Abstract
Several lines of evidence suggest that, within a lineage, particular genomic regions are subject to instability that can lead to specific types of chromosome rearrangements important in species incompatibility. Within family Macropodidae (kangaroos, wallabies, bettongs, and potoroos), which exhibit recent and extensive karyotypic evolution, rearrangements involve chiefly the centromere. We propose that centromeres are the primary target for destabilization in cases of genomic instability, such as interspecific hybridization, and participate in the formation of novel chromosome rearrangements. Here we use standard cytological staining, cross-species chromosome painting, DNA probe analyses, and scanning electron microscopy to examine four interspecific macropodid hybrids (Macropus rufogriseus × Macropus agilis). The parental complements share the same centric fusions relative to the presumed macropodid ancestral karyotype, but can be differentiated on the basis of heterochromatic content, M. rufogriseus having larger centromeres with large C-banding positive regions. All hybrids exhibited the same pattern of chromosomal instability and remodeling specifically within the centromeres derived from the maternal (M. rufogriseus) complement. This instability included amplification of a satellite repeat and a transposable element, changes in chromatin structure, and de novo whole-arm rearrangements. We discuss possible reasons and mechanisms for the centromeric instability and remodeling observed in all four macropodid hybrids.
IT has been noted since the 1970s that chromosome rearrangements within some plant and animal lineages are nonrandom. For example, within the primate lineage, fissions predominate within the Old World monkeys, pericentric inversions within the great apes and Robertsonian translocations within the lemurs (Dutrillaux 1979). In the human lineage, there is a striking correspondence between the position of evolutionary breakpoints conserved in mammals, human fragile site locations, and the distribution of tandem repeats (Ruiz-Herrera et al. 2006). Numerous studies (e.g., McClintock 1987 for Zea spp.) have shown that multiple chromosomal rearrangements of the same type can occur in different individuals simultaneously (reviewed in King 1993). These data suggest that “hot spots” in the genome are predisposed to instability and may be subject to genomic rearrangements.
The family Macropodidae exhibits recent and extensive chromosome evolution, in contrast with most other marsupials, which are generally karyotypically conservative (reviewed in Hayman 1990; Eldridge and Metcalfe 2006). Chromosome evolution within macropodids has been extensively studied; the majority of macropodids have been karyotyped and the evolutionary trajectory of chromosome rearrangements has been determined (Hayman 1990; Eldridge and Close 1993; Bulazel et al. 2007). Within the genus Macropus (kangaroos and wallabies), rearrangements involving the centromere predominate, in particular centric fusions (Rofe 1978; Hayman 1990) and whole-arm reciprocal translocations (WARTs) (reviewed in O'Neill et al. 2004). However, the mechanism responsible for the rapid karyotypic diversification within this group of mammals has not been fully explored.
Instances of rapid genomic change can result from an increased mutation rate caused by genome destabilizing events, such as inter- and/or intraspecific hybridization or exposure to environmental mutagens and stress (Fontdevila 1992). This increase in mutation rate has been observed to coincide with an increase in the local activity of transposable elements, retroelements, and other repeated DNAs (see Lim and Simmons 1994; O'Neill et al. 1998; Labrador et al. 1999; Fontdevila 2005; Ungerer et al. 2006). In addition to a bias in sequence classes involved in rearrangement, extensive genome sequence comparisons and comparative cytogenetics now point to specific chromosome features, such as centromeres, that can also influence the number, position, and type of rearrangement.
Our previous work in macropodid hybrids showed that a retroviral sequence located at the centromere had undergone demethylation and amplification, concomitant with chromosome remodeling (O'Neill et al. 1998). Subsequent analyses using cross-species chromosome painting of four other macropodid hybrid individuals (hybrids) showed that the rearrangements observed in these genomes were also restricted to centromeres (O'Neill et al. 2001).
In our previous work, it was not determined whether the observed rearrangements were shared in other hybrids of the same type or whether the rearrangements were the result of interchromosomal segmental duplications of centromeric sequences, non-allelic recombination between sequences at centromeres on different chromosomes, or whether they resulted from the transposition and/or amplification of mobile DNA or other repeated DNAs. In this study, the genomes of four interspecific hybrids from a cross between two macropodid species not previously studied, Macropus rufogriseus (maternal component) and Macropus agilis (paternal component), were assayed for chromosome rearrangements and genomic instability using standard cytological staining techniques, cross-species chromosome painting, DNA probe analyses, as well as ultrastructural analyses of centromeres via scanning electron microscopy.
These data show that the centromere is a significant contributing factor in chromosome aberration in all of the hybrid genomes examined. The current analyses show that, in these hybrids, some centromeres differ structurally from parental chromosomes and are the site of extensive genome rearrangements accompanied by DNA amplification of retroelement sequences and satellites. These rearrangements include a broad array of abnormalities typical of genomic instability, including fissions, isochromosomes, whole-arm reciprocal translocations, and minichromosomes. Remarkably, rearrangements were found only within the maternal complement and all were associated with the maternally derived centromeres. This study extends previous work and, for the first time, clearly defines the centromere as the site of genomic instability, anomalous chromosome structures, and structural variants.
MATERIALS AND METHODS
Animals and karyotypes:
Five M. rufogriseus × M. agilis hybrids were available. RA1190 is a normal XY male, and RA1122 and “new” RA are normal XX females. RA1118 is a XX animal with no pouch or penis and a small, empty but well-developed scrotum. RAX0 was a female with no pouch, a small, empty scrotum, rudimentary female reproductive tract, and large amounts of fat in the body cavity.
Three normal parental animals (A1843, R1188, and R3242) were examined. The male M. agilis (A1843) is the father of all hybrids. The M. rufogriseus animal, R1188, is the mother of RA1188. The mother of the remaining hybrids is from the same population as R1188 but could not be identified by microsatellite analysis (data not shown). An unrelated M. rufogriseus male (R3242) was also examined.
Cell culture:
Ear biopsies were obtained from animals held at Macquarie University, Fauna Park. Primary fibroblast cultures were established from ear biopsies and maintained using standard techniques (Cooper et al. 1977). Cultures were passaged no more than 10 times, with the exception of R3242 (>15 passages). Metaphases were prepared using standard techniques from all cultures. Cytogenetic techniques were performed on metaphases from the three parental animals (A1843, R1188, and R3242) and four hybrids (RA1190, RA1122, RA1188, and “new” RA).
C-banding:
Metaphases from each species were C banded using the technique of Sumner (1972) as modified by Eldridge (1991).
