Abstract
The house mouse is a well-established model organism, particularly for studying the genetics of complex traits. However, most studies of mice use classical inbred strains, whose genomes derive from multiple species. Relatively little is known about the distribution of genetic variation among these species or how variation among strains relates to variation in the wild. We sequenced intronic regions of five X-linked loci in large samples of wild Mus domesticus and M. musculus, and we found low levels of nucleotide diversity in both species. We compared these data to published data from short portions of six X-linked and 18 autosomal loci in wild mice. We estimate that M. domesticus and M. musculus diverged <500,000 years ago. Consistent with this recent divergence, some gene genealogies were reciprocally monophyletic between these species, while others were paraphyletic or polyphyletic. In general, the X chromosome was more differentiated than the autosomes. We resequenced classical inbred strains for all 29 loci and found that inbred strains contain only a small amount of the genetic variation seen in wild mice. Notably, the X chromosome contains proportionately less variation among inbred strains than do the autosomes. Moreover, variation among inbred strains derives from differences between species as well as from differences within species, and these proportions differ in different genomic regions. Wild mice thus provide a reservoir of additional genetic variation that may be useful for mapping studies. Together these results suggest that wild mice will be a valuable complement to laboratory strains for studying the genetics of complex traits.
THE house mouse presents an excellent mammalian model for studies of the genetic basis of complex traits, including many diseases. Dozens of inbred strains are available with sufficient genetic variability among them for linkage mapping and association studies (Paigen 2003a,b; Peters et al. 2007). A variety of molecular genetic tools are available for the mouse, making it possible to identify and functionally characterize candidate genes for some traits. The mouse genome has been sequenced (Waterston et al. 2002), patterns of expression in different tissues have been described for nearly all genes (e.g., Su et al. 2004), there is a large and growing set of knockouts, and phenotypes have been associated with >10% of all genes (Grimm 2006).
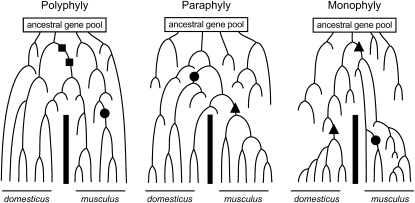
The classical inbred strains in which these resources have been developed, including the sequenced C57Bl/6J (Waterston et al. 2002), derive from matings among different species of the house mouse (Silver 1995; Wade et al. 2002). Thus, understanding the genetic variation among inbred strains requires understanding the evolutionary history of the species from which they were derived. The house mouse consists of three main lineages: Mus domesticus in Western Europe, Mus musculus in Eastern Europe and Asia, and Mus castaneus in Southeast Asia and India (also referred to as subspecies of Mus musculus: i.e., M. m. musculus, M. m. domesticus, and M. m. castaneus; Silver 1995). M. musculus and M. domesticus diverged between ∼350,000 and 1,000,000 years ago (She et al. 1990; Boursot et al. 1996; Suzuki et al. 2004). These species recently came into secondary contact following the spread of M. domesticus into Western Europe from the Middle East with the spread of agriculture over the last few thousand years (Cucchi et al. 2005). M. domesticus and M. musculus form a stable hybrid zone where they meet, and laboratory crosses between these species result in sterile hybrid males (Britton-Davidian et al. 2005). Classical lab strains of mice derive principally from M. domesticus and M. musculus, with a smaller contribution from M. castaneus (Silver 1995; Wade et al. 2002; Frazer et al. 2007; Yang et al. 2007). The partitioning of genetic variation among M. musculus and M. domesticus is largely unknown. In particular, the recent separation of these lineages raises the possibility that some loci will retain ancestral polymorphisms and that other loci will show fixed differences. From a genealogical perspective, some loci may be monophyletic within each species (i.e., all alleles within a species are more closely related to each other than to any alleles in the other species), while other loci may be paraphyletic or polyphyletic (i.e., some alleles within a species might be more closely related to alleles in the sister species than to other alleles within the same species; Figure 1).
Figure 1.—
Hypothetical examples of gene genealogies for alleles sampled from M. domesticus and M. musculus. As two species diverge, gene genealogies are expected initially to be polyphyletic (left), then paraphyletic (middle), and finally monophyletic (right). Because of the variance in evolutionary history among loci, all three patterns might be present in different genes in the genome at the same time, especially in the early stages of divergence. Without recurrent mutation, shared polymorphisms (squares) will be associated only with polyphyletic genealogies, fixed differences (triangles) will be associated only with paraphyletic or monophyletic genealogies, and polymorphisms that are present in just one species (circles) may be associated with any of these genealogies. This figure is modified from Avise (1994).
The relationship of classical inbred strains to wild mice is important for several reasons. First, it is likely that the different inbred strains capture a small amount of the naturally occurring variation, but the amount of variation in the wild is still unclear. What proportion of haplotypes and single nucleotide polymorphism (SNPs) found in wild mice are also present among inbred strains? Does this pattern differ between the X chromosome and the autosomes? Differences between the X chromosome and autosomes among inbred strains might arise if selection acted differently on the X and autosomes during the founding of these strains or if unequal numbers of males and females were used in the founding of these strains. Wild mice might provide a reservoir of additional genetic variation for studies that seek to understand the genetic basis of complex traits, but quantifying variation in the wild is a necessary first step. Second, some of the variation among inbred strains derives from fixed differences between species, while some of the variation reflects differences within one or several species (Wade et al. 2002). Epistatic interactions between alleles from different species are likely to have shaped the current variation among strains (Payseur and Hoekstra 2005; Petkov et al. 2005), and this is likely to affect the genetic architecture underlying complex traits.
Although several large-scale efforts have characterized variation among classical and wild-derived inbred strains of mice (Wade et al. 2002; Wiltshire et al. 2003; Frazer et al. 2004; Ideraabdullah et al. 2004; Petkov et al. 2004; Yalcin et al. 2004; Frazer et al. 2007; Yang et al. 2007), these studies do not adequately describe the amount of variation in natural populations for two reasons. First, and most importantly, <10 wild-derived inbred strains of each species (and often only 1) have been included in these studies, representing a very small portion of the geographic range of the wild house mice. Second, some of these studies have described variation among wild-derived inbred strains for polymorphisms that were previously ascertained among the classical inbred strains. This ascertainment bias may hide the true distribution of variation in natural populations (Boursot and Belkhir 2006).
To begin to address these issues, we compared variation among nine of the most commonly used classical inbred strains with variation among wild M. domesticus and M. musculus. First, we present data from five X-linked loci sequenced in relatively large samples of wild mice and we compare these data to previously published X-linked and autosomal data. Second, we sequenced eight inbred strains and analyzed them with the already sequenced C57Bl/6J for nearly all genes for which polymorphism data have been published from wild M. domesticus and M. musculus for a total of 11 X-linked and 18 autosomal loci. Finally, we compared all data to the publicly available SNP databases.
MATERIALS AND METHODS
Samples:
We sequenced five X-linked loci in large samples of wild mice and then resequenced eight classical inbred strains and one each M. spretus and M. caroli for 11 X-linked and 18 autosomal loci. These include the five X-linked loci as well as 24 loci for which wild mouse population data have already been published (Harr 2006a; Baines and Harr 2007).
Male M. domesticus and M. musculus were wild caught in Europe (Table 1). Males were used to obtain unambiguous haplotypes for X-linked loci. The standard karyotype of M. domesticus is 2n = 40, with all acrocentric chromosomes. However, M. domesticus has many chromosomal races with 2n < 40; all M. musculus have 2n = 40 (Pialek et al. 2005). To exclude chromosomal races, all M. domesticus were karyotyped as described previously (Nachman et al. 1994), and only mice with 2n = 40 were used. Genomic DNA representing the classical inbred strains 129S1/SvImJ, A/J, AKR/J, BALB/cByJ, C3H/HeJ, DBA/2J, FVB/NJ, and SJL/J, as well as one M. spretus and one M. caroli, was purchased from the Jackson Laboratories (Bar Harbor, ME). Sequence for C57Bl/6J was downloaded from NCBI or Ensembl (Build 36).
TABLE 1.
Sampling localities or origins of house mouse DNA samples
| Species | Locality or strain | N |
|---|---|---|
| M. domesticus | Mainland Italy | 34 |
| Sicily (Italy) | 11 | |
| Spain | 13 | |
| Greece | 6 | |
| M. musculus | Czech Republic | 19 |
| Serbia | 2 | |
| Denmark | 2 | |
| Austria | 3 | |
| M. spretus | Spain | 1 |
| M. caroli | Thailand | 1 |
| Classical inbred strains | 129S1/SvImJ | 1 |
| A/J | 1 | |
| AKR/J | 1 | |
| BALB/cByJ | 1 | |
| C3H/HeJ | 1 | |
| DBA/2J | 1 | |
| FVB/NJ | 1 | |
| SJL/J | 1 |
All classical inbred strain DNA and samples of M. spretus and M. caroli were ordered from the Jackson Labs in Bar Harbor, Maine. N, number of individuals.
