Abstract
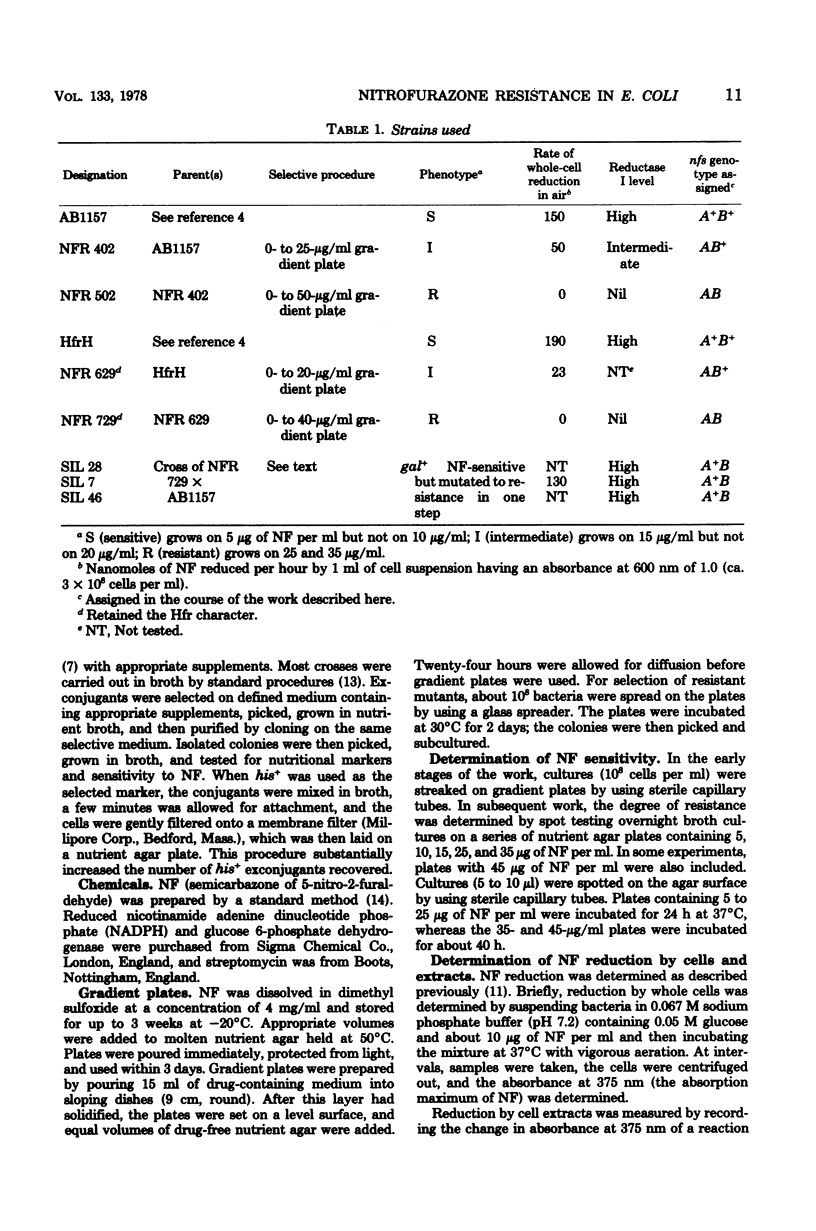
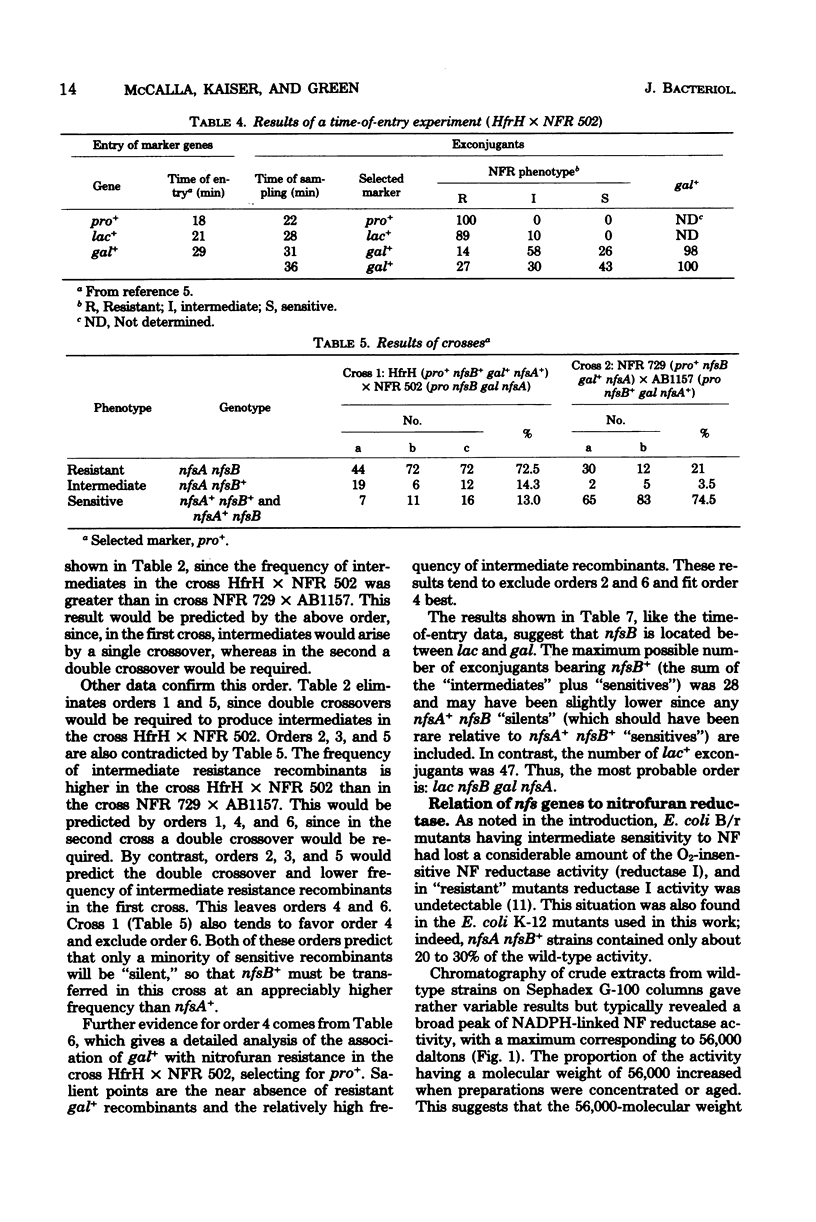
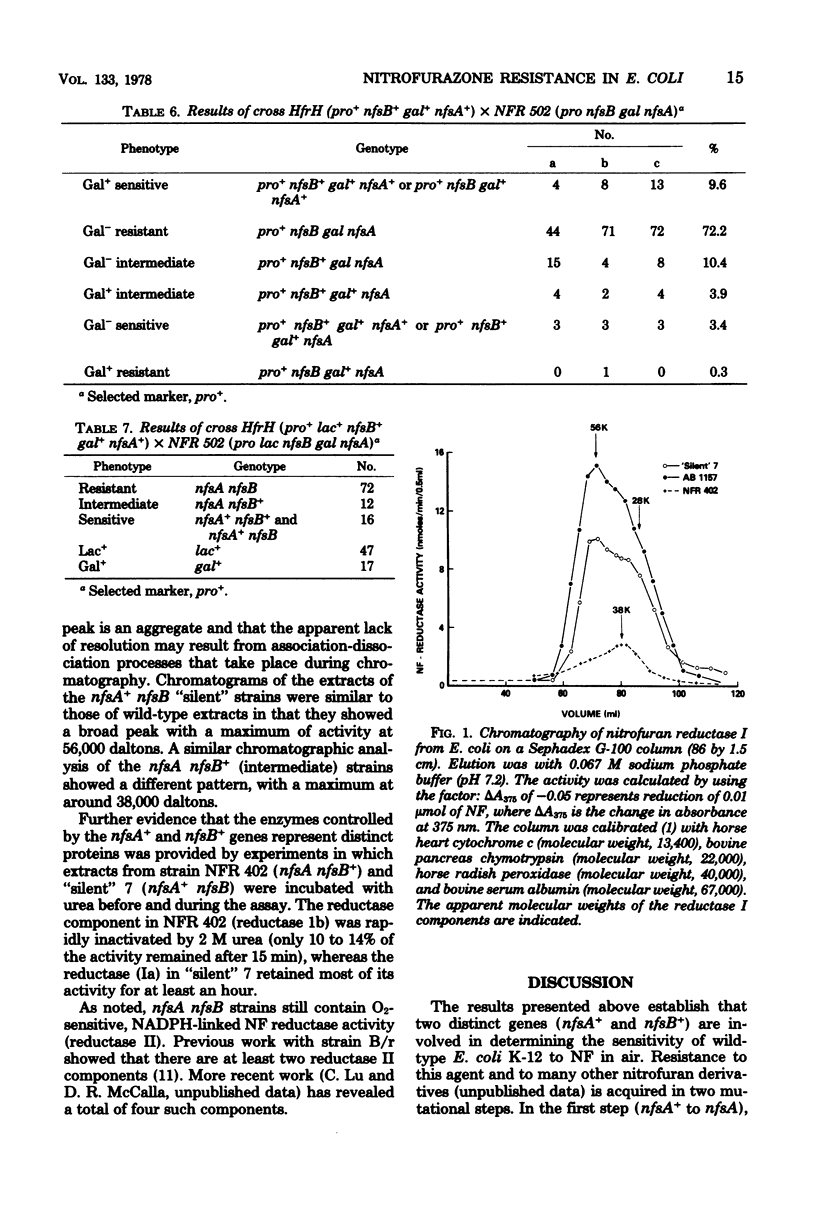
Wild-type Escherichia coli cells are sensitive to nitrofurazone (NF) and many other nitrofuran derivatives. A variety of evidence indicated that these compounds are converted to toxic “active” metabolites by reductases present in the bacteria. Sensitive E. coli K-12 acquired threefold-greater resistance to NF in one mutational step. These partially resistant mutants could undergo a second mutation that made them 10 times as resistant as the wild type. Mutation of wild-type strain K-12 to the higher level of resistance in a single step was not observed. The first mutational step was associated with partial loss of reduced nicotinamide adenine dinucleotide phosphate-linked, O2-insensitive NF reductase activity, and the second step was associated with loss of the remaining activity. The two-step mutants did, however, contain other NF reductases that were inhibited by O2 and reduced NF only under anaerobic conditions. We designated the genes that control reductase activity “nitrofuran sensitivity genes” (nfsA and nfsB). Thus, wild-type strains are nfsA+nfsB+, and the resistant double mutants are nfsA nfsB. A variety of crosses established that these genes are both located close to gal, that the most probable sequence