Scanning electron microscopy:
Chromosomes were isolated according to the “drop/cryo” technique (for details see Martin et al. 1994). Briefly, cell suspensions as per standard chromosome preparations were dropped onto cold laser-marked slides (Laser Marking, Fischen, Germany). Just prior to fixative evaporation, 20 μl of 45% acetic acid was applied, and the specimens were covered with a cover slip and frozen upside down on dry ice for 15 min. The cover slip was pried off, and specimens were immediately transferred into fixative (2.5% glutaraldehyde, 75 mm cacodylate buffer, 2 mm MgCl2, pH 7.0).
For DNA staining, chromosomes were incubated for 30 min at room temperature with platinum blue ([CH3CN]2Pt oligomer, 10 mm, pH 7.2) and washed with distilled water and then with 100% acetone prior to critical point drying (for details see Wanner and Formanek 1995).
Specimens were washed in distilled water, dehydrated in 100% acetone, and critical point dried from CO2. Slides were mounted onto stubs and carbon coated by evaporation. Specimens were examined at accelerating voltages from 15 to 30 kV with an Hitachi S-4100 field emission scanning electron microscope equipped with a yttrium aluminum garnet (YAG)-type backscattered electron (BSE) detector (Autrata). Secondary electron and BSE images were recorded simultaneously with Digiscan hardware (Gatan).
Preparation of chromosome paints and fluorescence in situ hybridization:
Flow sorting and chromosome paint production of Petrogale xanthopus (2n = 22 karyotype) chromosome paints were performed according to the protocol previously described (Rens et al. 1999). Highly pure chromosome samples were collected, with the exception of the Y chromosome, which was too small to be collected. Pools of individually isolated chromosome DNA were subjected to degenerate oligo primed (DOP)–PCR amplification for incorporation of either biotin– or digoxigenin (DIG)–dUTP (Roche) as per the manufacturer's instructions. Cross-species chromosome painting was performed as described by O'Neill et al. (1999) with the following modifications. P. xanthopus genomic DNA was sonicated to between 200 and 500 bp and was used for suppression during a preannealing step prior to hybridization. Biotin-labeled probes were detected using avidin–fluorescein-5-isothiocyanate (FITC) (Vector Labs) and digoxigenin probes were detected with antidigoxigenin rhodamine (Roche). Metaphases were captured on an Olympus AX70 microscope equipped with a Photometrics Sensys CCD camera and analyzed using the Genus Cytovision software (Applied Imaging).
Five chromosome paint “sets” were used corresponding to P. xanthopus chromosomes 1 and 10; 6, 9, and 7; 5 and 8; 3 and 4; and 2 and the X chromosome. The five paint sets correspond to M. rufogriseus and M. agilis chromosomes 1p and 1q; 6p, 6q and 7; 3p and 3q; 4 and 5; and chromosome 2 and the X chromosome, respectively. A total of 3094 metaphases were scored, 376–479 for each parent and hybrid individual. Cross-species chromosome painting was also performed and subject to analysis on an unrelated M. rufogriseus male to determine if observed hybrid-specific rearrangements were found within the parental species.
Dot-blot analyses of sat23 and KERV:
Dot-blot analyses were performed on each parent and hybrid animal, and serial dilutions of DNA concentrations ranging from 500 to 0.005 ng/μl were denatured for 5 min at 99°, snap cooled, and spotted onto Hybond N+ (Amersham, Piscataway, NJ) membrane and fixed according to the manufacturer's protocol. Blots were prehybridized and hybridized according to Ferreri et al. (2005) at 65° overnight, washed in 1× SSC, 0.1% SDS at 65°, and exposed to film overnight at −80°. Cytb probe was produced by PCR as per Bulazel et al. (2007) and hybridized as for sat23 and KERV.
Fluorescence in situ hybridization (sat23 and KERV):
The sat23 (pGEM-T vector containing 1.5 copies of the 178-bp satellite) and KERV-1 (pGEM-T vector containing 1.5 kb spanning the gag-pro-pol region of the retrovirus) probes were labeled with biotin–dUTP and digoxigenin–dUTP, respectively, through PCR amplification of insert DNA using 100 ng each of the plasmid primers SP6 and T7. PCR products were cleaned with a microcon 100, and 2 μl (of 10 μl) was resuspended in 8 μl of hybridization buffer (50% formamide, 10% dextran sulfate, 50 mm Na2HP04, 2× SSC) and denatured at 80° for 10 min. Following denaturation in 70% formamide/2× SSC at 70° for 2 min and processing through a serial ethanol series at −20°, slides were hybridized in the presence of probe overnight at 37°. For metaphases from hybrids, post-hybridization washes were performed as per Bulazel et al. (2006). For metaphases from M. agilis, low-stringency post-hybridization washes (three washes of 50% formamide/2× SSC for 5 min each; three washes of 2× SSC for 5 min each) were performed at room temperature. The biotinylated probe was detected with antibiotin rhodamine (Molecular Probes, Eugene, OR) and DIG-labeled probe was detected with FITC-anti-DIG antibody (Molecular Probes). Metaphases were counter stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were captured as described above.
RESULTS
Karyotypes:
A 2n = 22 karyotype is considered ancestral for macropodids (kangaroos and wallabies) since a homologous complement is found in representatives of four genera (Petrogale, Thylogale, Setonix, Dorcopsis) and the karyotypes of all other macropodid species can be readily derived from this 2n = 22 karyotype, principally via centric fusions (Eldridge and Metcalfe 2006). The karyotype of M. rufogriseus shares a suite of three centric fusions with two other Macropus species (Rofe 1978). M. agilis is presumed to also have the same complement, but only on the basis of conventional staining (Sharman 1961). Chromosome painting confirms that the M. agilis complement does have the same suite of chromosome rearrangements (data not shown).
Four hybrids produced from a cross between M. agilis and M. rufogriseus were confirmed as interspecific offspring on the basis of cytological examination of chromosome complements. In each case, the maternal and paternal complements are readily identifiable by X chromosome morphology differences and the marked centromere length differences for all chromosomes (Lowry et al. 1995; Bulazel et al. 2006).