Molecular methods:
Five X-linked loci were surveyed in wild mice: Maoa, Dmd, Msn, Dach2, and Amelx (supplemental Figure 1 at http://www.genetics.org/supplemental/). Loci were chosen so as to (i) be evenly distributed along the X chromosome, (ii) have human homologs for which population genetic data have been published (Hammer et al. 2004), and (iii) have at least one long intron (>5 kb). We focused on introns to capture a large amount of genetic variation. Our survey of Amelx included four small exons, which were excluded from analysis. All sequence information was based on Build 36 of the mouse genome.
Amplification and sequencing primers were designed using Primer3 (Rozen and Skaletsky 2000) on sequences prescreened using RepeatMasker (Smit et al. 1996–2004). Amplification primer sequences are given in supplemental Table 1 at http://www.genetics.org/supplemental/. Portions of Dach2, Amelx, Maoa, and Msn were PCR amplified in a single amplicon using high-fidelity Taq polymerase (Invitrogen, Carlsbad, CA). The following PCR conditions were used: initial denaturation (94° for 2 min) was followed by 35 cycles of 30 sec at 94°, 30 sec at 57°, and 5 min at 65°. High-fidelity (Invitrogen) and HotMaster (Eppendorf, Westbury, NY) Taq polymerases were used to amplify a portion of one intron of Dmd in multiple amplicons. PCR products were cleaned using either the QIAGEN (Valencia, CA) PCR clean-up kit or the 96-well format by the Genomic Analysis and Technology Core at the University of Arizona. Sequencing was performed on both strands using either an ABI 377 or a 3731 automated sequencer.
Data analysis:
Sequences were trimmed and assembled into contigs using Sequencher (Gene Codes, Ann Arbor, MI). These contigs have been deposited in GenBank under accession nos. EF067347–EF067807 and EU220489–EU220697. Alignments generated with Sequencher were manually edited using MacClade (Sinauer Associates, Sunderland, MA). Summary statistics describing the level and pattern of nucleotide variability were calculated manually or by DnaSP (Rozas and Rozas 1999). We also used DnaSP to calculate Hudson's minimum number of recombination events, Rm (Hudson and Kaplan 1985), and to describe linkage disequilibrium. Haplotype networks were drawn manually and neighbor-joining trees were generated using PAUP*, version 4.0b10 (Swofford 2003).
We performed several statistical tests of a neutral model of molecular evolution. Tajima's D (Tajima 1989), Fu and Li's D (Fu and Li 1993), and Fay and Wu's H (Fay and Wu 2000) all describe the allele-frequency spectrum by comparing different estimators of θ, the population mutation parameter (θ = 3Neμ for the X chromosome and θ = 4Neμ for the autosomes, where Ne is the effective population size and μ is the neutral mutation rate). We used several tests because they differ in their power to detect different perturbations from neutral equilibrium conditions (Braverman et al. 1995; Simonsen et al. 1995; Przeworski 2002). These tests were conducted using software available from Y. X. Fu (http://hgc.sph.uth.tmc.edu/fu) and from J. Fay (http://www.genetics.wustl.edu/jflab/htest.html). The HKA test compares the ratio of polymorphism to divergence among two or more genes (Hudson et al. 1987). Pairwise and multilocus HKA tests were performed using software developed by Jody Hey (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm#HKA). FST values and analysis of molecular variance (AMOVA) results were calculated using Arlequin (Schneider et al. 2000).
We also analyzed the data in the Mouse Phenome Database (MPD; http://www.jax.org/phenome) for the corresponding portions of the 11 X-linked and 18 autosomal loci. MPD includes a comprehensive database of all known mouse SNPs and it integrates SNPs from many sources, including the Broad Institute (Lindblad-Toh et al. 2000), Celera (e.g., Lemon et al. 2003), Perlegen (Frazer et al. 2007), The Jackson Laboratory (Petkov et al. 2004), The Wellcome Trust Centre for Human Genetics (e.g., Yalcin et al. 2004), The Genomics Institute of the Novartis Research Foundation (Pletcher et al. 2004), and others. This database contains >10 million SNPs representing >100 classical and wild-derived inbred strains of mice. We queried MPD for all SNPs in the regions that we sequenced. We conducted mouse-genome-specific BLAST searches using NCBI or basic local alignment tool (BLAT) searches with the University of California at Santa Cruz mouse genome server and identified sites that overlapped with our resequencing targets. All SNPs identified solely by differences from representatives of M. castaneus or M. molossinus (a hybrid between M. castaneus and M. musculus) were excluded to match the sampling used here.
RESULTS
Level and pattern of genetic variation at five X-linked genes:
We sequenced 4–5 kb of intronic DNA at each of five X-linked genes (supplemental Figure 1 at http://www.genetics.org/supplemental/) in 60–64 M. domesticus and 18–22 M. musculus. Levels of nucleotide variability were low in both species, with 1–29 segregating sites/locus (Table 2; Figure 2; supplemental Figures 2 and 3). In M. domesticus, nucleotide diversity ranged from 0.03 to 0.15% among the five genes, with an average value of 0.07%, almost identical to the average value for X-linked genes in humans (e.g., Hammer et al. 2004). In M. musculus, nucleotide diversity was considerably lower, ranging from 0 to 0.06% among genes, with an average value of 0.03%. We used the estimates of nucleotide diversity to estimate species-wide effective population sizes, assuming a mutation rate of 4 × 10−9 (Waterston et al. 2002). Thus, for M. domesticus, Ne = θ/3μ = (7 × 10−4)/(1.2 × 10−8) = 5.8 × 104, and for M. musculus, Ne = (3 × 10−4)/(1.2 × 10−8) = 2.5 × 104. To the extent that selection has reduced levels of variation on the X chromosome (Baines and Harr 2007), these will be underestimates.
TABLE 2.
Levels of polymorphism and divergence and tests of a neutral model of molecular evolution for five X-linked genes surveyed in M. domesticus and M. musculus
| Polymorphism
|
Divergencea
|
Allele-frequency-based tests
|
Pairwise HKA test χ2
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Sample | Sample size | Length (bp) | Base substitutions | Insertions/deletions | π (%) | θ (%) | Base substitutions | Insertions/deletions | D (%) | Tajima's D | Fu and Li's D | Fay and Wu's H | Maoa | Dmd | Msn | Dach2 | Amelx |
| Maoa | M. domesticus | 60 | 4026 | 17 | 4 | 0.06 | 0.10 | 128 | 16 | 3.20 | −1.002 | −0.766 | 6.176 | — | 0.014 | 0.058 | 0.255 | 2.221 |
| M. musculus | 22 | 4081 | 8 | 2 | 0.06 | 0.06 | 128 | 14 | 3.17 | 0.579 | 1.364 | 2.818 | — | 0.802 | 1.640 | 0.781 | 0.007 | |
| Laboratory strains | 8 | 4067 | 0 | 1 | 0.00 | 0.00 | 130 | 15 | 3.24 | — | — | — | ||||||
| Dmd | M. domesticus | 64 | 5147 | 24 | 2 | 0.05 | 0.10 | 189 | 24 | 3.69 | −1.463* | −2.486* | 6.363 | — | 0.019 | 0.169 | 3.054 | |
| M. musculus | 18 | 5130 | 6 | 4 | 0.03 | 0.03 | 195 | 25 | 3.82 | −0.817 | 1.261 | 4.961* | — | 0.249 | 0.004 | 0.643 | ||
| Laboratory strains | 8 | 5148 | 0 | 2 | 0.00 | 0.00 | 190 | 24 | 3.71 | — | — | — | ||||||
| Msn | M. domesticus | 61 | 3961 | 13 | 4 | 0.04 | 0.07 | 111 | 18 | 3.46 | −1.126 | −0.630 | 1.947 | — | 0.066 | 2.766 | ||
| M. musculus | 19 | 3931 | 1 | 3 | 0.00 | 0.01 | 113 | 19 | 3.21 | −1.511 | −2.022 | 0.506 | — | 0.263 | 1.471 | |||
| Laboratory strains | 8 | 3969 | 0 | 1 | 0.00 | 0.00 | 123 | 17 | 3.50 | — | — | — | ||||||
| Dach2 | M. domesticus | 61 | 3892 | 13 | 9 | 0.03 | 0.07 | 128 | 25 | 3.34 | −1.750* | −2.352* | 0.078 | — | 3.796 | |||
| M. musculus | 22 | 3901 | 4 | 4 | 0.03 | 0.03 | 129 | 23 | 3.37 | 0.036 | 0.958 | 1.736 | — | 0.639 | ||||
| Laboratory strains | 8 | 3904 | 6 | 5 | 0.08 | 0.06 | 128 | 22 | 3.34 | 1.554 | 1.314 | 2.179 | ||||||
| Amelx | M. domesticus | 61 | 4043 | 29 | 6 | 0.15 | 0.18 | 123 | 17 | 3.00 | −0.857 | −2.533* | 3.544 | — | ||||
| M. musculusb | 22 | 4033 | 7 | 7 | 0.05 | 0.05 | 119 | 20 | 2.80 | −0.037 | 0.627 | 0.961 | — | |||||
| Laboratory strains | 8 | 4066 | 0 | 2 | 0.00 | 0.00 | 119 | 17 | 2.89 | — | — | — | ||||||
| Mean | M. domesticus | 61.4 | 4235 | 20.2 | 5.6 | 0.07 | 0.10 | 135.80 | 20.00 | 3.34 | −1.240 | −1.753 | 3.622 | |||||
| M. musculus | 20.6 | 4237 | 5.4 | 4.0 | 0.03 | 0.03 | 136.80 | 20.20 | 3.27 | −0.350 | 0.509 | 1.505 | ||||||
| Laboratory strains | 10.8 | 4234 | 2.6 | 2.2 | 0.03 | 0.02 | 138.00 | 19.60 | 3.32 | — | — | — | ||||||
| Total | M. domesticus | 64 | 21177 | 101 | 28 | 679 | 100 | |||||||||||
| M. musculus | 22 | 21184 | 26 | 20 | 684 | 101 | ||||||||||||
*P < 0.05.
