Abstract
Repetitive minisatellite DNA tracts are stable in mitotic cells but unstable in meiosis, altering in repeat number and repeat composition. As relatively little is known about the factors that influence minisatellite stability, we isolated mutations that destabilize a minisatellite repeat tract in the ADE2 gene of Saccharomyces cerevisiae. One mutant class exhibited a novel color segregation phenotype, “blebbing,” characterized by minisatellite instability during stationary phase. Minisatellite tract alterations in blebbing strains consist exclusively of the loss of one 20-bp repeat. Timing experiments suggest that these tract alterations occur only after cells have entered stationary phase. Two complementation groups identified in this screen have mutations in either the high-affinity zinc transporter ZRT1 or its zinc-dependent transcriptional regulator ZAP1. The Δzrt1 mutant specifically affects the stability of minisatellite tracts; microsatellites or simple insertions in the ADE2 reading frame are not destabilized by loss of ZRT1. The Δzrt1 blebbing phenotype is partially dependent on a functional RAD50. Zinc is known for its role as an essential cofactor in many DNA-binding proteins. We describe possible models by which zinc can influence minisatellite stability. Our findings directly implicate zinc homeostasis in the maintenance of genomic stability during stationary phase.
DIFFERENT forms of repetitive DNA exist in eukaryotic genomes (Debrauwere et al. 1997; Niwa 2006). Microsatellite tracts consist of short identical sequences that are directly repeated. They can be destabilized by replication error during mitosis, but are relatively stable during meiosis. In contrast, minisatellite tracts have longer (16–100 bp) moderately variable repeats, which allows for complex arrangements of repeat types. Minisatellites are stable during vegetative growth, but can change in length and repeat-type composition during meiosis. Minisatellites have been implicated in transcriptional gene regulation and proper chromosome segregation and may act as fragile sites (Vergnaud and Denoeud 2000). To date, only a few of the factors mediating minisatellite stability have been identified.
To investigate mechanisms governing minisatellite stability, we previously developed a novel yeast minisatellite model system. We demonstrated that our model system, using the repetitive tract associated with the human HRAS1 oncogene, recapitulates in yeast all of the phenotypes associated with minisatellites in mammals (Jauert et al. 2002). We have shown that minisatellite stability during meiosis is dependent on the meiotic recombination-initiating protein Spo11p and on the DNA loop mismatch repair activity of Rad1p. In addition, other work has shown that some minisatellite tracts are affected during mitosis by mutations in genes that affect DNA replication: the loss of Rad27p, the FEN-1 endonuclease active in Okazaki fragment processing, or mutations in PCNA or POL3 lead to an increase in minisatellite tract alterations (Lopes et al. 2002; Maleki et al. 2002). The identification of minisatellite stability factors will provide us with insight into the mechanisms governing some types of genome rearrangements. This is an important advance as alterations in minisatellites have been linked to changes in transcription levels of nearby genes and to human disease phenotypes, including oncogenesis. Rare alleles of the minisatellite repeat tract adjacent to the HRAS1 oncogene in humans have been correlated with breast, colon, and urinary tract cancer and acute leukemia (Krontiris et al. 1993; Krontiris 1995a,b; Ding et al. 1999; Vega et al. 2001a,b). Tract changes in minisatellites associated with the cystatin B and IDDM2 genes may be responsible for progressive myoclonus epilepsy and insulin-dependent diabetes mellitus, respectively (Kennedy et al. 1995; Lafreniere et al. 1997; Virtaneva et al. 1997).
To identify novel factors involved in minisatellite stability, we conducted a screen for mutants that destabilized a minisatellite repeat construct inserted into the coding sequence of ADE2. We were able to efficiently identify mutants that increase the instability of the minisatellite tract by their color segregation phenotypes.
In this report, we describe a particularly interesting class of mutants that display a novel color segregation phenotype that we have designated “blebbing.” These mutants are characterized by minisatellite repeat loss that occurs exclusively during stationary phase. The majority of mutants that we isolated in this class fall into two complementation groups, with mutations in the genes encoding the high-affinity zinc transporter ZRT1 and the zinc-dependent transcription factor ZAP1. Interestingly, loss of the zinc transporter gene destabilized only minisatellite tracts; microsatellites and simple insertions were not similarly affected. While the mechanism responsible for zinc-mediated minisatellite stability is not yet fully elucidated, our results show that it is partially dependent on the recombination factor RAD50.
MATERIALS AND METHODS
Media, plasmids, and strains:
Standard media were used (Guthrie and Fink 1991). YPD + G418 media consisted of standard YPD solid media plus 200 mg/liter of G418 sulfate (geneticin). Strains were sporulated and dissected as described in Jauert et al. (2002).
To construct the plasmid pDTK123, used to integrate the ade2-min3 allele's 20-bp repeats into the ADE2 gene (see Figure 1), oligos containing the Min3 repeats with the XbaI flanking ends 5′-TCGA(CAACGCAATGCGTTGGATCT)3A-3′ and 5′-TCGAT(AGATCCAACGCATTGCGTTG)3-3′ were annealed and ligated into the XbaI site in the ADE2 region of the plasmid pEAS8 (Sia et al. 2001). Plasmids pEAS10 and pEAS19 were constructed in the same way, using 29 GT repeats and a 17-bp poly(C) sequence, respectively (Sia et al. 2001). The pDTK140 plasmid was constructed by digesting the pEAS8 plasmid with XbaI and filling in the ends with Klenow. The blunted ends were then ligated together, introducing a 4-bp insertion into the ADE2 region. All insertions were verified by sequencing.
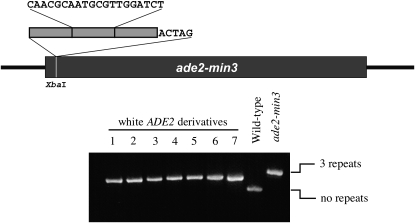
Figure 1.—
(Top) The location and sequence of the ade2-min3 allele. Three copies of the indicated sequence, plus one additional base, were tandemly inserted into an XbaI site, duplicating the four-base overhang of the site and changing the ADE2 reading frame. Loss of one repeat will restore the proper reading frame. (Bottom) Whole-colony PCR products generated from white Ade+ derivatives of the ade2-min3 parental strain compared to similar products from the ade2-min3 strain and the original ADE2 parental strain. All of the Ade+ derivatives have lost one repeat; this result was verified by sequencing of the PCR products.
The ZAP1 plasmid pPAJ199 was isolated from a yeast plasmid library of random DNA fragments, using a colony hybridization protocol. Approximately 4000 colony-forming units (CFUs) from the G418-resistant genomic library (Jauert et al. 2005) were grown on a 150-mm LB plate containing 100 mg/liter ampicillin. Colony lifts were then performed using the procedure given in the DIG application manual for filter hybridization (Roche Applied Science). Briefly, colonies were replica plated to a disc of Magnalift nylon membrane (Osmonics). The colonies were lysed and the DNA was UV crosslinked to the membrane. A probe was made by PCR, using digoxiginin-11-dUTP and oligos 5′-ACGGCTGGAGCAACTTCAACT-3′ and 5′-TGTGTTTGTGGGTTGGGGCAAT-3′ to amplify a 677-bp region of ZAP1. After hybridization to the filter, it was processed and exposed to film. Colonies containing library plasmids with ZAP1 inserts were identified and the ends of the inserts were sequenced. The plasmid pPAJ199 contained genomic sequences from 329,352 to 336,006 on chromosome X.