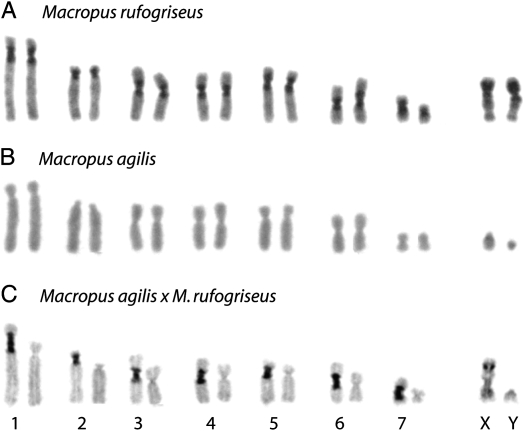
Previous studies of marsupial hybrids indicated a predominance of centromere-associated rearrangements within macropodid hybrids (O'Neill et al. 1998, 2001). To examine gross constitutive heterochromatin amounts at the centromeres of hybrids in this study, C-banding was performed. C-banding is a technique that specifically stains blocks of constitutive heterochromatin, typically at centromere locations. C-banding patterns (dark-staining regions) are markedly different between each parental animal (Figure 1, A–C), with extensive centromeric heterochromatin in M. rufogriseus and little detectable centromeric heterochromatin in M. agilis. The origin of each homologous chromosome in the hybrids was clearly identifiable by its C-banding pattern; the M. rufogriseus-derived autosomes within the hybrids are clearly identified as the chromosomes with extended, dark-staining centromere regions.
Figure 1.—
C-band karyotype of metaphase chromosomes from (A) M. rufogriseus, (B) M. agilis, and (C) M. rufogriseus × M. agilis (RA1120). Chromosome numbers are indicated below C.
Ultrastructural analyses:
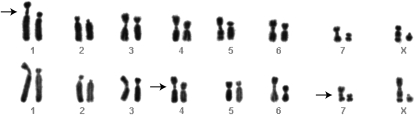
In preliminary fluorescent microscopic (DAPI) investigation of the chromosome morphology within the hybrid genomes, some centromeres appear elongated and segmented (Figure 2). In contrast to maternal centromeres, some hybrid centromeres appear to have two constrictions, lending the central chromatin a “knob-like” appearance (Figure 2). For structural analysis with higher resolution, hybrid chromosomes were investigated with scanning electron microscopy (SEM). Specific DNA staining for SEM with platinum blue (Pt blue) revealed that the DNA distribution is uneven along these hybrid chromosomes (Figure 3, A and B). DNA distribution in the centromere is similar, if narrower, to that of the chromosome arms. In the secondary electron (SE) image, chromatids are hardly distinguishable, but in the BSE images they are clearly distinguishable by a longitudinal “signal-free” space (Figure 3B). However, at the centromere the chromatids are joined in an area of compact chromatin, as indicated by an unseparated signal in the BSE image (Figure 3B). Chromosomes exhibited a generally loosened chromatin structure; parallel fibers and chromomeres are visible throughout the whole chromosome (Figure 3B).
Figure 2.—
Abnormal centromere structures in M. rufogriseus × M. agilis hybrids. DAPI-stained metaphase chromosomes from two cells of M. rufogriseus × M. agilis (RA1120) highlight the abnormal centromere structures of the M. rufogriseus-derived chromosomes (arrows indicate the “knob-like” structures) detected by gross karyotype analyses.
Figure 3.—
Scanning electron micrographs of M. rufogriseus × M. agilis hybrid chromosomes stained specifically for DNA with platinum blue. Some centromeres are elongated and exhibit an uneven DNA distribution similar, if narrower, to that on the chromosome arms (A). Parallel fibers and chromomeres are distributed throughout the chromosome, including the centromere (B, the SE image). DNA distribution shows distinguishable chromatids joined at a small region in the centromere (B, the BSE image); DNA distribution is uninterrupted longitudinally along the chromatid arms (B, the BSE image).
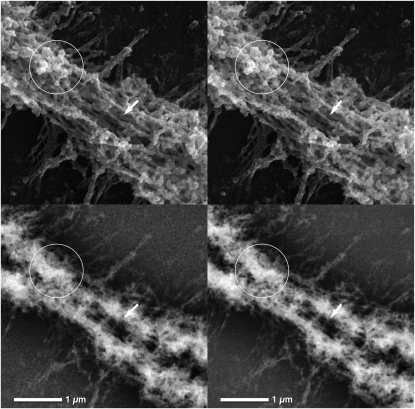
Three-dimensional ultrastructural analysis of elongated centromeres with high-resolution SEM showed that parallel fibers are interspersed with chromomeres of varying sizes (Figure 4). This diverges slightly from the centromere ultrastructure observed in other centric chromosomes in plants and animals, for which the centromere is characterized by exposed parallel matrix fibers (Wanner and Formanek 1995, 2000; Sumner 1998). Platinum blue staining revealed that DNA is rather heterogeneously distributed along the hybrid chromosomes in a network of varying local concentrations (Figure 4). Strong BSE signal regions correspond with chromomeres and compact structural regions (Figure 4). Centric chromosomes typically show distinctly less DNA content at the centromere, indicated by a significantly weaker BSE signal at the primary constriction with respect to the chromosome arms (Wanner and Formanek 2000). The DNA distribution for the hybrid chromosomes with elongated centromeres, however, is uninterrupted on each chromatid; the strong Pt blue signal reflects a high chromatin density in these chromosomal regions.
Figure 4.—
High-resolution stereo micrograph pairs of a M. rufogriseus × M. agilis hybrid chromosome centromere stained with platinum blue. (Top) SE images showing centromere topography. (Bottom) BSE images showing DNA distribution. Stereo viewing allows recognition of the spatial distribution of structural elements: chromomeres of varying sizes (top, circles) are interspersed with parallel fibers (top, arrows). Chromomeres (top, circles) correspond with local increases in DNA concentration (bottom, circles).
Centromere sequences in hybrid genomes:
We have previously isolated two sequences found specifically at active centromere locations within Macropus. The first sequence, sat23, is a 178-bp satellite that is capable of binding the centromere protein CENP-B both in vitro and in vivo and most likely represents the primary satellite dictating centromere function within this group of mammals (Bulazel et al. 2006). The second, KERV-1, is a retrovirus that is found in the genomes of a wide range of marsupials (Ferreri et al. 2005). It is found at higher copy number at the active centromere (O'Neill et al. 1998; Ferreri et al. 2004) as well as in low copy number at breaks of synteny between the conserved chromosome segments within marsupials (Ferreri et al. 2004).