is lac nfsB gal nfsA, and that the single-step mutants with an intermediate level of resistance are nfsA nfsB+. The nfsA+nfsB strains contained about 70 to 80% of the wild-type reductase I activity—apparently enough to confer wild-type sensitivity. This reductase activity was resistant to 2 M urea. The nfsA nfsB+ strains had only 20 to 30% of the wild-type activity, and this residual activity was sensitive to 2 M urea.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASNIS R. E. The reduction of Furacin by cell-free extracts of Furacin-resistant and parent-susceptible strains of Escherichia coli. Arch Biochem Biophys. 1957 Jan;66(1):208–216. doi: 10.1016/0003-9861(57)90551-9. [DOI] [PubMed] [Google Scholar]
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges B. A., Dennis R. E., Munson R. J. Differential induction and repair of ultraviolet damage leading to true revesions and external suppressor mutations of an ochre codon in Escherichia coli B-r WP2. Genetics. 1967 Dec;57(4):897–908. doi: 10.1093/genetics/57.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins S. T., Bennett P. M. Effect of mutations in deoxyribonucleic acid repair pathways on the sensitivity of Escherichia coli K-12 strains to nitrofurantoin. J Bacteriol. 1976 Mar;125(3):1214–1216. doi: 10.1128/jb.125.3.1214-1216.