Abstract
The inhibition of DPP-IV by saxagliptin has been proposed to occur through formation of a covalent but reversible complex. To evaluate further the mechanism of inhibition, we determined the X-ray crystal structure of the DPP-IV:saxagliptin complex. This structure reveals covalent attachment between S630 and the inhibitor nitrile carbon (C–O distance <1.3 Å). To investigate whether this serine addition is assisted by the catalytic His-Asp dyad, we generated two mutants of DPP-IV, S630A and H740Q, and assayed them for ability to bind inhibitor. DPP-IVH740Q bound saxagliptin with an ∼1000-fold reduction in affinity relative to DPP-IVWT, while DPP-IVS630A showed no evidence for binding inhibitor. An analog of saxagliptin lacking the nitrile group showed unchanged binding properties to the both mutant proteins, highlighting the essential role S630 and H740 play in covalent bond formation between S630 and saxagliptin. Further supporting mechanism-based inhibition by saxagliptin, NMR spectra of enzyme–saxagliptin complexes revealed the presence of three downfield resonances with low fractionation factors characteristic of short and strong hydrogen bonds (SSHB). Comparison of the NMR spectra of various wild-type and mutant DPP-IV:ligand complexes enabled assignment of a resonance at ∼14 ppm to H740. Two additional DPP-IV mutants, Y547F and Y547Q, generated to probe potential stabilization of the enzyme–inhibitor complex by this residue, did not show any differences in inhibitor binding either by ITC or NMR. Together with the previously published enzymatic data, the structural and binding data presented here strongly support a histidine-assisted covalent bond formation between S630 hydroxyl oxygen and the nitrile group of saxagliptin.
Keywords: DPP-IV; X-ray crystal structure; mutant; ITC; proton NMR; short, strong hydrogen bond; serine protease; saxagliptin
Dipeptidyl peptidase IV is a serine protease that modulates the biological activity of specific circulating peptide hormones, chemokines, cytokines, and neuropeptides (Lambeir et al. 2001) by specifically cleaving two N-terminal amino acids. DPP-IV attenuates postprandial blood glucose control through inactivation of GLP-1 (Ahren et al. 2002), suggesting that modulation of DPP-IV activity can serve as a potential treatment for diabetes. Toward that end, the pharmacological proof of concept for DPP-IV inhibitors in the treatment of type II diabetes is now well-established, and the first member of this class, sitagliptin (Kim et al. 2005), is now approved by the FDA; the second member, vildagliptin (Ahren et al. 2004, 2005), awaits approval.
DPP-IV is a prototypic serine protease whose substrate cleavage is driven by activation of a Ser-His-Asp triad. Numerous reports describing the discovery of structurally diverse DPP-IV inhibitors have appeared in the last decade (Hughes et al. 1999; Villhauer et al. 2002; Augeri et al. 2005; Kim et al. 2005). All known inhibitors reported to date occupy the S1–S2 pocket at the DPP-IV active site and make extensive hydrophobic, van der Waals, and hydrogen-bonding interactions with residues lining this pocket. Interestingly, cyanopyrrolidine derivatives were suggested to yield a covalent adduct with the active-site serine (S630) of DPP-IV, forming an imidate adduct (Hughes et al. 1999; Oefner et al. 2003). Mechanism-based enzyme–inhibitor complex formation between DPP-IV and saxagliptin (Fig. 1) has been extensively described in our previous publication (Kim et al. 2006). We suggested initial inhibitor binding is followed by serine addition to the inhibitor nitrile carbon, catalyzed by histidine. This suggestion was supported by the observation of an inverse solvent kinetic isotope effect (SKIE) for the onset of inhibition and by the NMR detection of a proton fractionating in a short and strong hydrogen bond in the DPP-IV:saxagliptin complex.
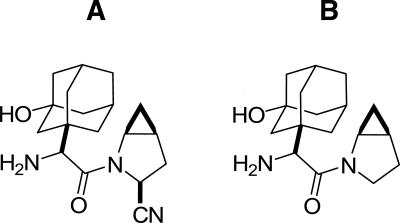
Figure 1.