Divergence values were calculated by comparing a random individual to M. caroli.
All values shown were calculated excluding one individual (MWN1379) whose haplotype is believed to be introgressed.
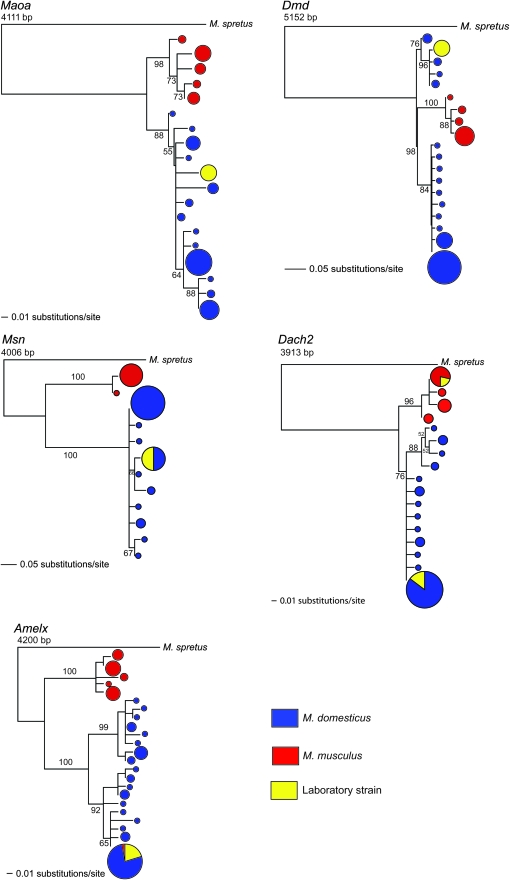
Figure 2.—
Neighbor-joining trees based on uncorrected distance matrices, rooted with M. spretus for five X-linked loci sequenced here. Circles represent haplotypes at each terminal node, with circle size proportional to the number of chromosomes and colors designating sample identities. Blue, M. domesticus; red, M. musculus; yellow, classical inbred strain. Numbers adjacent to branches indicate bootstrap values >50 (500 replicates).
Levels of divergence with respect to M. caroli were consistently ∼3% (Table 2), similar to previous observations (She et al. 1990). Average divergence between M. musculus or M. domesticus and M. spretus (∼1%) was also consistent with previous estimates (Nachman 1997). The average pairwise divergence (k) between alleles from M. musculus and alleles from M. domesticus was 0.43%. We can use this value to estimate the time of separation of these species. Under a neutral model, k = 2μt + 3Neμ for X-linked loci, where μ is the neutral mutation rate, t is the species divergence time in generations, and Ne is the effective population size of the ancestral population. We can estimate the ancestral value of 3Neμ as the average of current nucleotide diversity in M. musculus and M. domesticus [π = 3Neμ = (0.0003 + 0.0007)/2 = 0.0005]. Assuming a neutral mutation rate of 4 × 10−9 (Waterston et al. 2002), this leads to an estimate of t = (k − 3Neμ)/2μ = (0.0043 − 0.0005)/(8 × 10−9) = 475,000 generations. Although mice in captivity produce several generations per year, mice in the wild often breed seasonally and may produce only one or two generations per year. This suggests that these species diverged on the order of 237,500–475,000 years ago. This is a very rough estimate but is consistent with a previous estimate of 350,000 years based on DNA–DNA hybridization data (She et al. 1990).
We examined patterns of linkage disequilibrium (LD) both within and between genes. Within genes, we found no evidence for recombination: we never observed all four gametic types in pairwise comparisons between sites within a gene (Hudson's minimum number of recombination events, Rm, was 0 for each gene). Our sequences span 4–5 kb for each gene, suggesting that LD extends over distances greater than this for X-linked loci, similar to patterns seen for many X-linked loci in humans (e.g., Hammer et al. 2004). Between genes we found no LD. Of 4950 pairwise comparisons between all sites in our data set, 372 were found to be in significant LD using a Fisher's exact test (FET). However, after a Bonferroni correction for multiple tests, only 86 of these values were significant, and all of those described intralocus pairs of sites.
We also considered the relationship among haplotypes (Figure 2). Within M. domesticus, there was generally a single common haplotype and several rare haplotypes, often separated by one or a few mutational steps from the most common haplotype. One exception was Maoa, where several intermediate-frequency haplotypes were observed within M. domesticus. Within M. musculus, the pattern varied among loci, with some showing a single common haplotype (Msn) and others showing more intermediate-frequency haplotypes (Amelx, Maoa). Reciprocal monophyly between M. domesticus and M. musculus was unambiguously observed at Maoa, Dach2, and Msn (Figure 2). At Amelx, we observed two divergent lineages corresponding to M. musculus and M. domesticus, with one exception. A single M. musculus haplotype was identical to the most common M. domesticus haplotype over its entire 4-kb length, including all insertion/deletion (indel) variants, a microsatellite locus, and all nucleotide sites. Hybridization is known to occur between these two species not far from the sampling locality at which this mouse was trapped (Munclinger et al. 2002). Given the geographic origin of this mouse and its Amelx haplotype that is identical to haplotypes otherwise seen only in M. domesticus, this allele may represent recent introgression from M. domesticus into M. musculus. Excluding this individual, the haplotype network at Amelx is consistent with reciprocal monophyly. At Dmd, the haplotype network is unresolved, as the root (M. spretus) falls at a trichotomy, with one branch leading to all M. musculus haplotypes and two branches each leading to M. domesticus haplotypes. This pattern is not inconsistent with reciprocal monophyly between the species, but additional data are needed to resolve this trichotomy.
We performed several statistical tests of neutrality within M. domesticus and within M. musculus. First we performed tests based on the distribution of allele frequencies, including Tajima's D (Tajima 1989), Fu and Li's D (Fu and Li 1993), and Fay and Wu's H (Fay and Wu 2000). In M. domesticus, we generally observed negative values of Tajima's D and Fu and Li's D, consistent with an excess of rare polymorphisms, although most of these values were not significant and no locus showed significant values for all tests (Table 2). In M. musculus, both positive and negative values for Tajima's D and Fu and Li's D were observed, and none was significant. We also calculated Tajima's D for short and long indels within M. domesticus and within M. musculus, summed across loci. None of these values was significant (P > 0.05 for each). Second, we compared the ratio of polymorphism to divergence across multiple loci using the HKA test (Hudson et al. 1987). This test was applied in a pairwise manner and for all five loci simultaneously, using M. caroli as the outgroup. Neither the pairwise tests (Table 2) nor the multilocus test (sum of deviations = 6.389) rejected a neutral model.
Mice are known to live in highly structured demes at the local level (e.g., Delong 1967), but large-scale geographic structure was less evident when mapped onto a haplotype network for M. domesticus (supplemental Figure 4 at http://www.genetics.org/supplemental/). All geographic regions included the most common haplotype and several rare haplotypes. We calculated FST to assess the degree of population subdivision in M. domesticus. Values of FST obtained for each gene in comparisons among major geographic regions ranged from 0 to 0.5, with a mean value of 0.189 (Table 3). Some comparisons revealed significant structure. For example, the average FST value at Maoa was 0.312 and comparisons between geographic regions for this locus were all individually significant. An AMOVA analysis was carried out for a concatenated data set within M. domesticus. Consistent with a lack of strong population structure on a continental scale (across Western Europe), the greatest proportion of variation was found within populations (99.15%). Almost none of the variation was due to differences among regions (0.05%). In this multilocus analysis, FST was low and marginally significant (0.09, P = 0.06). Both φCT and φSC, measures of differentiation among populations and among regions, respectively, were low and nonsignificant (0.008, P = 0.09 and 0.0006, P = 0.95, respectively).
TABLE 3.