All Saccharomyces cerevisiae strains were derived from EAS28 (MATa his7-2 trp1-289 ura3-52) from Sia et al. (2001) (Table 1). DTK260 was constructed by transforming plasmid pNKY85 (Alani et al. 1987) into EAS28, disrupting LEU2 by insertion of the Δleu2∷HisG construct. The ade2-min3 strain DTK264 was made by transforming DTK260 with BglII-digested pDTK123. Ura− derivatives were selected on 5-fluoroorotic acid (5-FOA) plates. The arg8∷HisG construct was introduced into DTK264 using pDS27 (arg8∷HisG-URA3-HisG from T. Fox), resulting in strain DTK284. DTK264 was mating type switched to MATα using the pGal-HO plasmid (Herskowitz and Jensen 1991). Subsequently, transformants were grown on 5-FOA plates to identify MATα cells without the plasmid, resulting in strain DTK271. DTK260 was transformed with BglII-digested plasmids pEAS10, pEAS19, and pDTK140, creating strains DTK885, DTK887, and DTK889, respectively, and then Ura− derivatives were selected as described above, generating strains DTK886, DTK888, and DTK890, respectively.
TABLE 1.
Yeast strains
| Strain | Relevant genotype | Construction details |
|---|---|---|
| EAS28 | Wild type | MATahis7-2 trp1-289 ura3-52 (Sia et al. 2001) |
| DTK260 | leu2∷HisG | EAS28 with pNKY85 |
| DTK264 | ade2-min3 | DTK260 with pDTK123 |
| DTK271 | ade2-min3, MATα | DTK264 with pGal-HO (Herskowitz and Jensen 1991) |
| DTK284 | ade2-min3, arg8∷HisG | DTK264 with pDS27 |
| DTK886 | ade2-GT(29) | DTK260 with pEAS10 (Sia et al. 2001) |
| DTK888 | ade2-C(17) | DTK260 with pEAS19 (Sia et al. 2001) |
| DTK890 | ade2-CTAG | DTK260 with pDTK140 |
| DTK878 | ade2-min3, zrt1∷KAN | DTK271 with zrt1∷KANa |
| DTK900 | ade2-GT(29), zrt1∷KAN | DTK886 with zrt1∷KANa |
| DTK913 | ade2-C(17), zrt1∷KAN | DTK888 with zrt1∷KANa |
| DTK901 | ade2-CTAG, zrt1∷KAN | DTK890 with zrt1∷KANa |
| DTK904 | ade2-min3, zrt1∷LEU2 | DTK284 with zrt1∷LEU2a |
| DTK978 | ade2-min3, zrt2∷KAN | DTK271 with zrt2∷KANa |
| DTK977 | ade2-min3, zrt1∷LEU2, zrt2∷KAN | DTK904 with zrt2∷KANa |
| DTK902 | ade2-min3, zap1∷KAN | DTK284 with zap1∷KANa |
| DTK921 | ade2-min3, zrt1∷KAN, zap1∷KAN | DTK878 × DTK902, isolated spore |
| DTK1056 | ade2-min3, rad50∷KAN | DTK271 with rad50∷KANa |
| DTK1057 | ade2-min3, zrt1∷LEU2, rad50∷KAN | DTK904 with rad50∷KANa |
| DTK1033 | zrt3∷KAN | Spore isolated from Yeast Deletion Consortium strain dissection |
| DTK1068 | ade2-min3, zrt3∷KAN | DTK271 × DTK1033, isolated spore |
| DTK1077 | ade2-min3, yke4∷KAN | DTK271 with yke4∷KANa |
| DTK1078 | ade2-min3, zrc1∷KAN | DTK271 with zrc1∷KANa |
| DTK1082 | ade2-min3, cot1∷KAN | DTK271 with cot1∷KANa |
Indicates that the strain was made using a PCR-generated construct.
To construct strains DTK878 and DTK902, bearing deletions of the genes ZRT1 and ZAP1, respectively, PCR products were made using oligos 12966370 and 12966371 for ZRT1 and 14767981 and 14767982 for ZAP1 with genomic DNA as templates from the Δzrt1:KANMX4 or Δzap1:KANMX4 Yeast Deletion Consortium strains. To construct strains DTK1057, DTK1077, DTK1078, and DTK1082, bearing deletions in RAD50, YKE4, ZRC1, and COT1, respectively, PCR products were made using oligos 20619256 and 20619257 for Δrad50:KANMX4, 27016542 and 27016543 for Δcot1:KANMX4, 26910872 and 26910873 for Δyke4:KANMX4, and 27016544 and 27016545 for Δzrc1:KANMX4, with genomic DNA from the corresponding Yeast Deletion Consortium strains. These PCR products contained the G418 resistance gene flanked with 3′ and 5′ homology to the target sequence. Strains transformed with these products were grown for 4 hr in liquid YPD and then plated on YPD + G418 solid media to select for integration events. Transformants were checked by PCR. The ZRT1 deletion strain DTK904 was constructed in a similar manner. PCR products were made using oligos 14670543 and 14670544 with plasmid pRS305 (Sikorski and Hieter 1989) as a template. These PCR products contained the LEU2 gene flanked with 3′ and 5′ homology to the target sequence. Strains transformed with this product were plated on SD–leu solid media to select for integration events and then checked by PCR as above.
To construct strain DTK1068, the Yeast Deletion Consortium strain bearing homozygous Δzrt3∷KANMX4 alleles was sporulated and dissected as described in Jauert et al. (2002). DTK1033 is a MATa mating-type spore isolated from this dissection. DTK1033 was crossed to DTK271 and dissected as described, and a spore bearing the ade2-min3 allele and the ZRT3 deletion was isolated by color and viability on YPD+G418 sulfate media. This isolate was backcrossed to DTK271 and another ade2-min3, Δzrt3∷KANMX4 spore was identified as above. This second isolate was again backcrossed to DTK271 and dissected as above to generate DTK1068, an ade2-min3, Δzrt3∷KANMX4 spore isolate.
Mutagenesis:
Dilutions of strain DTK284 were plated on YPD solid media. Cells were UV irradiated with a dosage suitable for causing 85–90% lethality, as judged by comparison to nonirradiated control plates that were used for cell viability counts. Following irradiation, colonies were grown in the dark at 25° for 10 days and scored for phenotypes. A total of 505,000 colonies arising after mutagenesis were examined for phenotypes. Colonies that exhibited a sectoring or blebbing phenotype were struck for singles to ensure that they maintained the phenotype and stored as candidates. Candidates were backcrossed to strain DTK271 to identify recessive mutations and then sporulated. Tetrads were dissected and scored for sectoring or blebbing phenotype. Heterozygous strains in which all tetrads exhibited a segregation pattern with two wild type and two mutant spores were considered to harbor a single mutation that caused the phenotype. One blebbing or sectoring MATα spore from each candidate was saved for use in complementation testing. To group mutant alleles into complementation groups, candidates were systematically mated to each other. If these mated diploids blebbed or sectored, the parental haploids were considered to be in the same complementation group. One representative for each group was chosen to represent the group for subsequent analysis; these strains (Y195 and Y451) exhibited phenotypes that were consistent with the majority of the mutant strains within that group.