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCALLA D. R. NITROFURAN DERIVATIVES AS RADIOMIMETIC AGENTS: CROSS-RESISTANCE STUDIES WITH ESCHERICHIA COLI. Can J Microbiol. 1965 Apr;11:185–191. doi: 10.1139/m65-024. [DOI] [PubMed] [Google Scholar]
- McCalla D. R., Olive P., Tu Y., Fan M. L. Nitrofurazone-reducing enzymes in E. coli and their role in drug activation in vivo. Can J Microbiol. 1975 Oct;21(10):1484–1491. doi: 10.1139/m75-220. [DOI] [PubMed] [Google Scholar]
- McCalla D. R., Reuvers A., Kaiser C. Mode of action of nitrofurazone. J Bacteriol. 1970 Dec;104(3):1126–1134. doi: 10.1128/jb.104.3.1126-1134.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCalla D. R., Voutsinos D. On the mutagenicity of nitrofurans. Mutat Res. 1974 Feb;26(1):3–16. doi: 10.1016/s0027-5107(74)80065-5. [DOI] [PubMed] [Google Scholar]
- WOODY-KARRER P., GREENBERG J. RESISTANCE AND CROSS RESISTANCE OF ESCHERICHIA COLI S MUTANTS TO THE RADIOMIMETIC AGENT NITROFURAZONE. J Bacteriol. 1963 Jun;85:1208–1216. doi: 10.1128/jb.85.6.1208-1216.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahagi T., Nagao M., Hara K., Matsushima T., Sugimura T., Bryan G. T. Relationships between the carcinogenic and mutagenic or DNA-modifying effects of nitrofuran derivatives, including 2-(2-furyl)-3-(5-nitro-2-furyl)acrylamide, a food additive. Cancer Res. 1974 Sep;34(9):2266–2273. [PubMed] [Google Scholar]