Pairwise FST values among groups of M. domesticus
| Locus | Locality | Mainland Italy | Greece | Sicily (Italy) | Average |
|---|---|---|---|---|---|
| Maoa | Mainland Italy | — | 0.312 | ||
| Greece | 0.214** | — | |||
| Sicily (Italy) | 0.170** | 0.504** | — | ||
| Spain | 0.346** | 0.465** | 0.175** | ||
| Dmd | Mainland Italy | — | 0.119 | ||
| Greece | 0.044 | — | |||
| Sicily (Italy) | 0.288** | 0.149 | — | ||
| Spain | 0.166** | 0.064 | 0.000 | ||
| Dach2 | Mainland Italy | — | 0.151 | ||
| Greece | 0.226* | — | |||
| Sicily (Italy) | 0.000 | 0.207* | — | ||
| Spain | 0.158** | 0.228 | 0.088 | ||
| Msn | Mainland Italy | — | 0.249 | ||
| Greece | 0.000 | — | |||
| Sicily (Italy) | 0.000 | 0.000 | — | ||
| Spain | 0.448** | 0.520** | 0.525** | ||
| Amelx | Mainland Italy | — | 0.113 | ||
| Greece | 0.165 | — | |||
| Sicily (Italy) | 0.000 | 0.135 | — | ||
| Spain | 0.122 | 0.179 | 0.076 |
*P < 0.05; **P < 0.01.
Levels of variation and gene genealogies for 11 X-linked and 18 autosomal loci:
We compared levels of variation at the five X-linked loci that we resequenced to levels of variation at six X-linked loci previously published (Table 4). Mean π was slightly higher for the six loci in Baines and Harr (2007) than for the five loci sequenced here, both within M. domesticus (0.10% vs. 0.07%) and within M. musculus (0.07% vs. 0.03%). These differences might be due to sampling differences or stochastic variation in levels of π among loci. The mean length of the five loci sequenced here was 4234 bp while the mean length of the six loci in Baines and Harr (2007) was 523 bp. We also compared the number of polymorphisms and fixed differences among the five X-linked loci that we resequenced and the six loci in Baines and Harr (2007), and we found no significant differences (FET, P = 0.869). These data sets are therefore pooled in the analyses below. The average level of nucleotide variability on the X chromosome (π = 0.08%) was considerably lower than on the autosomes (π = 0.26%), as previously noted (Baines and Harr 2007).
TABLE 4.
Levels of variation and numbers of fixed differences and polymorphisms for 11 X-linked and 18 autosomal loci in M. musculus and M. domesticus
| Variable sites by category
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
N
|
Length (bp) | π (%)
|
Total Sa | Fixed differences (musculus vs. domesticus) | Shared polymorphisms | Polymorphism within M. musculus | Polymorphism within M. domesticus | ||||||
| Locus | Chromosome | musculus | domesticus | musculus | domesticus | Genealogyb | Reference | ||||||
| Amelxc | X | 22 | 61 | 4200 | 0.05 | 0.15 | 49 | 13 | 0 | 7 | 29 | Mono | This study |
| Arx | X | 16 | 28 | 365 | 0.00 | 0.10 | 4 | 0 | 0 | 0 | 4 | Unresolved | Baines and Harr (2007) |
| Dach2 | X | 22 | 61 | 3913 | 0.03 | 0.03 | 18 | 4 | 0 | 3 | 11 | Mono | This study |
| Dmd | X | 18 | 64 | 5152 | 0.03 | 0.05 | 40 | 10 | 0 | 6 | 24 | Mono or Para | This study |
| Fate1 | X | 16 | 25 | 574 | 0.11 | 0.03 | 5 | 0 | 0 | 3 | 2 | Unresolved | Baines and Harr (2007) |
| Gm719 | X | 16 | 27 | 353 | 0.09 | 0.14 | 3 | 0 | 0 | 1 | 2 | Unresolved | Baines and Harr (2007) |
| Hadh2 | X | 16 | 27 | 1025 | 0.08 | 0.04 | 6 | 2 | 0 | 2 | 2 | Mono | Baines and Harr (2007) |
| Lamp2 | X | 16 | 25 | 391 | 0.09 | 0.06 | 5 | 1 | 0 | 2 | 2 | Mono or Para | Baines and Harr (2007) |
| Maoa | X | 22 | 60 | 4111 | 0.06 | 0.06 | 31 | 6 | 0 | 8 | 17 | Mono | This study |
| Msn | X | 19 | 61 | 4006 | 0.00 | 0.04 | 44 | 30 | 0 | 1 | 13 | Mono | This study |
| Tex16 | X | 12 | 28 | 431 | 0.06 | 0.20 | 7 | 3 | 0 | 2 | 2 | Mono or Para | Baines and Harr (2007) |
| Sum (X loci) | 24308 | — | — | 212 | 69 | 0 | 35 | 108 | |||||
| Mean (X loci) | 2210 | 0.05 | 0.08 | 19 | 6 | 0 | 3 | 10 | |||||
| Atp6v1c1 | 15 | 12 | 16 | 661 | 0.05 | 0.15 | 11 | 7 | 0 | 2 | 2 | Mono | Harr (2006a) |
| Bnc1 | 7 | 14 | 16 | 639 | 0.19 | 0.16 | 13 | 3 | 0 | 5 | 5 | Mono | Harr (2006a) |
| Dusp27 | 1 | 14 | 16 | 678 | 0.13 | 0.28 | 12 | 6 | 0 | 2 | 4 | Mono | Harr (2006a) |
| Ggh | 4 | 32 | 46 | 485 | 0.04 | 0.20 | 5 | 0 | 0 | 1 | 4 | Para or Poly | Harr (2006a); Baines and Harr (2007) |
| Melk | 4 | 32 | 36 | 923 | 0.22 | 0.57 | 27 | 1 | 1 | 10 | 15 | Para | Harr (2006a); Baines and Harr (2007) |
| Mme | 3 | 14 | 16 | 686 | 0.54 | 0.73 | 24 | 3 | 1 | 8 | 12 | Mono | Harr (2006a) |
| Nkd1 | 8 | 32 | 48 | 1182 | 0.18 | 0.20 | 16 | 0 | 2 | 5 | 9 | Poly | Baines and Harr (2007) |
| Nr1d2 | 14 | 14 | 16 | 631 | 0.09 | 0.00 | 2 | 1 | 0 | 1 | 0 | Unresolved | Harr (2006a) |
| Nudt7 | 8 | 32 | 48 | 1413 | 0.07 | 0.47 | 39 | 3 | 0 | 5 | 31 | Mono | Baines and Harr (2007) |
| Pcm1 | 8 | 14 | 16 | 623 | 0.00 | 0.06 | 3 | 1 | 0 | 0 | 2 | Para | Harr (2006a) |
| Ppp1r3b | 8 | 14 | 16 | 679 | 0.02 | 0.52 | 11 | 1 | 0 | 1 | 9 | Para | Harr (2006a) |
| Pum1 | 4 | 32 | 48 | 1209 | 0.02 | 0.04 | 9 | 0 | 0 | 4 | 5 | Unresolved | Baines and Harr (2007) |
| Q61875_MOUSE | 8 | 12 | 16 | 712 | 0.11 | 0.07 | 16 | 12 | 0 | 2 | 2 | Mono | Harr (2006a) |
| Rims2 | 15 | 14 | 16 | 407 | 0.06 | 0.16 | 7 | 4 | 0 | 1 | 2 | Mono | Harr (2006a) |
| Sec63 | 10 | 14 | 16 | 680 | 0.05 | 0.00 | 4 | 3 | 0 | 1 | 0 | Mono | Harr (2006a) |
| Sfrp1 | 8 | 32 | 48 | 1174 | 0.34 | 0.21 | 39 | 6 | 1 | 15 | 17 | Mono | Baines and Harr (2007) |
| Wdt2 | 1 | 10 | 12 | 287 | 0.37 | 0.28 | 4 | 0 | 1 | 2 | 1 | Poly | Harr (2006a) |
| XP_620246 | 5 | 14 | 16 | 448 | 0.21 | 0.45 | 9 | 0 | 0 | 2 | 7 | Para or Poly | Harr (2006a) |
| Sum (autosomes) | 14018 | — | — | 271 | 57 | 6 | 71 | 137 | |||||
| Mean (autosomes) | 738 | 0.16 | 0.26 | 14 | 3 | 0 | 4 | 7 | |||||
Total number of segregating sites.
Gene genealogies were classified as reciprocally monophyletic (Mono) between M. musculus and M. domesticus, paraphyletic in one species with respect to the other (Para), or polyphyletic in both species (Poly). Some gene genealogies were completely unresolved, as indicated, while others could be resolved to one of two possible topologies (e.g., Mono or Para).
All values for Amelx were calculated excluding one individual (MWN1379), captured near the hybrid zone, whose haplotype is believed to be introgressed.