Identification of the sites of mutations:
One strain from each complementation group was transformed with a plasmid library containing 6- to 10-kb yeast genomic DNA inserts on a G418-resistant vector (Jauert et al. 2005). Transformants were plated on YPD+G418 and grown at 30° for 4 days and then at 25° for 4 days and scored for blebbing. Nonblebbing transformants were picked and the library plasmids were rescued from them. Rescued plasmids were retransformed into the mutant strain to verify phenotype suppression. The ends of the genomic insert of complementing plasmids were sequenced. Genes present on the genomic insert were then deleted in DTK271 using the G418 resistance KanMX4 cassette to identify the gene responsible for the blebbing or sectoring phenotype, resulting in strains DTK878 and DTK902 as described above. To verify that the original mutation mapped to the deleted gene, DTK878 and DTK902 were mated with the original mutants and the resulting diploid was examined for blebbing or sectoring phenotypes. The genes in the original mutant strain were subsequently sequenced to identify the specific alteration.
PCR primers:
The following PCR primers were used in this study:
Primer 12966370 (Zrt1F): TACGCACGGCATTAGCTC
Primer 12966371 (Zrt1R): ACTCGTAGATGGCACGGTC
Primer 14767981 (Zap1F): ACTTGCCGCCTACTTGGC
Primer 14767982 (Zap1R): AATGTCCTTCCCCCCCAC
Primer 20619256 (Rad50F): GCGGCTTTCAAGCTTTGATCT
Primer 20619257 (Rad50R): TGGCTAAGCAACAGAAGCGT
Primer 27016542 (Cot1F): CTACGTGGGAGCTCGAAAAGCATT
Primer 27016543 (Cot1R): ATCACCTGTTTTCGCTTTTCCTCT
Primer 26910872 (Yke4F): CATTCCCTTTCCATAGAACAGT
Primer 26910873 (Yke4R): GAAGCGTCGGTACCATATAAGG
Primer 27016544 (Zrc1F): GGGCTTGGCGCAGTAATTTT
Primer 27016545 (Zrc1R): GAATAACGCAGTTCTTGACTTAGG
Primer 14670543 (zrt1:LEU2F): TTACAAAAAAACTGCAAGTATAGACAATAAAACAACAGCACAAATATCAAAAAAGGAATTGATTGTACTGAGAGTGCACC
Primer 14670544 (zrt1:LEU2R): ATAAATAACTATAAAATATGAAATAGAATCTATATGGAACATGCAGAATTTCGCTTTGGTCTGTGCGGTATTTCGCACCG.
Blebbing time course:
DTK271 and DTK878 were streaked onto solid YPD medium and incubated at 30° for 48 hr. Single colonies of each strain were inoculated into 5 ml of liquid YPD medium and incubated at 30° for 8 hr with agitation. The 5-ml cultures of DTK271 and DTK878 were inoculated into 45 ml of fresh liquid YPD medium; this was designated as 0 hr from inoculation. The 50-ml cultures of DTK271 and DTK878 were grown at 30° with agitation. Beginning at 0 hr, the OD600 of each culture was measured and appropriate dilutions of each culture were plated onto solid YPD medium every 12 hr. The dilution plates of DTK271 and DTK878 for each time point were grown at 30° for ∼72 hr, after which the red Ade− and white Ade+ colonies were counted and the percentage of white Ade+ CFUs was determined. Three independent time courses were conducted.
RESULTS
Isolation of minisatellite tract stability mutants:
To screen for mutations that affect the stability of repetitive minisatellite tracts and influence the repair of large unpaired DNA loops, we developed a visual assay by inserting a minisatellite tract into the ADE2 gene on chromosome XV of S. cerevisiae (Figure 1, top). This insertion mutation, ade2-min3, consists of three 20-bp repeats plus five additional nucleotides, resulting in a disruption of ADE2. Cells without a functional ADE2 gene require adenine to grow, and they develop a red, rather than a white, colony color. The minisatellite sequence was chosen because its stability was previously shown to be unaffected by deletion of mismatch repair proteins required for microsatellite stability (Sia et al. 1997). Loss of a repeat in an ade2-min3 cell restores the correct reading frame, leading to a white sector in the red colony, an infrequent event in wild-type cells. Repeat number alterations presumably occur during mitotic replication via DNA polymerase slippage that leads to a looped intermediate, as failure to recognize and repair the loop will lead to an increase or decrease in the tract length. To verify this, PCR analysis of the ade2-min3 insertion in 110 white colonies showed that the Ade+ phenotype resulted from the loss of one of the three 20-bp repeats (Figure 1, bottom). This result was confirmed by sequencing five randomly chosen PCR products; all five tracts had lost one 20-bp repeat.
The haploid ade2-min3 strain DTK284 was mutagenized with ultraviolet radiation to ∼10% survival. Of ∼505,000 colonies, we identified 97 strains with a phenotype. A majority of the colonies exhibited an unusual phenotype when grown on rich YPD medium, developing white papillations (“blebs”) instead of the standard sectoring phenotype (Figure 2). Each mutant strain was backcrossed to DTK271, an isogenic parental strain of the opposite mating type. The resulting diploid strains were examined for a blebbing phenotype; all were wild type, indicating that all of the mutant alleles were recessive. The diploids were sporulated and the tetrads were dissected to identify those strains in which the blebbing phenotype was due to mutation at a single locus.
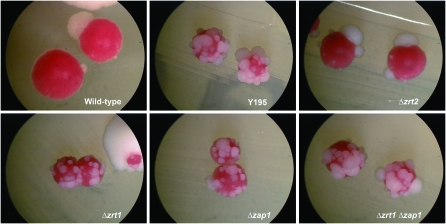
Figure 2.—
Colony morphology in ade2-min3 strains. Strains were grown at 30° for 3 days and then incubated at room temperature for an additional 3 days. The wild type is the parental ade2-min3 strain DTK271, while Y195 is a UV-induced mutant strain identified by its blebbing phenotype. The remaining strains were constructed by transformation: Δzrt1 (DTK878), Δzap1 (DTK902), Δzrt2 (DTK978), and Δzrt1 Δzap1 (DTK921).
Thirty mutant strains with a strong blebbing phenotype that was due to mutation at a single locus were placed into four complementation groups by pairwise matings, followed by examination of the diploids for a blebbing phenotype. The Y195 group has 17 alleles and the Y451 group has 11, while Y797 and Y857 are each represented by a single allele. The remainder of this article will focus on the Y195 and Y451 complementation groups.
Identification of the genes altered in the Y195 and Y451 complementation groups:
The blebbing phenotype is apparent only on rich YPD medium; mutant strains failed to bleb on all synthetic complete media formulations that we tested. Therefore, to identify the nature of the mutations in these strains, we constructed a new yeast genomic library (Jauert et al. 2005). This library uses a plasmid that confers resistance to geneticin, as extant yeast genomic libraries all utilize auxotrophic markers, which are not suited for use with rich media. The plasmid library was introduced into the mutant strains, and the resulting transformants were screened for loss of the blebbing phenotype.