Chemical structures of (A) saxagliptin and (B) BMS-538305.
In this report, we further investigate the mechanism of inhibition of DPP-IV by saxagliptin. The structure of the DPP-IV:saxagliptin complex was determined by X-ray crystallography. To understand more fully the origin of the downfield NMR resonances and to elaborate the involvement of the active-site residues in the inhibitor binding, we performed mutagenesis studies targeting residues in the active site of the enzyme. Human DPP-IV mutants DPP-IVS640A, DPP-IVH740Q, DPP-IVY547Q, and DPP-IVY547F were expressed in baculovirus, purified to homogeneity, and tested for activity in a dipeptide (Gly–Pro–pNA) cleavage assay. The effect of the mutations on saxagliptin binding was characterized by NMR, ITC, and TSE. The role of the nitrile moiety was assessed by monitoring the binding of BMS-538305, an analog of saxagliptin lacking the nitrile group (Fig. 1).
Results
Characterization of mutant proteins
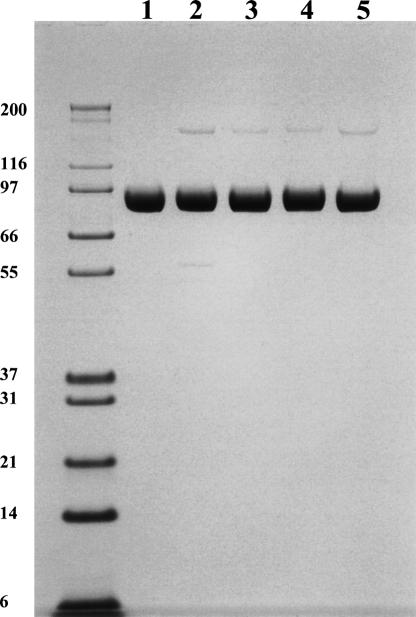
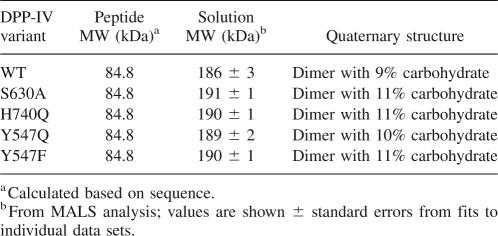
All mutant proteins were expressed to the same level and purified to the same purity (Fig. 2). As expected, all were found to be devoid of catalytic activity when measured at 10-fold excess of enzymes and 10-fold higher substrate concentration compared with the wild-type protein. Hence, specific activity could not be used as a gauge of protein quality. Instead, various biophysical methods were used to assess whether the proteins were properly folded. TSE and CD thermal denaturation data indicated that the melting temperatures (T m) for DPP-IVWT, DPP-IVS630A, DPP-IVY547F, and DPP-IVY547Q were identical (70°C), whereas the T m for DPP-IVH740Q was ∼10°C lower (60°C). Comparison of the CD spectra for wild-type and mutant DPP-IV proteins revealed that the secondary structure was unperturbed by the mutations (data not shown). Similarly, the upfield region of the 1H NMR spectra were indistinguishable, indicating the hydrophobic cores of the proteins were unperturbed (data not shown). To evaluate the quaternary states of the proteins, SEC-MALS studies were performed. All of the DPP-IV mutants were found to be dimeric with the same solution MW as wild type (Table 1), providing further evidence that the mutants were well folded and suitable for binding studies. Finally, ITC studies demonstrated that all proteins were competent for binding inhibitor (see below).
Figure 2.
SDS-PAGE of DPP-IVWT (lane 1), DPP-IVH740Q (lane 2), DPP-IVS630A (lane 3), DPP-IVY547F (lane 4), and DPP-IVY547Q (lane 5). The 4%–12% NuPAGE gel was run under nonreducing conditions. Each lane contains 5 μg of protein.
Table 1.