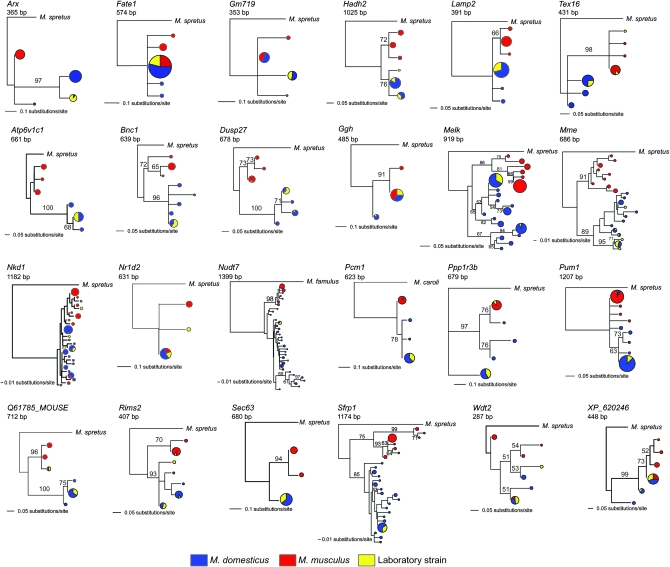
Neighbor-joining trees for the 11 X-linked and 18 autosomal loci are shown in Figures 2 and 3, and the numbers of fixed differences, shared polymorphisms, and exclusive polymorphisms are given in Table 4. All three kinds of genealogies that are depicted in Figure 1 can be seen among these 29 loci (Figures 2 and 3). Some loci showed reciprocal monophyly between M. musculus and M. domesticus (e.g., Bnc1), while others were paraphyletic (e.g., Melk) or polyphyletic (e.g., Nkd1). Some gene genealogies are unresolved due to an absence of informative sites, especially on the X chromosome, consistent with its low level of variation. Nonetheless, there are some interesting differences between the X chromosome and the autosomes. Excluding unresolved genealogies, the X chromosome included five monophyletic, no paraphyletic, and no polyphyletic gene genealogies, while the autosomes included nine monophyletic, three paraphyletic, and two polyphyletic gene genealogies. Thus, there was unambiguous evidence for paraphyly and polyphyly only at autosomal loci. Similarly, the proportion of fixed differences was greater on the X chromosome (69/212 = 33%) than on the autosomes (57/271 = 21%; Table 4). We compared the number of fixed differences, shared polymorphisms, polymorphisms within M. musculus, and polymorphisms within M. domesticus on the X chromosome (the counts in each category, respectively, are 69, 0, 35, and 108) and on the autosomes (57, 6, 71, and 137; Table 4). We used a Monte Carlo procedure (Lewontin and Felsenstein 1965) to ask whether the counts are significantly different between the X chromosome and the autosomes in this 2 × 4 contingency table. We generated 100,000 random tables with marginal sums equal to the observed data, using a program kindly provided by Bill Engels, and found that the observed table is highly unlikely (P = 0.0005). Similarly, the ratio of fixed differences to total polymorphisms was greater on the X (69:143) than on the autosomes (57:214; FET, P = 0.004). Thus, both the gene genealogies and the distribution of polymorphic and fixed nucleotide sites support the notion that the X chromosome is more differentiated than the autosomes between M. musculus and M. domesticus.
Figure 3.—
Neighbor-joining trees based on uncorrected distance matrices, rooted with M. spretus (except as noted), and based on data from Harr (2006a), Baines and Harr (2007), and nine classical inbred strains sequenced in this study (Top row) X-linked loci. (Other rows) Autosomal loci. Circles represent haplotypes at each terminal node, with circle size proportional to the number of chromosomes and colors designating sample identities. Blue, M. domesticus; red, M. musculus; yellow, classical inbred strain. Numbers adjacent to branches indicate bootstrap values >50 (500 replicates).
To further investigate the differentiation of the X chromosome compared to the autosomes, we analyzed data from the Wellcome Trust Center for Human Genetics in which ∼8500 SNPs were typed in seven wild-derived M. domesticus and eight wild-derived M. musculus (http://gscan.well.ox.ac.uk/gs/strains.cgi). Perl scripts were written to parse X-linked and autosomal variation. On the autosomes, we observed roughly equal numbers of fixed differences and shared polymorphisms, while on the X chromosome there were 112 fixed differences but only one shared polymorphism (Table 5). The relatively greater differentiation of the X chromosome in the Wellcome Trust data compared to the data shown in Table 4 may reflect bias in the ascertainment of SNPs in the Wellcome Trust data (Boursot and Belkhir 2006; Harr 2006a,b).
TABLE 5.
Fixed differences and polymorphisms among 15 wild-derived inbred strains from the Wellcome Trust database
| Fixed differences | Shared polymorphisms | Polymorphisms within M. domesticus | Polymorphisms within M. musculus | |
|---|---|---|---|---|
| Autosomes | 929 | 989 | 5753 | 561 |
| X chromosome | 112 | 1 | 133 | 5 |
Amount of wild variation captured by classical inbred strains:
The haplotypes seen in the nine classical inbred strains are shown in Figures 2 and 3. The number and origin of SNPs found among inbred strains for the 29 loci surveyed here are given in Table 6. These nine classical inbred strains capture only a small proportion of the variation seen in nature. For example, the strains were invariant at 12 of the 29 loci. Similarly, the inbred strains contained only 87 SNPs (Table 6), compared to 483 SNPs among wild mice at these same loci (Table 4).
TABLE 6.
Number and origin of SNPs among nine classical inbred strains for 11 X-linked and 18 autosomal loci
| Origin of SNP among inbred strains
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Chromosome | N | Length (bp) | Total Sa | Fixed differences (musculus vs. domesticus) | Shared polymorphisms | Polymorphism within M. musculus | Polymorphism within M. domesticus | Unsampled polymorphism | Species origin of inbred strain haplotype(s) |
| Amelx | X | 9 | 4067 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Arx | X | 9 | 365 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Dach2 | X | 9 | 3904 | 6 | 6 | 0 | 0 | 0 | 0 | Both species |
| Dmd | X | 9 | 5148 | 0 | 0 | 0 | 0 | 0 | 0 | Unsampled |
| Fate1 | X | 9 | 574 | 0 | 0 | 0 | 0 | 0 | 0 | Unresolved |
| Gm719 | X | 9 | 353 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Hadh2 | X | 9 | 1025 | 2 | 0 | 0 | 0 | 1 | 1 | domesticus and unsampled |
| Lamp2 | X | 9 | 391 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Maoa | X | 9 | 4081 | 0 | 0 | 0 | 0 | 0 | 0 | Unsampled |
| Msn | X | 9 | 3969 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Tex16 | X | 9 | 431 | 5 | 3 | 0 | 0 | 1 | 1 | Both species and unsampled |
| Sum (X loci) | 24308 | 13 | 9 | 0 | 0 | 2 | 2 | |||
| Mean (X loci) | 2210 | 1 | 1 | 0 | 0 | 0 | 0 | |||
| Atp6v1c1 | 15 | 8 | 661 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Bnc1 | 7 | 9 | 639 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Dusp27 | 1 | 9 | 678 | 7 | 6 | 0 | 0 | 1 | 0 | Both species |
| Ggh | 4 | 9 | 485 | 2 | 0 | 0 | 0 | 2 | 0 | Unresolved and domesticus |
| Melk | 4 | 9 | 923 | 11 | 0 | 1 | 0 | 10 | 0 | domesticus |
| Mme | 3 | 9 | 686 | 11 | 1 | 1 | 0 | 8 | 1 | domesticus and unsampled |
| Nkd1 | 8 | 7 | 1182 | 6 | 0 | 0 | 3 | 1 | 2 | domesticus and unsampled |
| Nr1d2 | 14 | 9 | 631 | 1 | 0 | 0 | 0 | 0 | 1 | Unresolved and unsampled |
| Nudt7 | 8 | 7 | 1413 | 7 | 0 | 0 | 0 | 5 | 2 | domesticus and unsampled |
| Pcm1 | 8 | 9 | 623 | 0 | 0 | 0 | 0 | 0 | 0 | Both species |
| Ppp1r3b | 8 | 9 | 679 | 7 | 1 | 0 | 0 | 6 | 0 | Both species |
| Pum1 | 4 | 8 | 1209 | 1 | 0 | 0 | 0 | 1 | 0 | Both species |
| Q61875_MOUSE | 8 | 9 | 712 | 13 | 11 | 0 | 1 | 1 | 0 | Both species |
| Rims2 | 15 | 9 | 407 | 5 | 3 | 0 | 0 | 1 | 1 | Both species and unsampled |
| Sec63 | 10 | 9 | 680 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Sfrp1 | 8 | 9 | 1174 | 0 | 0 | 0 | 0 | 0 | 0 | domesticus |
| Wdt2 | 1 | 9 | 287 | 2 | 0 | 1 | 0 | 0 | 1 | Unresolved and unsampled |
| XP_620246 | 5 | 9 | 448 | 1 | 0 | 0 | 0 | 1 | 0 | domesticus and unresolved |
| Sum (autosomes) | 13517 | 74 | 22 | 3 | 4 | 37 | 8 | |||
| Mean (autosomes) | 751 | 4 | 1 | 0 | 0 | 2 | 0 | |||
Total number of segregating sites.