One plasmid, 1b1q, was capable of suppressing the blebbing phenotype of Y451. Sequencing showed that the 1b1q genomic fragment consisted of nucleotides 20,180 to 24,995 on chromosome VII. One gene contained in this interval is ZRT1. The deletion of ZRT1 in the parental strain exhibited a blebbing phenotype (DTK878; Figure 2); this phenotype was suppressed by introduction of the 1b1q plasmid. We crossed DTK878 to Y451 and Y195. Surprisingly, the zrt1 deletion complemented the blebbing mutation in Y451. However, it did not complement the Y195 mutation, indicating that Y195 (but not Y451) was likely to have a mutation in ZRT1. We sequenced the ZRT1 locus in Y195 and identified a mutation in the coding sequence at amino acid 329 that converts a leucine to a stop codon. From these data we conclude that the blebbing phenotype in Y195 is due to a mutation in ZRT1 and that extra copies of ZRT1 can suppress the mutation in Y451.
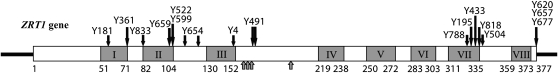
The Y195 complementation group consists of 17 independent isolates. We sequenced the ZRT1 gene in these strains. All 17 contained mutations in ZRT1 (Table 2); 3 strains contained two mutations. All mutations were point mutations that altered an amino acid, altered the wild-type stop codon, or introduced a novel stop codon.
TABLE 2.
Sequence changes in blebbing mutants
| Allele | Nucleotide change | Amino acid change |
|---|---|---|
| ZRT1 | ||
| Y4 | G 450 A | Trp 150 Stop |
| Y181 | T 167 G | Val 56 Gly |
| Y195 | T 986 A | Leu 329 Stop |
| Y361 | T 211 C | Ser 71 Pro |
| Y433 | C 995 T | Thr 332 Ile |
| Y491 | A 491 G | Asp 164 Gly |
| A 494 T | Glu 165 Val | |
| Y504 | T 1010 C | Leu 337 Pro |
| Y522 | T 308 C | Leu 103 Ser |
| Y599 | T 308 C | Leu 103 Ser |
| Y620 | T 1129 C | Stop 377 Gln |
| Y654 | G 338 A | Gly 113 Asp |
| G 368 A | Trp 123 Stop | |
| Y657 | T 1129 C | Stop 377 Gln |
| Y659 | C 304 T | His 102 Tyr |
| Y677 | T 1129 C | Stop 377 Gln |
| Y788 | C 974 T | Ser 325 Phe |
| Y818 | G 1003 A | Val 335 Asn |
| T 1004 A | ||
| Y833 | T 246 G | Tyr 82 Stop |
| A 261 C | Ala 87 Ala | |
| ZAP1 | ||
| Y53 | G 1709 TT | Frameshift after amino acid 570 |
| Y76 | 2245 +T | Frameshift after amino acid 749 |
| Y451 | G 1150 T | Glu 384 Stop |
ZRT1 encodes the high-affinity zinc transporter for the plasma membrane (Zhao and Eide 1996a) (Figure 3); a low-affinity transporter is encoded by the ZRT2 gene (Zhao and Eide 1996b). ZAP1 encodes a zinc-regulated protein that induces transcription of the ZRT1 gene under low-zinc conditions (Zhao and Eide 1997). We constructed deletion alleles of both of these genes. The Δzap1 strain blebbed at approximately the same level as the Δzrt1 strain. The Δzrt2 strain also exhibited the blebbing phenotype, but at a significantly lower level (Figure 2).
Figure 3.—
ZRT1 gene with mutation locations. The ZRT1 gene is shown as an open rectangle; numbers underneath the rectangle indicate amino acid positions within the 377-amino-acid protein, while numbered shaded boxes indicate the location of putative transmembrane domains and shaded arrows indicate the zinc-binding residues. The location of each of the mutations identified in the ade2-min3 screen is shown by solid arrows, with the name of the mutant strain above the arrow. Details of the nature of each mutation are given in Table 2.
We crossed the Δzap1 strain to Y451, and the resulting diploid exhibited a blebbing phenotype. Sequencing of the ZAP1 locus identified a G-to-T mutation at nucleotide 1150 that converts a glutamine to a stop codon in Y451. We isolated a ZAP1-containing plasmid from our yeast genomic library by colony hybridization using a ZAP1 probe. This plasmid, pPAJ199, contained genomic sequences from 329,352 to 336,006 on chromosome X. Introduction of pPAJ199 into Y451 suppressed the blebbing phenotype completely (data not shown). The ZAP1 locus in two other strains from the Y451 complementation group was sequenced; both contained point mutations that led to frameshifts in the coding sequence of ZAP1 (Table 2).
As described above, the 1b1q plasmid containing a wild-type copy of the ZRT1 gene suppresses the blebbing phenotype of the Y451 zap1 mutation, indicating that an increased level of the ZRT1 high-affinity zinc transporter can bypass the loss of the ZAP1-encoded zinc-responsive transcription factor. To determine if the reciprocal is true, we introduced the pPAJ199 plasmid bearing a wild-type copy of ZAP1 into the zrt1 strain Y195. No suppression of the Y195 blebbing phenotype was observed, indicating that excess ZAP1 does not bypass the loss of ZRT1.
The ZRT1 and ZRT2 proteins are part of a group of related metal transporters, the ZIP family (Eng et al. 1998). S. cerevisiae contains other ZIP family members that have been demonstrated to transport zinc. Yke4p regulates zinc transport in the endoplasmic reticulum (Kumanovics et al. 2006), while Zrt3 serves as the zinc influx pump for the vacuole (MacDiarmid et al. 2000). Zrc1p and Cot1p are the major and minor vacuolar zinc efflux pumps (MacDiarmid et al. 2000); these proteins are members of another group of transporters, the CDF family. Strains bearing deletions of these genes do not exhibit a blebbing phenotype (data not shown), indicating that they are not involved in maintenance of minisatellite stability.
Analysis of the role of ZRT1 and zinc in minisatellite stability:
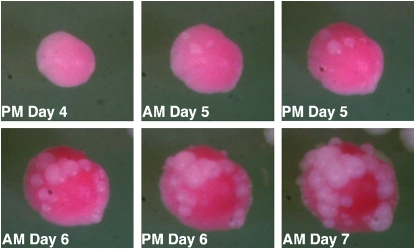
In Y195 and Y451, the white blebs on the surface of the red colonies result from the loss of one 20-bp repeat from the minisatellite tract inserted into the ADE2 gene; PCR of DNA from blebbing cells showed that the ade2-min3 tract lost one copy of the repeat (data not shown). These tract alterations do not occur stochastically during the growth of the colony, as that would lead to a sectored colony. If both the red Ade− cells and the white Ade+ cells were dividing, a wedge-shaped sector of white would be detected at the margin of the red cells; this colony morphology is never observed in the blebbing strains. Instead, the blebbing phenotype indicates that the alterations are occurring very late in the growth cycle of the colony; blebs are not visible until 72–96 hr after cells begin forming colonies. Figure 4 is a time course of bleb formation. This analysis of the blebbing colony morphology indicates that the minisatellite tract alterations underlying the appearance of the blebs occur only in growth-arrested cells. Appearance of the blebs on the surface of the colony can be delayed, but not prevented, by supplementing the growth medium with additional adenine. This additional adenine also delays the formation of the red colony coloration.