Quaternary structure of DPP-IV variants by SEC/MALS
Crystal structure of wild-type DPP-IV complex with saxagliptin
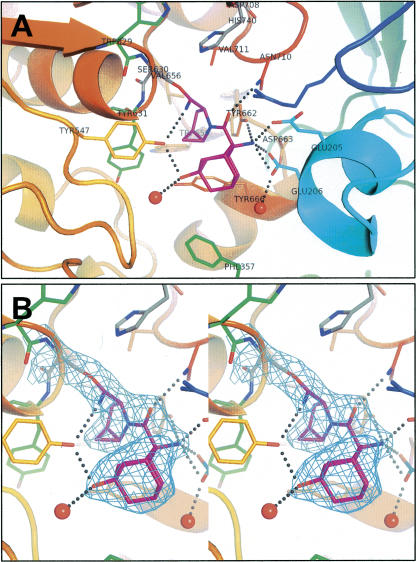
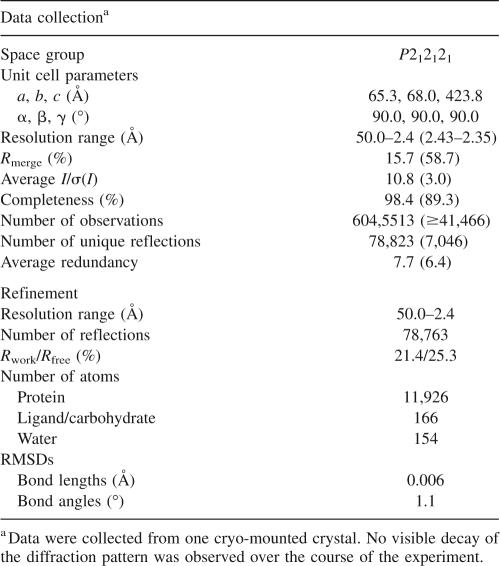
The three-dimensional structure of DPP-IV:saxagliptin complex was determined by X-ray crystallography (Fig. 3). Statistics for the data refinement are provided in Table 2. The structure revealed that saxagliptin is covalently bound to the DPP-IV active site, with the nitrile forming a covalent imidate adduct with the hydroxyl of the active-site serine, S630 (Fig. 3B), resulting in a stable trigonal complex. The 4,5-methanopyrrolidine ring is buried in the hydrophobic S1 pocket, next to the catalytic serine, where it forms van der Waals interactions with the side-chain residues that form the pocket: V711, V656, Y662, Y666, W659, and Y547. In the S2 pocket, the side-chain NH2 of N710 hydrogen bonds with the carbonyl oxygen of saxagliptin. The primary amine of saxagliptin, which is a critical functionality for potent inhibitory activity in this series, participates in a symmetrical hydrogen-bonding network with Y662 and glutamic acids, E205 and E206. The adamantane ring extends into the S2 pocket, enabling two hydrogen bonds with the side-chain hydroxyl of Y547: one with the adamantane hydroxyl and the second with the imidate nitrogen. Although DPP-IV exists as a dimer in the unit cell, only monomer A is shown in Figure 3 for simplicity because the protein–inhibitor interactions are much the same for both molecules in the asymmetric unit. One notable difference is the hydrogen bond between the imidate nitrogen and the side-chain oxygen of Y547 (distance of 3.1 Å). This hydrogen bond is seen in monomer A but not in monomer B. In monomer B, the imidate nitrogen points toward R125 and may interact with this residue though water-mediated hydrogen bonds (unmodeled; data not shown).
Figure 3.
Illustrations of X-ray co-crystal structure of saxagliptin (magenta) complexed with DPP-IV. (A) Protein colored from blue (N terminus) to red (C terminus). Catalytic residues (Ser630, His740, and Asp708) are shown in gray. All atoms are shown for Trp629, Ser630, and Tyr631 (green but for Ser630) to outline the oxyanion hole. Other active-site residues are colored per the ribbons trace. Selected hydrogen-bonded water molecules are shown as red spheres. (B) Same as above in stereo and zoomed in on BMS-477118. The electron density (2F o − F c, 1σ, cyan) is consistent with covalent attachment to Ser630. Figure created with PyMOL (DeLano Scientific).
Table 2.