To test for a correlation between the copy numbers of these two centromere-predominant sequences and the abnormally extended centromeres observed within the hybrid genomes, dot-blot analyses were performed to assay for copy-number variance in relation to the parent from which the maternal complement was derived (M. rufogriseus). The titration dilution used in this assay was necessarily low due to the extreme high copy number of these sequences at the centromeres within M. rufogriseus (Bulazel et al. 2006) compared with M. agilis. Thus, as M. agilis carries both KERV-1 and sat23 at its centromeres in significantly lower copy number than M. rufogriseus (Bulazel et al. 2006, 2007), all copies in the former species were essentially diluted to nondetectable levels in this assay. Dot-blot hybridization levels were normalized with cytB and intensities were examined on the Bio-Rad (Hercules, CA) Gel Doc EQ, Quantity One software package. Table 1 shows that each hybrid exhibited an average approximately twofold increase over the M. rufogriseus parent in copy number of both sat23 and KERV-1 sequences.
TABLE 1.
Copy number quantitation of centromere sequences: dot-blot analyses of sat23 and KERV-1 in M. rufogriseus, M. agilis, and M. rufogriseus × M. agilis hybrids
| sat23
|
KERV-1
|
|||
|---|---|---|---|---|
| Normalized value | Fold change | Normalized value | Fold change | |
| Parentals | ||||
| M. rufogriseus (R1188) | 1892.8 | 2323.3 | ||
| M. agilis (A1843) | 0 | 0 | ||
| Hybrids | ||||
| RA 1190 | 2398.5 | 2.534 | 2850.7 | 2.454 |
| RA 1122 | 1887.9 | 1.995 | 1217.2 | 1.048 |
| RA 1118 | 2327.3 | 2.459 | 2067.2 | 1.779 |
| New RA | 2346.5 | 2.479 | 2533 | 2.181 |
| Average hybrids | 2.37 | 1.87 | ||
Normalized values and fold change compared to M. rufogriseus are indicated.
The most likely location of the hybrid-specific amplified copies is within the centromeres of the M. rufogriseus complement as no homogenously staining regions outside of the active centromere were observed within the hybrid karyotypes. This was confirmed by fluorescence in situ hybridization of both DNA probes (sat23 and KERV-1) to metaphase chromosomes of each hybrid (Figure 5A). On hybrid metaphases, sat23 was observed at all of the centromeres of M. rufogriseus-derived chromosomes, while KERV-1 was observed at all of the centromeres of M. rufogriseus-derived chromosomes, albeit in virtually undetectable copy numbers on the X chromosome. Interestingly, a subset of cells in each hybrid displayed minichromosomes containing sat23 DNA (Figure 5B). It is not known whether these are capable of forming kinetochores or are capable of stable mitotic division.
Figure 5.—
sat23 and KERV-1 FISH to metaphase chromosomes of M. rufogriseus × M. agilis hybrids. (A) Hybridization (from left) of sat23 (red), KERV-1 (green) to metaphase chromosomes (inverted DAPI) of RA1118 (merge). (B) Minichromosome from “new RA” containing both sat23 (red) and KERV-1 (green) DNA (merge).
Due to the extremely low copy number for both sat23 and KERV-1 at the centromeres of M. agilis (Bulazel et al. 2007), low stringency conditions were required to detect these sequences in the hybrid genomes on the paternal complement (data not shown). These hybridization conditions resulted in spurious signal throughout both complements and were thus uninformative.
Cross-species chromosome painting:
To test whether there were other structural abnormalities present within the karyotypes of these hybrids, cross-species chromosome painting was performed using two-color fluorescence in situ hybridization and dual-peak localization of chromosome paints derived from P. xanthopus. This species was used for cross-species analyses as it carries the ancestral macropodid karyotype (Eldridge et al. 1991, 1992). Each paint probe was used in a two-probe hybridization to delineate de novo rearrangements in each parent, each of the four hybrids, and an unrelated male M. rufogriseus cell line that had undergone a high number of passages.
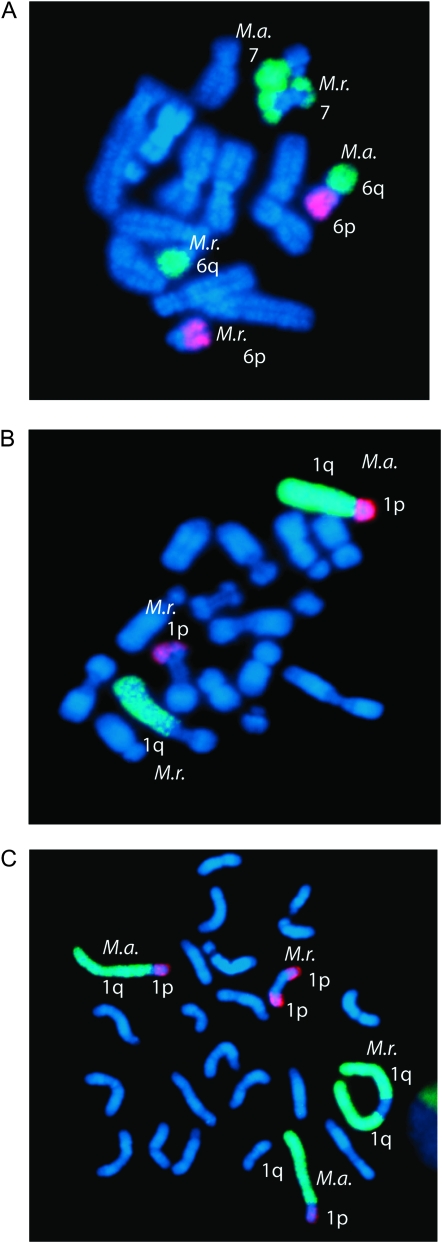
No rearrangements for any chromosome paint pair were observed in either of the two parental animals or in the unrelated M. rufogriseus male (Table 2A). In contrast, each hybrid carried a mosaic suite of rearrangements at varying frequencies (Table 2B), with most rearrangements classified as unique (Table 2C). Table 2 lists the frequency of each observed rearrangement by chromosome paint pair. A χ2 test was used to determine whether the number of rearrangements observed in the hybrids was statistically significant. The number of rearrangements was found to be significantly different in hybrids compared with the parents (χ2 = 21, d.f. = 1, P = 3.214 × 10−6). The M. rufogriseus-derived chromosome in the hybrids was easily identified both by inverse DAPI, where the M. rufogriseus chromosome showed extensive staining at the centromere, and by a lack of hybridization of the chromosome paints at the M. rufogriseus-derived centromere. Each of the observed rearrangements in the hybrids involved the M. rufogriseus-derived centromere and included isochromosomes, WARTs, and fissions (Figure 6). However, each hybrid cell line was mosaic for cells with abnormal chromosomes derived from independent rearrangement events (i.e., de novo for each cell) and karyotypically normal cells. While only one tissue (fibroblast) was available for this study, this level of mosaicism within one tissue type implies that the instability may manifest late in development and is likely continuing at a low frequency.