Figure 4.—
A time-lapse photo progression of bleb formation. Cells were placed on rich medium at 30° for 4 days until colonies had formed and begun to exhibit typical Ade− red coloration. Colonies were photographed twice a day with ∼12 hr between each session. No sectoring is observed in this strain; only blebbing is detected.
White Ade+ cells arise throughout the colony, rather than just on the surface. We suspended colonies in liquid growth medium when the first blebs appeared on the surface of the colony, after ∼3 days incubation at 30°. Dilutions of this suspension were spread on solid growth media and incubated for 4 days, and the relative number of white and red colonies were determined. The proportion of white colonies to red colonies was higher than expected from the proportion of white cells in the nascent blebs to the red cells composing the majority of the colony, indicating that white cells were also present within the colony.
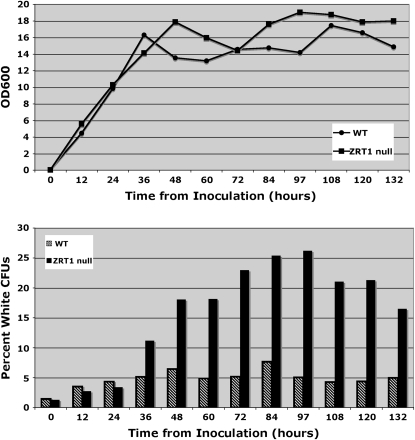
To further characterize the timing of the minisatellite tract alteration, we compared the growth curves and relative number of Ade+ CFUs in the Δzrt1 strain DTK878 to those of the parental DTK271. Figure 5 shows that growth of both strains arrests between 36 and 48 hr after inoculation, a period that is considered post-diauxic. Although Ade+ CFUs are present in both strains from time zero, the Δzrt1 strain shows a significant increase in Ade+ CFUs compared to the parental beginning at 48 hr after inoculation. This result indicates that the increase in minisatellite tract alterations displayed by ZRT1 mutants does indeed take place after the culture has undergone a growth arrest and entered the stationary phase.
Figure 5.—
An increase in minisatellite tract alteration takes place after a growth arrest in the Δzrt1 strain. (Top) Wild-type (WT) vs. Δzrt1 growth curves. The zrt1 null strain DTK878 and its parent DTK271 were grown in rich liquid media at 30° for 132 hr. The OD600 was measured (circles for DTK271, squares for DTK878) every 12 hr. Data represent an average of three independent experiments. (Bottom) Frequency of white CFUs in WT vs. Δzrt1. Cultures from the growth curves (top) were diluted and plated every 12 hr as described above. Data shown represent an average of three independent experiments. Chi-squared analysis showed a significant difference between the percentage of white CFUs in the zrt1 deletion strain (solid bars) and the wild-type parental strain (striped bars) for all time points from 48 hr on (P < 0.01).
Alterations in zinc homeostasis specifically affect minisatellite tracts:
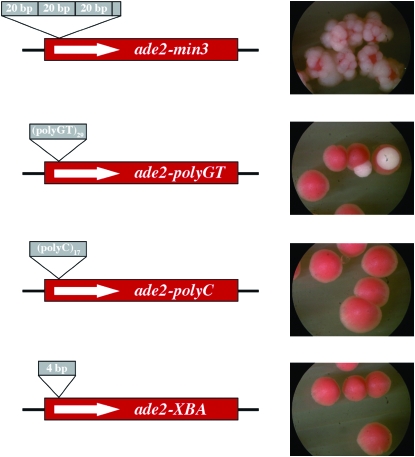
We introduced three other sequences into the XbaI restriction site used for the ade2-min3 minisatellite tract insertion. The first allele, ade2-polyGT, contains 29 copies of a GT dinucleotide; the second allele, ade2-polyC, contains 17 cytosines; and the third allele, ade2-XBA, is a duplication of the four central bases in the XbaI restriction site. The ZRT1 gene was deleted in strains containing these alleles, and the resulting strains were examined for a blebbing phenotype. As can be seen in Figure 6, only the strain with the original ade2-min3 minisatellite insertion exhibited a blebbing phenotype. Colonies of the ade2-polyGT strain occasionally had a single bleb on the surface of the colony. Figure 6 shows two such colonies; these were the only two colonies that exhibited blebbing on the entire plate.
Figure 6.—
The zrt1-dependent blebbing phenotype is observed only with minisatellite insertion alleles of ADE2. Three novel alleles of ADE2 were constructed: ade2-polyGT contains an insertion of 29 copies of a GT dinucleotide into the XbaI site used for the ade2-min3 allele, ade2-polyC contains 17 cytosines, while ade2-XBA is a fill-in of the four-base overhang of the XbaI site. These alleles were introduced into yeast, and then Δzrt1 derivatives were constructed. None of the novel alleles exhibited the severe blebbing phenotype associated with the ade2-min3 Δzrt1 strains.
Deletion of RAD50 reduces the frequency of blebbing:
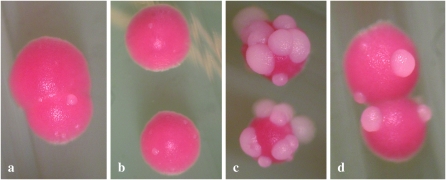
We used a directed approach to identify genes required for bleb formation in the Δzrt1 strain. Rad50p is a central factor in recombination and double-strand break repair (DSBR), the only one that contains a zinc-binding domain. We deleted RAD50 in the zrt1 deletion strain DTK904 to generate DTK1057. This zrt1 rad50 strain exhibits a diminished blebbing phenotype (Figure 7), indicating that Rad50p is involved in at least a portion of the minisatellite tract alteration events in the Δzrt1 strain. We counted the number of blebs on 50 colonies after 10 days of growth: DTK904 had an average of 24.9, while DTK1057 had 11.7. The 99.9% confidence intervals for the two data sets do not overlap, indicating that the difference is significant.
Figure 7.—
Deletion of RAD50 in an ade2-min3 Δzrt1 strain reduces the blebbing frequency. (a) DTK271, wild type. (b) DTK1056, Δrad50. (c) DTK904, Δzrt1. (d) DTK1057, Δzrt1 Δrad50. The wild-type ade2-min3 strain and the derivative Δrad50 strain do not exhibit a blebbing phenotype. The Δzrt1 strain has a strong blebbing phenotype; this is reduced significantly in the Δzrt1 Δrad50 strain.