There are notable differences between the X chromosome and the autosomes in the levels of variation captured in the inbred strains. The inbred strains were invariant at 8/11 = 73% of loci on the X chromosome and 4/18 = 22% of loci on the autosomes, a difference that was significant (FET, P = 0.02). This can also be seen in the number of SNPs on the X chromosome and on the autosomes; the inbred strains contained 6.1% of the SNPs present in the wild on the X chromosome and 26.6% of the SNPs present in the wild on the autosomes, a difference that was also significant (Table 7). Not only did the X chromosome contain less variation than the autosomes, but also the origin of the SNP variation on the X chromosome was different from the autosomes. The ratio of fixed differences to polymorphisms on the X (9/4) was significantly greater than on the autosomes (22/52; Table 7). In other words, much of the SNP variation among inbred strains on the X chromosome corresponds to differences between species, while most of the SNP variation among inbred strains on the autosomes corresponds to differences within species. It is important to bear in mind that this conclusion derives from consideration of only 29 loci in nine strains; sequencing of additional loci in samples of wild and laboratory mice will be needed to fully describe the origin of genetic variation among lab mice.
TABLE 7.
Comparison between the autosomes and the X chromosome for inbred strain SNPs
| Chromosome | No. of SNPs in wild mice | No. of SNPs in inbred strains | % variation captured by inbred strains | Fixed differences between species | Polymorphisms within species |
|---|---|---|---|---|---|
| Autosomes | 271 | 74 | 27.3 | 22 | 52 |
| X | 212 | 13 | 6.1 | 9 | 4 |
Inbred strains contain significantly fewer SNPs on the X chromosome than on the autosomes compared to wild mice (FET, P < 10−6). Inbred strains contain significantly more fixed differences on the X chromosome than on the autosomes compared to polymorphisms within species (FET, P < 10−2).
Our data allowed us to identify the species origin of individual haplotypes among inbred strains. We surveyed nine strains at 29 loci, representing 261 gene copies (supplemental Table 2 at http://www.genetics.org/supplemental/). Of these, 16.5% were of uncertain origin, usually because the corresponding haplotype was shared between M. musculus and M. domesticus, 5.4% were of M. musculus origin, and 75.1% were of M. domesticus origin. Thus, for these 29 loci, these inbred strains are predominantly of M. domesticus origin, consistent with other recent studies (Frazer et al. 2007; Yang et al. 2007).
Comparisons to MPD:
We were interested in asking more generally how SNP variation in our wild sample compares to SNP variation in existing databases, including all laboratory strains (both classical and wild derived). This is important since these laboratory strains represent the existing tools for most current mapping efforts. We were interested in asking two questions: (1) How many of the SNPs in existing databases were found in our survey of wild mice? and (2) How many of the SNPs in our sample of wild mice were found in existing databases? The MPD is the central repository for SNPs among all laboratory strains of mice. A total of 117 SNPs were found in MPD for the regions that we sequenced, and 107 of these were observed in wild mice (supplemental Table 3 at http://www.genetics.org/supplemental/). Thus, nearly all of the SNPs known from laboratory strains were captured in these wild samples. A total of 483 SNPs were identified in the samples of wild mice and 107 of these (∼22%) were found in MPD. Thus, wild mice provide a large reservoir of additional genetic variation not currently captured in laboratory strains.
DISCUSSION
We studied DNA sequence variation at 11 X-linked and 18 autosomal loci in wild and inbred mice. We draw four main conclusions:
Levels of genetic variation in wild mice are generally low, although slightly higher than in humans. Effective population sizes for mice are on the order of 105.
M. musculus and M. domesticus diverged recently, and many gene genealogies are reciprocally monophyletic between these species, while others are paraphyletic or polyphyletic. In general, the X chromosome is more differentiated than the autosomes.
Nine commonly used inbred strains contain only a small amount of the genetic variation seen in wild mice. The X chromosome contains proportionately less variation among inbred strains than do the autosomes. SNP variation on both the X chromosome and the autosomes derives from differences between species as well as differences within species, although the proportion of SNPs deriving from interspecific variation was greater on the X than on the autosomes for the genes that we surveyed.
Public SNP databases for the mouse, which include variants from all inbred strains, still contain only a small fraction of the SNPs seen in wild mice.
Below we discuss each of these conclusions in turn.
Levels and patterns of genetic variation in wild mice:
We resequenced five X-linked loci and found generally low levels of nucleotide variation and little geographic structure on a continental scale. Previous studies of genetic variation in natural populations of house mice have included work on allozymes (e.g., Selander et al. 1969a,b; Hunt and Selander 1973; Britton-Davidian et al. 1989), mtDNA (e.g., Nachman et al. 1994; Prager et al. 1996; Tryfonopoulos et al. 2005), MHC alleles (e.g., Arden and Klein 1982; Potts et al. 1991), t-haplotypes (e.g., Ardlie and Silver 1998; Dod et al. 2003), chromosomal polymorphisms (e.g., Pialek et al. 2005), microsatellites (e.g., Ihle et al. 2006), and DNA sequence variation at nuclear genes (e.g., Nachman and Aquadro 1994; Nachman 1997; Karn and Nachman 1999; Harr 2006a; Ihle et al. 2006; Baines and Harr 2007). Our results are concordant with some of these earlier studies in several respects. First, the average level of nucleotide diversity reported here for introns of five X-linked genes in M. domesticus (π = 0.07%) matches very nearly the value previously reported for 6022 bp of X-linked intron sequence from a sample of 10 M. domesticus (π = 0.08%; Nachman 1997) and 6 X-linked genes in six to eight mice from across the range of M. domesticus (π = 0.10%; Baines and Harr 2007). Second, studies of mtDNA and nuclear genes documented higher levels of genetic variation in M. domesticus than in M. musculus (Prager et al. 1996; Baines and Harr 2007), and this is corroborated by each of the five X-linked loci that we studied. Third, studies of allozymes (Britton-Davidian et al. 1989), mtDNA (Nachman et al. 1994), and nuclear DNA (Baines and Harr 2007) found relatively little geographic structure across Western Europe in M. domesticus, comparable to our findings. The lack of strong geographic structure for M. domesticus is consistent with an archaeological record indicating that this species expanded its range into Western Europe from the Fertile Crescent within the past 10,000 years (Auffray et al. 1990; Cucchi et al. 2005). The generally negative values of Tajima's D and Fu and Li's D suggest a population expansion, and this may have accompanied the known range expansion.
The observed levels of variation at X-linked loci suggest a long-term effective population size for mice of 58,000 for M. domesticus and 25,000 for M. musculus. As pointed out by Baines and Harr (2007), however, levels of variation on the X chromosome may be reduced by selection, relative to levels of variation on the autosomes. The average level of nucleotide variation on the autosomes (Baines and Harr 2007) suggests an effective population size of 160,000 for M. domesticus and 100,000 for M. musculus [M. domesticus: Ne = θ/4μ = (2.6 × 10−3)/(1.6 × 10−8) = 1.6 × 105; M. musculus, Ne = (1.6 × 10−3)/(1.6 × 10−8) = 105]. Given uncertainties in the estimates of mutation rate, these values should be taken as rough approximations; however, it seems that M. domesticus has a long-term species-wide Ne on the order of 105.
This study allows us to compare patterns of DNA sequence variation in mice to patterns seen in humans, the only other mammalian species for which extensive population samples of DNA sequence variation have been obtained. Levels of variation on the X chromosome in mice are nearly identical to levels of nucleotide diversity at X-linked introns in humans (π = 0.07%; Hammer et al. 2004), although levels of variation on the autosomes appear to be roughly twice as high in mice (π = 0.26%: Table 4) as in humans (π = 0.11%; Li and Sadler 1991; Aquadro et al. 2001). Similarly negative values of Tajima's D are seen in non-African populations of humans and in European populations of mice at X-linked loci (Hammer et al. 2004), probably consistent with population expansions associated with range expansions in both species. Levels of intralocus LD are also similarly high in both species (e.g., Hammer et al. 2004). Recombination rates in mice (∼0.5 cM/Mb; Shifman et al. 2006) are roughly half as high as in humans (∼1 cM/Mb; Kong et al. 2002); however, the larger population size of mice suggests that the population recombination rate (i.e., 4Nec) may be roughly similar in both species. Recent work has shown that the decay of LD occurs over similar genomic distances in mice and humans (Laurie et al. 2007). The similarities between mice and humans suggest that wild mice will provide a useful comparison to humans for understanding the forces governing genetic variation in nature. In addition, wild mice might serve as useful models for genetic association studies.
Divergence between M. musculus and M. domesticus:
Our results suggest that M. musculus and M. domesticus diverged <500,000, or ∼5Ne, generations ago. The average coalescence time for all alleles in a neutral genealogy is 4Ne generations; however, the variance in the coalescent process is large, and even after 5Ne generations, reciprocal monophyly is not expected for all loci between diverging taxa (Tajima 1983). Consistent with these theoretical expectations, we observed all three patterns in Figure 1 in our data (Figures 2 and 3). Nonetheless, 14/19 = 74% of all unambiguous genealogies were reciprocally monophyletic between M. domesticus and M. musculus. These data highlight the fact that these groups are genetically well differentiated.
Despite this overall high level of differentiation, the X chromosome appears to be more differentiated than the autosomes. Excluding the unresolved genealogies, all X-linked loci were reciprocally monophyletic, while only 64% of autosomal loci were reciprocally monophyletic. We found clear evidence for paraphyly or polyphyly only at autosomal loci. Similarly, the ratio of polymorphic to fixed nucleotide differences was significantly lower on the X chromosome than on the autosomes. The greater differentiation of the X chromosome compared to the autosomes is also seen in our analysis of the Wellcome Trust data (Table 5), although these SNPs were ascertained in classical inbred strains and then typed in wild-derived inbreds and thus may contain some bias (Harr 2006a,b; Boursot and Belkhir 2006).