TABLE 2.
Summary of chromosome rearrangements in M. rufogriseus × M. agilis hybrids
| Chromosome 1p and 1q
|
Chromosome 6p, 6q 7
|
Chromosome 3p and 3q
|
Chromosome 4 and 5
|
Chromosome 2 and X
|
Totals
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scored | Abnormal | Scored | Abnormal | Scored | Abnormal | Scored | Abnormal | Scored | Abnormal | Scored | Abnormal | |
| A. Parentals | ||||||||||||
| R1188 | 93 | 0 | 91 | 0 | 100 | 0 | 100 | 0 | 79 | 0 | 463 | 0 |
| A1843 | 100 | 0 | 93 | 0 | 107 | 0 | 50 | 0 | 101 | 0 | 451 | 0 |
| R3242 | 86 | 0 | 74 | 0 | 100 | 0 | 55 | 0 | 100 | 0 | 415 | 0 |
| Totals | 279 | 0 | 258 | 0 | 307 | 0 | 205 | 0 | 280 | 0 | 1329 | 0 |
| Grand total | ||||||||||||
| Scored | 1329 | |||||||||||
| Rearranged | 0 | |||||||||||
| B. Hybrids—all rearrangements | ||||||||||||
| RA1118 | 104 | 0 | 85 | 0 | 98 | 0 | 142 | 3 | 50 | 0 | 479 | 3 |
| RA1122 A | 102 | 9 | 91 | 4 | 100 | 0 | 88 | 0 | 82 | 1 | 463 | 14 |
| RA1190 | 102 | 2 | 87 | 0 | 79 | 0 | 79 | 0 | 100 | 0 | 447 | 2 |
| “New” RA | 89 | 0 | 80 | 6 | 70 | 0 | 100 | 2 | 37 | 2 | 376 | 10 |
| Totals | 397 | 11 | 343 | 10 | 347 | 0 | 409 | 5 | 269 | 3 | 1765 | 29 |
| Grand total | ||||||||||||
| Scored | 1765 | |||||||||||
| Rearranged | 29 | |||||||||||
| C. Hybrids—unique rearrangements only | ||||||||||||
| RA1118 | 104 | 0 | 85 | 0 | 98 | 0 | 142 | 3 | 50 | 0 | 479 | 3 |
| RA1122 A | 102 | 4 | 91 | 3 | 100 | 0 | 88 | 0 | 82 | 1 | 463 | 8 |
| RA1190 | 102 | 2 | 87 | 0 | 79 | 0 | 79 | 0 | 100 | 0 | 447 | 2 |
| “New” RA | 89 | 0 | 80 | 1 | 70 | 0 | 100 | 2 | 37 | 0 | 376 | 3 |
| Totals | 397 | 6 | 343 | 4 | 347 | 0 | 409 | 5 | 269 | 1 | 1765 | 16 |
| Grand total | ||||||||||||
| Scored | 1765 | |||||||||||
| Rearranged | 16 | |||||||||||
Scored cells containing abnormal (i.e., rearranged) chromosomes in both parents (R1188, A1843) and a high passage cell line for M. rufogriseus (R3242) (A) and four M. rufogriseus × M. agilis hybrids (as indicated) (B and C). Hybrid data are divided into the total number of rearrangements observed (B) and those that are unique (i.e., different types of de novo rearrangements within a particular hybrid). Each chromosome or chromosome pair targeted by chromosome paints from P. xanthopus are indicated. Minichromosomes were not scored. All rearrangements observed in the hybrids involved the centromere of the M. rufogriesus-derived chromosome (see results).
Figure 6.—
De novo chromosome aberrations in M. rufogriseus × M. agilis hybrids. Examples of chromosome rearrangements identified through cross-species chromosome painting in RA1122: (A) fission, (B) translocation, and (C) isochromosome. The parental origin is labeled as M.r. (M. rufogriseus) and M.a. (M. agilis).
DISCUSSION
Modified centromeres are correlated with chromosome aberrations:
The most remarkable feature of the chromosome instability manifested in all four M. rufogriseus × M. agilis marsupial hybrids is that the instability was confined to the centromeres of the maternally derived, i.e., M. rufogriseus, chromosomes. While the hybrids that we examined were of the same type of cross and generated from the same M. agilis male parent, they did not share the same M. rufogriseus female parent, excluding parental instability as a contributing factor to the chromosome aberrations observed in the hybrid genomes. All hybrids displayed a low frequency of de novo rearrangements involving a M. rufogriseus-derived centromere, specifically with chromosome arms of the M. rufogriseus complement. While we cannot exclude the possibility that the observed instability may be due partially to a maternal effect, we propose that this instability may also be an inherent feature of the structure of the M. rufogriseus centromere in a hybrid or otherwise destabilizing environment.
The increased frequency of chromosome rearrangements in these hybrids is specific only to the centromere that within the parental species (M. rufgogriseus) contains higher copy numbers of both centromeric sequences, sat23 and KERV, as determined by fluorescence in situ hybridization (FISH) (Bulazel et al. 2006, 2007) (Figure 5) and dot-blot analyses (Table 1). These rearrangements consist of a broad spectrum of karyotypic instabilities, including WARTs, fissions, isochromosomes, and minichromosomes. FISH localization of both the KERV-1 element and the sat23 satellite repeat suggests that, in the hybrids, amplification has occurred at the M. rufogriseus-derived centromere, supporting the SEM data. The amplification of KERV-1 in all four of these hybrids is analogous to that previously observed in another macropodid hybrid (O'Neill et al. 1998) and implies a general mechanism for genomic instability involving retroelement amplification in marsupial hybrid karyotypic instability specific to centromere locations.