DISCUSSION
We have isolated a novel class of mutants that destabilize a minisatellite repeat tract in S. cerevisiae. These mutants are characterized by blebbing, a previously unreported color segregation phenotype. Alterations in the minisatellites in these mutants consist exclusively of the deletion of one repeat unit; these alterations take place during stationary phase. Three strains with blebbing phenotypes have mutations in genes that influence zinc homeostasis: the high-affinity zinc transporter ZRT1, its zinc-dependent transcriptional regulator ZAP1, and the low-affinity zinc transporter ZRT2. Mutations in these genes specifically affect minisatellite sequences; other repeat types or a simple insertion into ADE2 do not exhibit the blebbing phenotype. The blebbing phenotype is partially dependent on a functional RAD50. This work directly implicates zinc homeostasis in genomic instability during stationary phase.
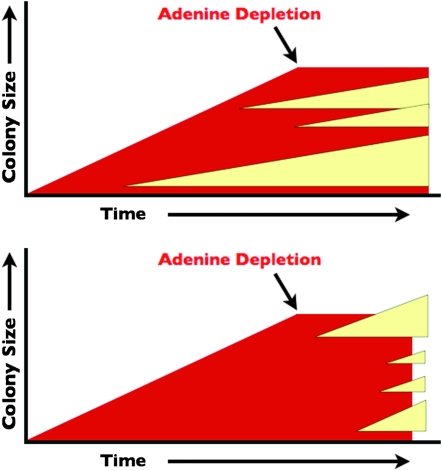
The blebbing phenotype is a highly novel minisatellite instability phenotype characterized by white Ade+ papillations forming on the surface of a red Ade− colony. Blebbing is observed in a yeast strain in which three 20-bp repeats and a short linker have been inserted into a restriction site in the ADE2 gene, disrupting both the restriction site and the reading frame of the gene, leading to a red colony color (Figure 1, top). When one repeat is lost from this minisatellite tract, the cells become white again (Figure 1, bottom). The red cells, which cannot produce their own adenine because of the ADE2 minisatellite insertion, grow normally, forming a colony, and then undergo a growth arrest. We hypothesize that this growth arrest is due to a depletion of local adenine in the growth media (Figure 8). After the red Ade− cells have stopped growing, some lose a minisatellite repeat, restoring the ADE2 reading frame. This subpopulation is now capable of producing adenine, allowing cells to resume growth, eventually forming the white surface papillations. As shown in Figure 7, the blebbing phenotype is in sharp contrast to the standard yeast sectoring phenotype.
Figure 8.—
Models for stationary-phase bleb formation and sectoring in ade2 strains. Two models are shown for the appearance of white Ade+ cells within a colony of red Ade− cells during two stages of growth: an exponential phase and subsequent stationary phase upon loss of a rate-limiting nutrient such as adenine. (Top) If the Ade+ cells appear during exponential growth, white sectors will form within the colony, with the size of the sector depending on how early in the formation of the colony the Ade− to Ade+ transition occurred. (Bottom) The Ade−-to-Ade+ transitions do not occur during exponential growth, and so no white sectors are present. However, once the cells go into stationary phase, in this case due to adenine depletion, an Ade−-to-Ade+ transition will allow the Ade+ cells to resume growth, resulting in a white microcolony (bleb) within or on top of the existing colony.
The observations that blebbing occurs only after the red colonies have stopped growing and that no white sectors are observed in the red colonies imply that the underlying minisatellite alteration occurs during stationary phase and is dependent upon a stationary-phase-specific mechanism. In sectoring cells, loss of a minisatellite repeat would occur while the cells in the colony are actively growing, with continued division of the resulting Ade+ cell forming a white wedge-shaped sector in the colony; such sectors are never observed in blebbing colonies. The data shown in Figure 5 confirm that the minisatellite alteration does indeed take place after the cells have undergone a growth arrest and entered a nonproliferating state.
Our work has shown that stationary-phase mutational mechanisms can destabilize a minisatellite tract. Stationary phase, also known as quiescence or G0, is not well understood in spite of the fact that most eukaryotic cells spend the majority of their life span in this state. Minisatellites are known to be stable during mitosis and unstable during meiosis, but their stability during stationary phase has not been investigated. Previous studies on stationary-phase genome stability in Escherichia coli demonstrate that adaptive mutations can occur in stationary-phase cells that are under stress, allowing the cells to become better equipped for survival and growth (Harris et al. 1999; Bull et al. 2001; Ponder et al. 2005). This stationary-phase-specific mutational mechanism is dependent on recombination proteins and error-prone polymerases. Stationary-phase mutations may play a significant role in the evolution of microbes, including the development of drug resistance by pathogens. This poorly understood process could be a significant source of somatic mutation in humans, as the majority of our somatic cells are quiescent. In mammals, the acquisition of mutations during stationary phase could bypass growth control, leading to oncogenesis and influencing tumor progression. Thus, the minisatellite stability assay system described here may allow us to gain a better understanding of stationary-phase mutation in eukaryotes.
Interestingly, the mutations that we have identified as elevating the rate of stationary-phase minisatellite instability reside in the zinc homeostasis-control genes ZRT1, ZRT2, and ZAP1. Zrt1p is a high-affinity zinc transporter found at the plasma membrane in S. cerevisiae cells (Zhao and Eide 1996a). Transcription of ZRT1 is activated under low-zinc conditions. After zinc levels increase, Zrt1p is removed from the plasma membrane through endocytosis and degraded in the vacuole (Gitan et al. 1998). Zrt2p is a low-affinity zinc transporter that is expressed under both zinc-limiting and zinc-replete conditions. (Zhao and Eide 1996b). Zap1p is a zinc-responsive transcription factor that controls the transcription of both ZRT1 and ZRT2 (Zhao and Eide 1997). Zap1p senses the zinc levels in the cell through its zinc-binding motifs (Zhao et al. 1998). Mutations affecting Zrt1p, Zrt2p, or Zap1p could easily alter the activity of some of the many zinc-binding proteins found in the cell.
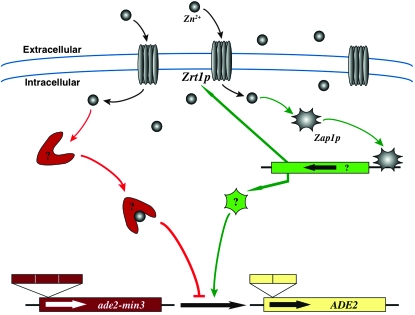
Our results suggest that perturbation of intracellular zinc levels is responsible for the blebbing phenotype. We have developed a model for zinc-dependent minisatellite alteration mediated by Zrt1p and/or Zap1p (Figure 9). This model incorporates two possibly interdependent pathways. In the first pathway, intracellular zinc transported through Zrt1p binds a zinc-dependent factor responsible for stabilizing the ADE2 minisatellite construct. Thus, when mutation of Zrt1p perturbs the intracellular zinc level, this stabilizing activity no longer functions properly and minisatellite stability is compromised. Alternatively, Zap1p function could be affected by intracellular zinc levels, activating expression of a factor that destabilizes the minisatellite tract. However, overexpression of ZRT1 in a ZAP1 mutant rescues the blebbing phenotype, indicating that the major role of ZAP1 in development of the blebbing phenotype is to influence the level of ZRT1 expression.