The greater differentiation of the X chromosome compared to the autosomes may be due to several factors. One is the difference in effective population size. The average coalescence time for all alleles is 3Ne generations for the X chromosome and 4Ne generations for autosomes, and thus the X is expected to achieve reciprocal monophyly more quickly. Another possible explanation comes from levels of gene flow across the musculus–domesticus hybrid zone. A number of studies have documented reduced introgression of the X chromosome compared to the autosomes (Tucker et al. 1992; Dod et al. 1993; Prager et al. 1997), and reproductive incompatibilities map to the X chromosome (Oka et al. 2004; Storchova et al. 2004). Third, if positive selection is more frequent on the X chromosome than on the autosomes (e.g., Charlesworth et al. 1987), then the X chromosome would be expected to exhibit shallower gene genealogies and reciprocal monophyly more often than the autosomes. Finally, it is possible that additional sampling will uncover X-linked genes with shared polymorphism.
Amount and pattern of genetic variation among inbred strains:
This study is the first to compare variation at multiple loci in the classical inbred strains of mice to variation in large samples of wild mice. Three important patterns emerge.
First, the inbred strains of mice capture a small percentage of the variation seen in nature. The inbred strains were invariant at 41% of the loci surveyed (12/29) and contained 85 SNPs compared to 483 in wild mice. The lack of variation among inbred strains has important implications for their use in identifying genes underlying traits of interest. There are good mouse models for many complex diseases (Peters et al. 2007). If laboratory strains are used to identify the genetic architecture of disease phenotypes and if laboratory strains contain a small fraction of the variation seen in nature, then some genes of importance may go undetected. For example, the recent “collaborative cross” uses a set of eight inbred lines that will be intercrossed and then used to produce a set of 1000 recombinant inbred lines (Churchill et al. 2004). This panel will provide a powerful tool for mapping genes to ∼1 cM resolution. The total amount of variation present in this panel, however, is limited by the variation present in the initial founders. It is important to recognize that inbred strains of mice may contain a sufficient number of SNPs to “tag” nearly every gene in the genome. However, the reduction in variation in lab strains compared to wild mice is likely to have excluded many functionally important alleles, especially if they were rare in the wild. Wild mice could provide an additional source of functional genetic variation for mapping efforts. Yang et al. (2007) note that the genomes of laboratory mice contain large regions of extremely low diversity and that these represent “blind spots” for the study of complex traits. Our results suggest that wild mice could fill in these genetic blind spots.
Second, despite the overall low level of variation seen among inbred strains, there is significantly less variation on the X compared to autosomes, and this difference is greater than expected on the basis of levels of variation seen in the wild (Table 7). Thus, X chromosome variation is underrepresented in lab strains of mice. This could be caused by a small ratio of females to males in the founding of inbred strains (Ferris et al. 1982). Another possibility is that the X chromosome contained a large number of incompatibilities in the crosses that were used to establish lab strains, resulting in selection against many X-linked alleles. The fact that hybrid male sterility maps to the X chromosome (Oka et al. 2004; Storchova et al. 2004) is consistent with this view.
Third, among inbred strains, most of the SNPs on the X derive from differences between species, while most of the SNPs on the autosomes derive from differences within species at the genes that we surveyed (Table 7). The genes contributing to fixed differences on the X in our study, Tex16 and Dach2, lie in a large region also identified by Yang et al. (2007) as deriving from different species (on the basis of a single inbred mouse from each species). However, for both the X and the autosomes, a nontrivial proportion of SNPs derive from fixed differences between species. Such species-specific alleles have not been tested by natural selection in combination with species-specific alleles at other loci, and this may give rise to epistatic incompatibilities (e.g., Payseur and Hoekstra 2005). The extent to which such interactions underlie phenotypes of interest in mouse models is unknown, but it is clear that such trans-species polymorphisms do not serve as an accurate model for most human genetic variation.
SNP databases include a small fraction of the variation in nature:
MPD includes a database of all mouse SNPs compiled from many sources. The database contains >10 million SNPs representing >100 classical and wild-derived inbred strains of mice. It is not comprehensive in the sense that not all strains have been sequenced for all genomic regions, but it probably contains a large fraction of all SNPs among all strains since it includes the >8 million Perlegen SNPs derived from resequencing the genomes of 15 major inbred strains (Frazer et al. 2007). The Perlegen data are based on oligonucleotide arrays that cover ∼58% of the reference genome (C57Bl/6J) with a high false-negative rate (∼40%). Although there are many genomic gaps in these data, including many of the regions that we studied here, they represent a very large catalog of SNPs among mouse strains. Nonetheless, we identified only 117 SNPs in MPD at these 29 autosomal and X-linked loci, corresponding to 24% of the SNPs discovered among wild mice (supplemental Table 3 at http://www.genetics.org/supplemental/). This underrepresentation of variation supports the conclusion that studying even a diverse collection of inbred strains of mice may miss important alleles underlying complex traits. Fixed differences between M. domesticus and M. musculus constitute a large proportion of the SNPs in MPD at these 29 loci (supplemental Table 3) on both the X (62%) and the autosomes (40%), which further suggests that the genetic architecture of traits mapped using these strains may not correspond to the architecture of the same traits in human populations.
Acknowledgments
P. Basset, M. D. Dean, J. M. Good, E. Kelleher, L. Kent, M. Sans-Fuentes, M. A. Saunders, and G. Wlasiuk provided useful discussion and suggestions. We also thank D. J. Begun and three anonymous reviewers for helpful comments on the manuscript, J. G. Krenz and M. A. D'Urso for technical assistance, and M. D. Dean and M. Carneiro for computational assistance. T.S. was supported by a fellowship from the National Science Foundation (NSF) Integrative Graduate Education and Research Traineeship Program in Evolutionary, Functional and Computational Genomics at the University of Arizona. A.G. was supported by a fellowship from the Fundacao para a Ciencia e a Tecnologia (SFRH/BPD/24743/2005). This work was supported by NSF and National Institutes of Health grants to M.W.N.
References
- Aquadro, C. F., V. B. DuMont and F. A. Reed, 2001. Genome-wide variation in the human and fruitfly: a comparison. Curr. Opin. Genet. Dev. 11: 627–634. [DOI] [PubMed] [Google Scholar]
- Arden, B., and J. Klein, 1982. Biochemical comparison of major histocompatibility complex molecules from different subspecies of Mus musculus: evidence for trans-specific evolution of alleles. Proc. Natl. Acad. Sci. USA 79: 2342–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie, K. G., and L. M. Silver, 1998. Low frequency of t haplotypes in natural populations of house mice (Mus musculus domesticus). Evolution 52: 1185–1196. [DOI] [PubMed] [Google Scholar]
- Auffray, J. C., F. Vanlerberghe and J. Britton-Davidian, 1990. The house mouse progression in Eurasia: a paleontological and archaeozoological approach. Biol. J. Linn. Soc. 41: 13–25. [Google Scholar]
- Avise, J. C., 1994. Molecular Markers, Natural History, and Evolution. Chapman & Hall, New York.