Transposable elements (TEs) have been shown to be directly responsible for chromosome rearrangements in both plants and insects (Zhang and Peterson 1999; Evgen'ev et al. 2000; Caceres et al. 2001). In Drosophila, for example, there is a high correlation between the presence of a particular TE and chromosome rearrangements (Lim 1988; Evgen'ev et al. 2000). A more detailed study of genome rearrangements in maize showed that a duplication/deletion event can be produced by a single transposition event involving a full-length TE and a TE fragment (Zhang and Peterson 1999). However, there are fewer examples of direct involvement of TEs in mammalian genome remodeling. Inversions in primates are known to be induced by recombination of some transposable elements (Schwartz et al. 1998; Kehrer-Sawatzki et al. 2002). Gerbils of the genus Taterillus have undergone rapid and extensive chromosome repatterning with concomitant amplification of LINE-1 elements. A clear lack of correlation between LINE-1 accumulation and chromosomal breakpoints, however, suggests that these elements were not directly involved in the chromosomal changes (Dobigny et al. 2004). The authors suggest that during intense genome repatterning some epigenetic features, such as DNA methylation, are relaxed, allowing TE amplification (Dobigny et al. 2004). The amplified KERV-1 element at the hybrid centromere may therefore be directly involved in the various chromosome rearrangements observed or, alternatively, the chromosome rearrangements observed may be concomitant with genomic stress during which epigenetic controls, such as DNA methylation or histone modification, are relaxed. In the Macropus hybrids, it is unclear whether epigenetic restructuring of the centromere allowed the KERV-1 and repeat amplification observed or whether the amplifications resulted in chromatin remodeling.
Hybrid centromeres are structurally modified:
Knobs were first described in maize by McClintock 1929). In maize, they appear in pachytene cells as darkly staining heterochromatic regions at intercalary positions and are composed of repeat elements and retrotransposable elements (Ananiev et al. 1998). Knobs represent a structural manifestation in meiosis of these chromosomal elements. In rice, a correlation has been observed between the size of the knob and the amount of repeat DNA, suggesting that a minimum amount of tandemly repeated DNA may be required to induce knob-like heterochromatic structure that can be visualized by microscopic methods (Cheng et al. 2001). In plants, centromeres are characterized by extremely large blocks of tandem repeats and retrotransposons (Houben and Schubert 2003). It is interesting that, in this respect, the centromeres of these marsupial species and their hybrids resemble plant chromosomes. To our knowledge, however, there are no reports of elongated or “segmented” plant centromeres.
The observed segmented or “knob-like” structures in mitotic metaphase chromosomes at the M. rufogriseus-derived centromeres in the hybrids were visible within elongated centromeres and not consistently found in the same position on a particular chromosome. This may indicate that the instability at these centromeric loci is not fixed but is in a dynamic state of chromatin remodeling. Three-dimensional ultrastructural analysis shows that, although these centromeres do not contain “knobs” in the dimension suggested by light microscopic data, they not only are elongated with respect to other chromosomes in their complement, but also are characterized by more chromomeres and a denser chromatin configuration with respect to other plant and animal mitotic centromeres (Wanner and Formanek 1995, 2000; Sumner 1998). Similar centromere elongations in human chromosomes have been described for cases of cytosine hypomethylation such as observed with 5-azacytidine (methylase antagonist) treatment and ICF (immunodeficiency, centromeric instability, and facial abnormalities) syndrome (Sumner 2003; Miniou et al. 1994) and have been attributed to decondensation of centromeric heterochromatin. Our results showing that the elongated hybrid centromeres are DNA rich support an amplification event rather than a decondensation event. ISH and dot-blot analysis confirm that the elongated centromeres are indeed manifestations of a significant increase in DNA repeats. Further immunodetection experiments are necessary to determine the degree of methylation in the hybrid centromeres.
Centromere evolution and hybrid dysgenesis:
Recent observations with regards to the rapid evolution of a centromeric protein in plants and animals (Malik and Henikoff 2001; Talbert et al. 2004) suggest that genomic conflict in the form of centromere drive (Henikoff et al. 2001; Henikoff and Malik 2002; Malik and Henikoff 2002) may be responsible for the centromere instability at the M. rufogriseus-derived centromeres. Centromere protein C (CENP-C) is a large protein that binds to vertebrate centromeric DNA (Sugimoto et al. 1994) with low sequence conservation except for a CENP-C motif (Talbert et al. 2004). The observation that both centromeres and CENP-C are rapidly evolving (Talbert et al. 2004), even between closely related species, suggests that a disconnect between CENP-C and centromeric sequence between the two parental centromeres may be responsible for the centromere instability observed in the marsupial hybrids (O'Neill et al. 1998, 2001; this study). However, it is not clear why this instability should be confined to the M. rufogriseus-derived centromere.
The most obvious difference between the M. rufogriseus and M. agilis karyotypes is the difference in the amount of constitutive heterochromatin at the centromeres, as evidenced by C-banding. The large amounts of heterochromatin at the M. rufogriseus centromeres may be the result of past centromeric drive activity. Talbert et al. (2004) propose a centromeric drive model, based on karyotypic drive in female meiosis (Pardo-Manuel De Villena and Sapienza 2001), to explain the apparent anomaly of rapidly evolving sequence but conserved function in centromeres and centromeric proteins. They suggest that, because only one product of female meiosis ends up in the final meiotic product, centromeres “compete” during female meiosis. A centromere that is “stronger” will have a greater chance of ending up in the single female meiotic product. However, differences in centromeric drive in male meiosis, where all products end up in the final meiotic product, may result in reduced male fertility. Mutations in CENP-C that restore centromere parity in male meiosis will therefore be selected for in males. Recurrent cycles of selection for “stronger” centromeres in females and “restoring” CENP-C in males would result in the observed rapid evolution of CENP-C and centromeres (Talbert et al. 2004). They further suggest that addition of satellite repeats at the centromere would attract more centromere histones (CenH3 or CENP-A), thus binding more microtubules during cell division and creating a “stronger” centromere (Talbert et al. 2004).
Genome repatterning in the four marsupial hybrids is limited to the M. rufogriseus centromere and correlates with the presence of the larger amounts of heterochromatin at these locations. Previous studies indicated that large heterochromatic blocks at centromeres of this species contain CENP-B-binding sequences. However, these centromeres do not form a large kinetochore region spread across these heterochromatic sequences, but, contrary to the centromere drive model, form a small, confined kinetochore akin to that of species with small centromere regions (Bulazel et al. 2006). The apparent restructuring of chromatin fibers at hybrid M. rufogriseus centromeres may be indicative of more extensive remodeling at these loci, although kinetochore size and CENP-A affinity have not been assayed. However, one backcross hybrid has been previously examined (Lowry et al. 1995), the karyotype of which did not indicate preferential inheritance of the M. rufogriseus-derived centromere.