Figure 9.—
A model for minisatellite stability that incorporates zinc homeostasis. Bleb formation is the phenotypic consequence of a reduction in ade2-min3 minisatellite repeat number that restores the proper ADE2 reading frame. This reduction does not occur frequently in cells with a wild-type ZRT1 or ZAP1 gene. Loss of the high-affinity zinc transporter Zrt1p reduces the ability of the cells to acquire zinc, a necessary cofactor in many DNA-interacting proteins. Loss of the zinc cofactor could potentially lead to a reduction in active protein; one such protein may maintain the stability of the ade2-min3 minisatellite sequence. ZAP1 encodes a zinc-regulated transcription factor, one of which is the ZRT1 gene. Loss of ZAP1 could lead to minisatellite instability through its effect on ZRT1 transcription or due to reduction in the transcription of another gene whose product acts in minisatellite stability maintenance during stationary phase. As excess copies of ZRT1 suppress the blebbing phenotype of a zap1 mutant, we favor the former explanation. The intermediate factors are unknown, although the phenotype of the Δrad50 mutant indicates that the RAD50 protein may be involved, as it is known that Rad50p binds zinc (Hopfner et al. 2002; Wiltzius et al. 2005).
Importantly, mutation of ZRT1 specifically affects minisatellite tracts. As described above, substituting different repeat types or a simple insertion in place of the minisatellite tract in ADE2 does not lead to a blebbing phenotype in a Δzrt1 strain (Figure 6). This result indicates that the mechanism for tract alteration that is active in zrt1 blebbing mutants is specific to minisatellite repeats. Thus, intracellular zinc levels may specifically affect minisatellite stability and not genomic stability in general.
Loss of zinc transporters whose function is to move zinc between compartments within the yeast cell does not influence minisatellite stability. Deletion of the YKE4, ZRT3, ZRC1, or COT1 genes does not lead to a blebbing phenotype. These genes encode transporters that function in the endoplasmic reticulum (Kumanovics et al. 2006) and vacuole (MacDiarmid et al. 2000). Our data indicate that the absolute level of zinc within the cytoplasm of the cell (presumably present as a cofactor to zinc-binding proteins) is the important determinant for minisatellite stability, rather than the compartmental localization of the zinc ions once they have been imported into the cell.
Zinc is an essential cofactor in many important cellular proteins. These include a number of nucleic-acid-binding proteins involved in replication, DNA repair, recombination, transcription, and translation (Ho 2004). In these proteins, zinc is often found in DNA-binding motifs known as zinc fingers, zinc knuckles, and zinc hooks. The DSBR and recombination protein Rad50p contains a zinc-hook domain, which is critical for the function of the Mre11-Rad50-Xrs1 complex in DSBR (Hopfner et al. 2002; Wiltzius et al. 2005). Loss of Rad50p in a Δzrt1 cell reduces the frequency of blebbing, showing that recombination or DSBR plays a role in the stationary-phase-dependent alteration of the ade2-min3 minisatellite tract. Recombination previously has been demonstrated to be involved in stationary-phase mutagenesis in bacteria (Bull et al. 2001).
In addition to its vital role as a cofactor for DNA-binding proteins, zinc is required for the activity of many other metalloproteins, including the essential kinetochore component CEP3 (Lechner 1994), and also acts during apoptosis (Martin et al. 1991; Truong-Tran et al. 2000; Fraker 2005). Zinc protects cells from DNA damage both by acting as an antioxidant and through Sod1p, the zinc-dependent superoxide dismutase (Ho 2004).
Zinc homeostasis plays a significant role in human health. Hereditary zinc deficiency, acrodermatitis enteropathica (AE), results from loss of the zinc transporter SLC39A4 (Wang et al. 2002), a member of the ZIP zinc transporter family, as are ZRT1 and ZRT2 in S. cerevisiae. Without zinc supplement therapy, AE is usually fatal (Sehgal and Jain 2000). Dietary zinc deficiency is a serious problem in economically underprivileged parts of the world, leading to retarded growth, neuropathy, appetite suppression, diarrhea, dermatitis, alopecia, and hypotension. Even in the United States, 10% of the population consumes less than half of the recommended dietary allowance of zinc (Ho 2004).
Suboptimal zinc intake has also been linked to a variety of disease states. Low zinc intake can lead to a dramatic loss in immune defense capacity (Fernandes et al. 1979) through apoptosis of precursor T- and B-cells, which results in lymphopenia (Fraker 2005). Zinc levels have been linked to cancer, particularly cancer of the prostate (Dhar et al. 1973; Costello and Franklin 1998, 2000; Platz and Helzlsouer 2001; Bataineh et al. 2002; Ho 2004), and to neurodegenerative disease, including amyotrophic lateral sclerosis (Dupuis et al. 2004; Ho 2004). Lesser effects include airway inflammation and asthma (Murgia et al. 2006) and potentially depression (Nowak et al. 2005).
In conclusion, our data show that a minisatellite tract in the yeast S. cerevisiae can be destabilized during stationary phase by mutation of zinc homeostasis genes. Identification of the novel yeast blebbing phenotype has allowed us to establish for the first time that minisatellites undergo fluctuations in repeat number during stationary phase and that these alterations are zinc dependent. One factor contributing to minisatellite alterations is a functional recombination or DSBR system. It is vital to understand this process, since both zinc homeostasis and minisatellite stability have been implicated in a variety of human diseases.
Acknowledgments
This work was funded by a grant from the Leukemia and Lymphoma Society (3806-99) and by a Basil O'Connor Starter Scholar Research Award from the March of Dimes (5-FY00-573) and is currently sponsored by a grant from the National Institutes of Health (5RO1-GM072598).