- Baines, J. F., and B. Harr, 2007. Reduced X-linked diversity in derived populations of house mice. Genetics 175: 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot, P., and K. Belkhir, 2006. Mouse SNPs for evolutionary biology: beware of ascertainment biases. Genome Res. 16: 1191–1192. [DOI] [PubMed] [Google Scholar]
- Boursot, P., W. Din, R. Anand, D. Darviche, B. Dod et al., 1996. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J. Evol. Biol. 9: 391–415. [Google Scholar]
- Braverman, J. M., R. R. Hudson, N. L. Kaplan, C. H. Langley and W. Stephan, 1995. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140: 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian, J., J. H. Nadeau, H. Croset and L. Thaler, 1989. Genic differentiation and origin of Robertsonian populations of the house mouse (Mus musculus domesticus Rutty). Genet. Res. 53: 29–44. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian, J., F. Fel-Clair, J. Lopez, P. Alibert and P. Boursot, 2005. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol. J. Linn. Soc. 84: 379–393. [Google Scholar]
- Charlesworth, B., J. A. Coyne and N. H. Barton, 1987. The relative rates of evolution of sex-chromosomes and autosomes. Am. Nat. 130: 113–146. [Google Scholar]
- Churchill, G., D. C. Airey, H. Allayee, J. M. Angel, A. D. Attie et al., 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Cucchi, T., J. D. Vigne and J. C. Auffray, 2005. First occurrence of the house mouse (Mus musculus domesticus Schwarz & Schwarz, 1943) in the Western Mediterranean: a zooarchaeological revision of subfossil occurrences. Biol. J. Linn. Soc. 84: 429–445. [Google Scholar]
- Delong, K. T., 1967. Population ecology of feral house mice. Ecology 48: 611. [Google Scholar]
- Dod, B., L. S. Jermiin, P. Boursot, V. H. Chapman, J. T. Nielsen et al., 1993. Counterselection on sex-chromosomes in the Mus musculus European hybrid zone. J. Evol. Biol. 6: 529–546. [Google Scholar]
- Dod, B., C. Litel, P. Makoundou, A. Orth and P. Boursot, 2003. Identification and characterization of t haplotypes in wild mice populations using molecular markers. Genet. Res. 81: 103–114. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, S. D., R. D. Sage and A. C. Wilson, 1982. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature 295: 163–165. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., C. M. Wade, D. A. Hinds, N. Patil, D. R. Cox et al., 2004. Segmental phylogenetic relationships of inbred mouse strains revealed by fine-scale analysis of sequence variation across 4.6 Mb of mouse genome. Genome Res. 14: 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K. A., E. Eskin, H. M. Kang, M. A. Bogue, D. A. Hinds et al., 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448: 1050–1053. [DOI] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, D., 2006. A mouse for every gene. Science 312: 1862–1866. [DOI] [PubMed] [Google Scholar]
- Hammer, M. F., D. Garrigan, E. Wood, J. A. Wilder, Z. Mobasher et al., 2004. Heterogeneous patterns of variation among multiple human X-linked loci: the possible role of diversity-reducing selection in non-Africans. Genetics 167: 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr, B., 2006. a Genomic islands of differentiation between house mouse subspecies. Genome Res. 16: 730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr, B., 2006. b Regions of high differentiation—worth a check. Genome Res. 16: 1193–1194. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguade, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, W. G., and R. K. Selander, 1973. Biochemical genetics of hybridization in European house mice. Heredity 31: 11–33. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah, F. Y., E. de la Casa-Esperon, T. A. Bell, D. A. Detwiler, T. Magnuson et al., 2004. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res. 14: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle, S., I. Ravaoarimanana, M. Thomas and D. Tautz, 2006. An analysis of signatures of selective sweeps in natural populations of the house mouse. Mol. Biol. Evol. 23: 790–797. [DOI] [PubMed] [Google Scholar]
- Karn, R. C., and M. W. Nachman, 1999. Reduced nucleotide variability at an androgen-binding protein locus (Abpa) in house mice: evidence for positive natural selection. Mol. Biol. Evol. 16: 1192–1197. [DOI] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Laurie, C. C., D. A. Nickerson, A. D. Anderson, B. S. Weir, R. J. Livingston et al., 2007. Linkage disequilibrium in wild mice. PLoS Genet. 3: e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon, W. J., C. H. Swinton, M. Wang, N. Berbari, Y. Wang et al., 2003. Single nucleotide polymorphism (SNP) analysis of mouse pulmonary adenoma susceptibility loci 1–4 for identification of candidate genes. J. Med. Genet. 40: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin, R. C., and J. Felsenstein, 1965. Robustness of homogeneity tests in 2xn tables. Biometrics 21: 19. [Google Scholar]
- Li, W.-H., and L. A. Sadler, 1991. Low nucleotide diversity in man. Genetics 129: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh, K., E. Winchester, M. J. Daly, D. G. Wang, J. N. Hirschhorn et al., 2000. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24: 381–386. [DOI] [PubMed] [Google Scholar]
- Munclinger, P., E. Bozikova, M. Sugerkova, J. Pialek and M. Macholan, 2002. Genetic variation in house mice (Mus, Muridae, Rodentia) from the Czech and Slovak republics. Folia Zool. 51: 81–92. [Google Scholar]
- Nachman, M. W., 1997. Patterns of DNA variability at X-linked loci in Mus domesticus. Genetics 147: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., and C. F. Aquadro, 1994. Polymorphism and divergence at the 5′ flanking region of the sex-determining locus, Sry, in mice. Mol. Biol. Evol. 11: 539–547. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., S. N. Boyer, J. B. Searle and C. F. Aquadro, 1994. Mitochondrial DNA variation and the evolution of Robertsonian chromosomal races of house mice, Mus domesticus. Genetics 136: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, A., A. Mita, N. Sakurai-Yamatani, H. Yamamoto, N. Takagi et al., 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen, K., 2003. a One hundred years of mouse genetics: an intellectual history. I. The classical period (1902–1980). Genetics 163: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen, K., 2003. b One hundred years of mouse genetics: an intellectual history. II. The molecular revolution (1981–2002). Genetics 163: 1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur, B. A., and H. E. Hoekstra, 2005. Signatures of reproductive isolation in patterns of single nucleotide diversity across inbred strains of mice. Genetics 171: 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, L. L., R. F. Robledo, C. J. Bult, G. A. Churchill, B. J. Paigen et al., 2007. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 8: 58–69. [DOI] [PubMed] [Google Scholar]
- Petkov, P. M., Y. M. Ding, M. A. Cassell, W. D. Zhang, G. Wagner et al., 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14: 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov, P. M., J. H. Graber, G. A. Churchill, K. DiPetrillo, B. L. King et al., 2005. Evidence of a large-scale functional organization of mammalian chromosomes. PLoS Genet. 1: 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pialek, J., H. C. Hauffe and J. B. Searle, 2005. Chromosomal variation in the house mouse. Biol. J. Linn. Soc. 84: 535–563. [Google Scholar]
- Pletcher, M. T., P. McClurg, S. Batalov, A. I. Su, S. W. Barnes et al., 2004. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2: 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts, W. K., C. J. Manning and E. K. Wakeland, 1991. Mating patterns in seminatural populations of mice influenced by MHC genotype. Nature 352: 619–621. [DOI] [PubMed] [Google Scholar]
- Prager, E. M., H. Tichy and R. D. Sage, 1996. Mitochondrial DNA sequence variation in the eastern house mouse, Mus musculus: comparison with other house mice and report of a 75-bp tandem repeat. Genetics 143: 427–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager, E. M., P. Boursot and R. D. Sage, 1997. New assays for Y chromosome and p53 pseudogene clines among East Holstein house mice. Mamm. Genome 8: 279–281. [DOI] [PubMed] [Google Scholar]
- Przeworski, M., 2002. The signature of positive selection at randomly chosen loci. Genetics 160: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Misener and S. A. Krawetz. Humana. Press, Totowa, NJ. [DOI] [PubMed]
- Schneider, S., D. Roessli and L. Excoffier, 2000. Arlequin: a software for population genetics data analysis 2.0. University of Geneva, Geneva.
- Selander, R. K., W. G. Hunt and S. Y. Yang, 1969. a Protein polymorphism and genic heterozygosity in 2 European subspecies of house mouse. Evolution 23: 379–390. [DOI] [PubMed] [Google Scholar]
- Selander, R. K., S. Y. Yang and W. G. Hunt, 1969. b XVI. Polymorphism in esterases and hemoglobin in wild populations of the house mouse (Mus musculus). Studies in Genetics V. Univ. Texas Publ. 6918: 271–338. [Google Scholar]
- She, J. X., F. Bonhomme, P. Boursot, L. Thaler and F. Catzeflis, 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linn. Soc. 41: 83–103. [Google Scholar]
- Shifman, S., J. T. Bell, R. R. Copley, M. S. Taylor, R. W. Williams et al., 2006. A high resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 4: 2227–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, L. M., 1995. Mouse Genetics: Concepts and Applications. Oxford University Press, New York.
- Simonsen, K. L., G. A. Churchill and C. F. Aquadro, 1995. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141: 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley and P. Green, 1996. –2004 RepeatMasker Open-3.0. http://www.repeatmasker.org/.
- Storchova, R., S. Gregorova, D. Buckiova, V. Kyselova, P. Divina et al., 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524. [DOI] [PubMed] [Google Scholar]
- Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching et al., 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101: 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., T. Shimada, M. Terashima, K. Tsuchiya and K. Aplin, 2004. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 33: 626–646. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 3b10. Sinauer Associates, Sunderland, MA.
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryfonopoulos, G. A., B. R. Chondropoulos and S. E. Fraguedakis-Tsolis, 2005. Mitochondrial DNA polymorphisms of the house mouse Mus musculus domesticus from Greece, focusing on the Robertsonian chromosomal system of north-west Peloponnese. Biol. J. Linn. Soc. 84: 643–651. [Google Scholar]
- Tucker, P. K., R. D. Sage, J. Warner, A. C. Wilson and E. M. Eicher, 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146–1163. [DOI] [PubMed] [Google Scholar]
- Wade, C. M., E. J. Kulbokas, III, A. W. Kirby, M. C. Zody, J. C. Mullikin et al., 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420: 574–578. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril et al., 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Wiltshire, T., M. T. Pletcher, S. Batalov, S. W. Barnes, L. M. Tarantino et al., 2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. USA 100: 3380–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin, B., J. Fullerton, S. Miller, D. A. Keays, S. Brady et al., 2004. Unexpected complexity in the haplotypes of commonly used inbred strains of laboratory mice. Proc. Natl. Acad. Sci. USA 101: 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., T. A. Bell, G. A. Churchill and F. Pardo-Manuel de Villena, 2007. On the subspecific origin of the laboratory mouse. Nat. Genet. 39: 1100–1107. [DOI] [PubMed] [Google Scholar]