An alternative hypothesis for the chromatin remodeling and destabilization of these centromeres is that there is an incompatibility within the RNA interference (RNAi) machinery. RNA silencing or RNA interference occurs in a wide variety of eukaryotic organisms (Meister and Tuschl 2004) and is involved in gene regulation, transposable element mobilization suppression, and epigenetic regulation of heterochromatin. RNA interference appears to be involved in the regulation of centromere function and architecture, in particular with the epigenetic maintenance of heterochromatic regions (Grewal and Jia 2007). While it has been better studied in yeast (Volpe et al. 2002, 2003) and Drosophila (Pal-Bhadra et al. 2004), there is evidence that RNAi is also involved in maintenance of heterochromatic and centromeric regions in mammals (Fukagawa et al. 2004; Kanellopoulou et al. 2005; Bouzinba-Segard et al. 2006). It has been proposed that incompatibility between short interfering RNAs (Lippman and Martienssen 2004) or piwi-interacting RNAs (Brennecke et al. 2007) between maternal and paternal species within a hybrid genome may lead to transposon activation and/or disruption of heterochromatic structure and ultimately to hybrid dysgenesis. Such activation could lead to copy-number increases and chromosome instability, as well as chromatin remodeling, all features of our hybrid system. The difference seen between M. rufogriseus- and M. agilis-derived centromeres may simply be an effect of a larger region in M. rufogriseus in which disruption of epigenetic maintenance could occur.
Conclusions:
Here we have identified centromeric instability and remodeling in hybrids within a marsupial genus known for rapid chromosome evolution predominately involving centromere-involved rearrangements. We observed the same instability in all four hybrids, which was confined to the maternally derived centromeres containing large blocks of heterochromatin that are expanded relative to the paternally derived centromere. The inherent instability that we have identified in this region of the genome may have a role in the rapid chromosome evolution and speciation within macropodids.
Acknowledgments
We thank Macquarie University, Fauna Park, for care of animals; the University of Connecticut Mentor Connection for participation of summer high school students in aspects of this research; Patricia C. M. O'Brien for flow sorting chromosomes; Sabine Steiner for excellent technical assistance; and M. J. O'Neill for critical evaluation of this manuscript. We also thank two anonymous reviewers for their helpful comments. C.J.M., K.B., G.C.F., and R.J.O. were supported by a National Science Foundation CAREER award (MCB-0093250). M.D.B.E. was supported by the Australian Research Council and Macquarie University, Sydney, Australia. W.R. was supported with funding from the Wellcome Trust.
References
- Ananiev, E. V., R. L. Phillips and H. W. Rines, 1998. A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: Are chromosome knobs megatransposons? Proc. Natl. Acad. Sci. USA 95: 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzinba-Segard, H., A. Guais and C. Francastel, 2006. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA 103: 8709–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., A. A. Aravin, A. Stark, M. Dus, M. Kellis et al., 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Bulazel, K., C. Metcalfe, G. Ferreri, J. Yu, M. Eldridge et al., 2006. Cytogenetic and molecular evaluation of centromere-associated DNA sequences from a marsupial (Macropodidae: Macropus rufogriseus) X chromosome. Genetics 172: 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulazel, K. V., G. C. Ferreri, M. D. Eldridge and R. J. O'Neill, 2007. Species-specific shifts in centromere sequence composition are coincident with break point reuse in karyotypically divergent lineages. Genome Biol. 8: R170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, M., M. Puig and A. Ruiz, 2001. Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res. 11: 1353–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., R. M. Stupar, M. Gu and J. Jiang, 2001. A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma 110: 24–31. [DOI] [PubMed] [Google Scholar]
- Cooper, D. W., C. Edwards, E. James, G. B. Sharman, J. L. VandeBerg et al., 1977. Studies on metatherian sex chromosomes. VI. A third state of an X-linked gene: partial activity for the paternally derived Pgk-A allele in cultured fibroblasts of Macropus giganteus and M. parryi. Aust. J. Biol. Sci. 30: 431–443. [Google Scholar]
- Dobigny, G., C. Ozouf-Costaz, P. D. Waters, C. Bonillo, J. P. Coutanceau et al., 2004. LINE-1 amplification accompanies explosive genome repatterning in rodents. Chromosome Res. 12: 787–793. [DOI] [PubMed] [Google Scholar]
- Dutrillaux, B., 1979. Chromosomal evolution in primates: tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum. Genet. 48: 251–314. [DOI] [PubMed] [Google Scholar]
- Eldridge, M. D. B., 1991. Chromosomal rearrangements and speciation in rock wallabies, Petrogale. Ph.D. Thesis, Macquarie University, Sydney.
- Eldridge, M. D. B., and R. L. Close, 1993. Radiation of chromosome shuffles. Curr. Opin. Genet. Dev. 3: 915–922. [DOI] [PubMed] [Google Scholar]
- Eldridge, M. D. B., and C. J. Metcalfe, 2006. Marsupial chromosomes, pp. 9–62 in Atlas of Mammalian Chromosomes, edited by S. J. O'Brien, J. Menninger and W. Nash. Wiley, New York.
- Eldridge, M. D. B., R. L. Close and P. G. Johnston, 1991. Chromosomal rearrangements in rock wallabies Petrogale (Marsupialia: Macropodidae). IV. G-banding analysis of the Petrogale lateralis complex. Aust. J. Zool. 39: 621–628. [Google Scholar]
- Eldridge, M. D. B., P. G. Johnston and R. L. Close, 1992. Chromosomal rearrangements in rock wallabies, Petrogale (Marsupialia: Macropodidae). VI. Determination of the plesiomorphic karyotype: G-banding comparison of Thylogale with Petrogale persephone, P. xanthopus, and P. l. lateralis. Cytogenet. Cell Genet. 61: 29–33. [DOI] [PubMed] [Google Scholar]
- Evgen'ev, M. B., H. Zelentsova, H. Poluectova, G. T. Lyozin, V. Veleikodvorskaja et al., 2000. Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc. Natl. Acad. Sci. USA 97: 11337–11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreri, G. C., M. Marzelli, W. Rens and R. J. O'Neill, 2004. A centromere-specific retroviral element associated with breaks of synteny in macropodine marsupials. Cytogenet. Genome Res. 107: 115–118. [DOI] [PubMed] [Google Scholar]
- Ferreri, G. C., D. M. Liscinsky, J. A. Mack, M. D. Eldridge and R. J. O'Neill, 2005. Retention of latent centromeres in the mammalian genome. J. Hered. 96: 217–224. [DOI] [PubMed] [Google Scholar]
- Fontdevila, A., 1992. Genetic instability and rapid speciation: Are they coupled? Genetica 86: 247–258. [Google Scholar]
- Fontdevila, A., 2005. Hybrid genome evolution by transposition. Cytogenet. Genome Res. 110: 49–55. [DOI] [PubMed] [Google Scholar]
- Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki et al., 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6: 784–791. [DOI] [PubMed] [Google Scholar]
- Grewal, S. I., and S. Jia, 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Hayman, D. L., 1990. Marsupial cytogenetics. Aust. J. Zool. 37: 331–349. [Google Scholar]
- Henikoff, S., and H. S. Malik, 2002. Centromeres: selfish drivers. Nature 417: 227. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Houben, A., and I. Schubert, 2003. DNA and proteins of plant centromeres. Curr. Opin. Plant Biol. 6: 554–560. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou, C., S. A. Muljo, A. L. Kung, S. Ganesan, R. Drapkin et al., 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki, H., B. Schreiner, S. Tanzer, M. Platzer, S. Muller et al., 2002. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am. J. Hum. Genet. 71: 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, M., 1993. Species Evolution: The Role of Chromosome Change. Cambridge University Press, Cambridge, UK.
