase, then that advantage will not be properly quantified.
In conclusion, the analytical approach presented here allows a qualitative and quantitative interpretation of the behavior of many aspects of competing asexual populations without the need to perform detailed numerical simulations. Analytical expressions for the narrowing of the range of effective selection coefficients and for various characteristic times during the adaptation of a population to new selective conditions are presented. The stochastic behavior of the population is accurately described, and the critical aspects of the description are highlighted to allow a simplified quantitative interpretation of experimental data. This analysis may be useful in considering alternate experimental designs that enhance probing of specific aspects of the adaptation process.
References
- Hegreness, M., N. Shoresh, D. Hartl and R. Kishony, 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]