References
- Alani, E., L. Cao and N. Kleckner, 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataineh, Z. M., I. H. Bani Hani and J. R. Al-Alami, 2002. Zinc in normal and pathological human prostate gland. Saudi Med. J. 23: 218–220. [PubMed] [Google Scholar]
- Bull, H. J., M. J. Lombardo and S. M. Rosenberg, 2001. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc. Natl. Acad. Sci. USA 98: 8334–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello, L. C., and R. B. Franklin, 1998. Novel role of zinc in the regulation of prostate citrate metabolism and its implications in prostate cancer. Prostate 35: 285–296. [DOI] [PubMed] [Google Scholar]
- Costello, L. C., and R. B. Franklin, 2000. The intermediary metabolism of the prostate: a key to understanding the pathogenesis and progression of prostate malignancy. Oncology 59: 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrauwere, H., C. G. Gendrel, S. Lechat and M. Dutreix, 1997. Differences and similarities between various tandem repeat sequences: minisatellites and microsatellites. Biochimie 79: 577–586. [DOI] [PubMed] [Google Scholar]
- Dhar, N. K., T. C. Goel, P. C. Dube, A. R. Chowdhury and A. B. Kar, 1973. Distribution and concentration of zinc in the subcellular fractions of benign hyperplastic and malignant neoplastic human prostate. Exp. Mol. Pathol. 19: 139–142. [DOI] [PubMed] [Google Scholar]
- Ding, S., G. P. Larson, K. Foldenauer, G. Zhang and T. G. Krontiris, 1999. Distinct mutation patterns of breast cancer-associated alleles of the HRAS1 minisatellite locus. Hum. Mol. Genet. 8: 515–521. [DOI] [PubMed] [Google Scholar]
- Dupuis, L., J. L. Gonzalez de Aguilar, H. Oudart, M. de Tapia, L. Barbeito et al., 2004. Mitochondria in amyotrophic lateral sclerosis: a trigger and a target. Neurodegener. Dis. 1: 245–254. [DOI] [PubMed] [Google Scholar]
- Eng, B. H., M. L. Guerinot, D. Eide and M. H. Saier, Jr., 1998. Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J. Membr. Biol. 166: 1–7. [DOI] [PubMed] [Google Scholar]
- Fernandes, G., M. Nair, K. Onoe, T. Tanaka, R. Floyd et al., 1979. Impairment of cell-mediated immunity functions by dietary zinc deficiency in mice. Proc. Natl. Acad. Sci. USA 76: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraker, P. J., 2005. Roles for cell death in zinc deficiency. J. Nutr. 135: 359–362. [DOI] [PubMed] [Google Scholar]
- Gitan, R. S., H. Luo, J. Rodgers, M. Broderius and D. Eide, 1998. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273: 28617–28624. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin et al., 1999. Mismatch repair is diminished during stationary-phase mutation. Mutat. Res. 437: 51–60. [PubMed] [Google Scholar]
- Herskowitz, I., and R. E. Jensen, 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194: 132–146. [DOI] [PubMed] [Google Scholar]
- Ho, E., 2004. Zinc deficiency, DNA damage and cancer risk. J. Nutr. Biochem. 15: 572–578. [DOI] [PubMed] [Google Scholar]
- Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui et al., 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418: 562–566. [DOI] [PubMed] [Google Scholar]
- Jauert, P. A., S. N. Edmiston, K. Conway and D. T. Kirkpatrick, 2002. RAD1 controls the meiotic expansion of the human HRAS1 minisatellite in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauert, P. A., L. E. Jensen and D. T. Kirkpatrick, 2005. A novel yeast genomic DNA library on a geneticin-resistance vector. Yeast 22: 653–657. [DOI] [PubMed] [Google Scholar]
- Kennedy, G. C., M. S. German and W. J. Rutter, 1995. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat. Genet. 9: 293–298. [DOI] [PubMed] [Google Scholar]
- Krontiris, T. G., 1995. a Minisatellites and human disease. Science 269: 1682–1683. [DOI] [PubMed] [Google Scholar]
- Krontiris, T. G., 1995. b Oncogenes. N. Engl. J. Med. 333: 303–306. [DOI] [PubMed] [Google Scholar]
- Krontiris, T. G., B. Devlin, D. D. Karp, N. J. Robert and N. Risch, 1993. An association between the risk of cancer and mutations in the HRAS1 minisatellite locus. N. Engl. J. Med. 329: 517–523. [DOI] [PubMed] [Google Scholar]
- Kumanovics, A., K. E. Poruk, K. A. Osborn, D. M. Ward and J. Kaplan, 2006. YKE4 (YIL023C) encodes a bidirectional zinc transporter in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 281: 22566–22574. [DOI] [PubMed] [Google Scholar]
- Lafreniere, R. G., D. L. Rochefort, N. Chretien, J. M. Rommens, J. I. Cochius et al., 1997. Unstable insertion in the 5′ flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy type 1, EPM1. Nat. Genet. 15: 298–302. [DOI] [PubMed] [Google Scholar]
- Lechner, J., 1994. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 13: 5203–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, J., H. Debrauwere, J. Buard and A. Nicolas, 2002. Instability of the human minisatellite CEB1 in rad27Delta and dna2-1 replication-deficient yeast cells. EMBO J. 21: 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid, C. W., L. A. Gaither and D. Eide, 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19: 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki, S., H. Cederberg and U. Rannug, 2002. The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41: 333–341. [DOI] [PubMed] [Google Scholar]
- Martin, S. J., G. Mazdai, J. J. Strain, T. G. Cotter and B. M. Hannigan, 1991. Programmed cell death (apoptosis) in lymphoid and myeloid cell lines during zinc deficiency. Clin. Exp. Immunol. 83: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia, C., C. J. Lang, A. Q. Truong-Tran, D. Grosser, L. Jayaram et al., 2006. Zinc and its specific transporters as potential targets in airway disease. Curr. Drug Targets 7: 607–627. [DOI] [PubMed] [Google Scholar]
- Niwa, O., 2006. Indirect mechanisms of genomic instability and the biological significance of mutations at tandem repeat loci. Mutat. Res. 598: 61–72. [DOI] [PubMed] [Google Scholar]
- Nowak, G., B. Szewczyk and A. Pilc, 2005. Zinc and depression: an update. Pharmacol. Rep. 57: 713–718. [PubMed] [Google Scholar]
- Platz, E. A., and K. J. Helzlsouer, 2001. Selenium, zinc, and prostate cancer. Epidemiol. Rev. 23: 93–101. [DOI] [PubMed] [Google Scholar]
- Ponder, R. G., N. C. Fonville and S. M. Rosenberg, 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19: 791–804. [DOI] [PubMed] [Google Scholar]
- Sehgal, V. N., and S. Jain, 2000. Acrodermatitis enteropathica. Clin. Dermatol. 18: 745–748. [DOI] [PubMed] [Google Scholar]
- Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell and T. D. Petes, 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17: 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, E. A., M. Dominska, L. Stefanovic and T. D. Petes, 2001. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 21: 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong-Tran, A. Q., L. H. Ho, F. Chai and P. D. Zalewski, 2000. Cellular zinc fluxes and the regulation of apoptosis/gene-directed cell death. J. Nutr. 130: 1459S–1466S. [DOI] [PubMed] [Google Scholar]
- Vega, A., F. Barros, M. E. Lleonart, S. Ramon y Cajal and A. Carracedo, 2001. a HRAS1 minisatellite alleles in colorectal carcinoma: relationship to microsatelite instability. Anticancer Res. 21: 2855–2860. [PubMed] [Google Scholar]
- Vega, A., M. J. Sobrido, C. Ruiz-Ponte, F. Barros and A. Carracedo, 2001. b Rare HRAS1 alleles are a risk factor for the development of brain tumors. Cancer 92: 2920–2926. [PubMed] [Google Scholar]
- Vergnaud, G., and F. Denoeud, 2000. Minisatellites: mutability and genome architecture. Genome Res. 10: 899–907. [DOI] [PubMed] [Google Scholar]
- Virtaneva, K., E. D'Amato, J. Miao, M. Koskiniemi, R. Norio et al., 1997. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat. Genet. 15: 393–396. [DOI] [PubMed] [Google Scholar]
- Wang, K., B. Zhou, Y. M. Kuo, J. Zemansky and J. Gitschier, 2002. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am. J. Hum. Genet. 71: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius, J. J., M. Hohl, J. C. Fleming and J. H. Petrini, 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12: 403–407. [DOI] [PubMed] [Google Scholar]
- Zhao, H., E. Butler, J. Rodgers, T. Spizzo, S. Duesterhoeft et al., 1998. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273: 28713–28720. [DOI] [PubMed] [Google Scholar]
- Zhao, H., and D. Eide, 1996. a The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93: 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., and D. Eide, 1996. b The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271: 23203–23210. [DOI] [PubMed] [Google Scholar]
- Zhao, H., and D. J. Eide, 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]