- Labrador, M., M. Farre, F. Utzet and A. Fontdevila, 1999. Interspecific hybridization increases transposition rates of Osvaldo. Mol. Biol. Evol. 16: 931–937. [DOI] [PubMed] [Google Scholar]
- Lim, J. K., 1988. Intrachromosomal rearrangements mediated by hobo transposons in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 85: 9153–9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. K., and M. J. Simmons, 1994. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. BioEssays 16: 269–275. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., and R. Martienssen, 2004. The role of RNA interference in heterochromatic silencing. Nature 431: 364–370. [DOI] [PubMed] [Google Scholar]
- Lowry, P. S., M. D. B. Eldridge and P. G. Johnston, 1995. Genetic analysis of a female macropodid hybrid (Macropus agilis × M. rufogriseus) and her backcross offspring. Aust. Mammal. 18: 79–82. [Google Scholar]
- Malik, H. S., and S. Henikoff, 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157: 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2002. Conflict begets complexity: the evolution of centromeres. Curr. Opin. Genet. Dev. 12: 711–718. [DOI] [PubMed] [Google Scholar]
- Martin, R., W. Busch, R. G. Herrmann and G. Wanner, 1994. Efficient preparation of plant chromosomes for high-resolution scanning electron microscopy. Chromosome Res. 2: 411–415. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1929. Chromosome morphology in Zea Mays. Science 69: 629. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1987. The Discovery and Characterization of Transposable Elements. Garland Publishing, New York.
- Meister, G., and T. Tuschl, 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349. [DOI] [PubMed] [Google Scholar]
- Miniou, P., M. Jeanpierre, V. Blanquet, V. Sibella, D. Bonneau et al., 1994. Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients. Hum. Mol. Genet. 3: 2093–2102. [DOI] [PubMed] [Google Scholar]
- O'Neill, R. J., M. J. O'Neill and J. A. Marshall Graves, 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393: 68–72. [DOI] [PubMed] [Google Scholar]
- O'Neill, R. J., M. D. B. Eldridge, R. Toder, M. A. Ferguson-Smith, P. C. O'Brien et al., 1999. Chromosome evolution in kangaroos (Marsupialia: Macropodidae): cross species chromosome painting between the tammar wallaby and rock wallaby spp. with the 2n = 22 ancestral macropodid karyotype. Genome 42: 525–530. [DOI] [PubMed] [Google Scholar]
- O'Neill, R. J., M. D. B. Eldridge and J. A. M. Graves, 2001. Chromosome heterozygosity and de novo chromosome rearrangements in mammalian interspecies hybrids. Mamm. Genome 12: 256–259. [DOI] [PubMed] [Google Scholar]
- O'Neill, R. J., M. D. B. Eldridge and C. J. Metcalfe, 2004. Centromere dynamics and chromosome evolution in marsupials. J. Hered. 95: 375–381. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and C. Sapienza, 2001. Female meiosis drives karyotypic evolution in mammals. Genetics 159: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens, W., P. C. M. O'Brien, F. Yang, J. A. M. Graves and M. A. Ferguson-Smith, 1999. Karyotype relationships between four distantly related marsupials revealed by reciprocal chromosome painting. Chromosome Res. 7: 461–474. [DOI] [PubMed] [Google Scholar]
- Rofe, R. H., 1978. G-banded chromosomes and the evolution of the Macropodidae. Aust. Mammal. 2: 53–63. [Google Scholar]
- Ruiz-Herrera, A., J. Castresana and T. Robinson, 2006. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 7: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, A., D. Chan, L. Brown, R. Alagappan, D. Pettay et al., 1998. Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum. Mol. Genet. 7: 1–11. [DOI] [PubMed] [Google Scholar]
- Sharman, G. B., 1961. The mitotic chromosomes of marsupials and their bearing on taxonomy and phylogeny. Aust. J. Zool. 9: 38–60. [Google Scholar]
- Sugimoto, K., H. Yata, Y. Muro and M. Himeno, 1994. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J. Biochem. 116: 877–881. [DOI] [PubMed] [Google Scholar]
- Sumner, A., 1972. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75: 304–306. [DOI] [PubMed] [Google Scholar]
- Sumner, A., 1998. The structure of the centromeric region of CHO chromosomes. Cell Biol. Int. 22(2): 127–130. [DOI] [PubMed] [Google Scholar]
- Sumner, A., 2003. Chromosomes: Organisation and Structure. Blackwell Science, Malden, MA.
- Talbert, P. B., T. D. Bryson and S. Henikoff, 2004. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer, M. C., S. C. Strakosh and Y. Zhen, 2006. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 16: R872–R873. [DOI] [PubMed] [Google Scholar]
- Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. S. Grewal et al., 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Volpe, T., V. Schramke, G. L. Hamilton, S. A. White, G. Teng et al., 2003. RNA interference is required for normal centromere function infission yeast. Chromosome Res. 11: 137–146. [DOI] [PubMed] [Google Scholar]
- Wanner, G., and H. Formanek, 1995. Imaging of DNA in human and plant chromosomes by high-resolution scanning electron microscopy. Chromosome Res. 3: 368–374. [DOI] [PubMed] [Google Scholar]
- Wanner, G., and H. Formanek, 2000. A new chromosome model. J. Struct. Biol. 132: 147–161. [DOI] [PubMed] [Google Scholar]
- Zhang, J., and T. Peterson, 1999. Genome rearrangements by nonlinear transposons in maize. Genetics 153: 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]