Wild-type DPP-IV complex with saxagliptin data collection and refinement statistics
Binding studies by ITC and TSE
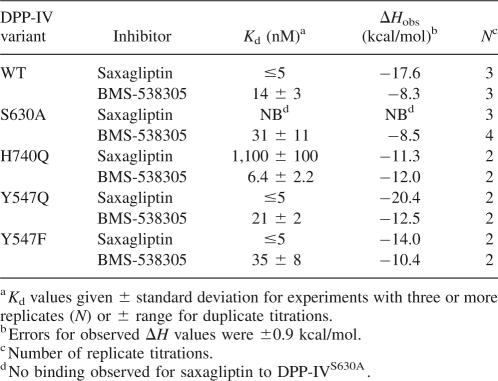
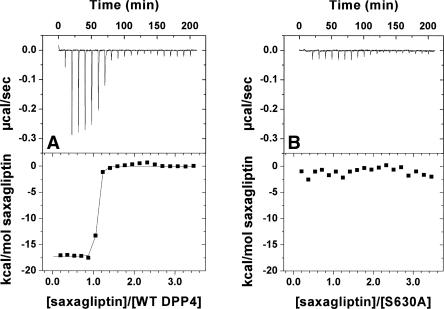
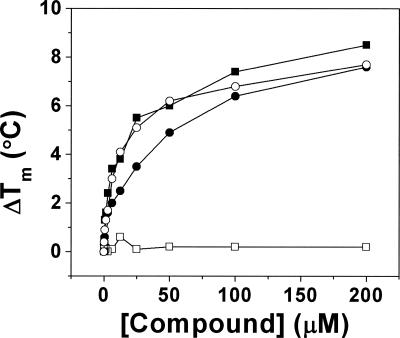
To evaluate the ability of the mutant proteins to bind saxagliptin and BMS-538305, we performed isothermal calorimetry and thermal stability enhancement studies. Saxagliptin bound DPP-IVWT very tightly by ITC (K d ≪ 5 nM, Table 3) but did not bind to DPP-IVS630A (Fig. 4A,B; Tables 3, 4). This result was verified by thermal stability enhancement experiments (Fig. 5), in which no change in T m was observed at saturating saxagliptin concentrations relative to protein alone. In contrast, the saxagliptin analog BMS-538305 bound tightly to both DPP-IVWT and the DPP-IVS630A. Saxagliptin also bound poorly (K d > 1 μM) to DPP-IVH740Q, whereas BMS-538305 showed almost unaltered affinity (K d = 6.4 ± 2.2 nM). Binding of both compounds was essentially unaffected by mutation of Y547 (Q or F). In all ITC experiments in which binding was observed, the apparent stoichiometry was approximately one compound molecule per DPP-IV monomer (observed molar ratio 0.93 ± 0.10, N = 23 titrations), providing further evidence that the inactive variants were properly folded.
Table 3.
Thermodynamic parameters measured by ITC for inhibitor binding to DPP-IV variants
Figure 4.
Binding of saxagliptin to (A) DPP-IVWT and (B) DPP-IVS630A by ITC.
Table 4.
Pairwise comparison of binding enthalpies measured by ITC for inhibitor binding to DPP-IV variants
Figure 5.
Effect of saxagliptin (squares) and BMS-538305 (circles) on the melting temperature of DPP-IVWT (filled symbols) and DPP-IVS630A (open symbols).
Detection of SSHB by NMR
When serine proteases bind mechanism-based inhibitors capable of forming Ser adducts analogous to the tetrahedral intermediate in catalysis, the Asp···His H-bond of the catalytic triad frequently becomes a SSHB (Cassidy et al. 1997; Lin et al. 1998). The proton in this hydrogen bond, the His nitrogen (Nδ1) proton, located between the imidazole and aspartate groups, becomes unusually stable and can often be detected in the low-field region of the 1H NMR spectrum, typically in the range of 14–20 ppm (Robillard and Schulman 1972). We had previously noticed that the NMR spectrum for DPP-IVWT showed a resonance at 16.1 ppm, even in the absence of bound inhibitor. By comparison to literature reports, we had assigned this resonance to the H740 Nδ1 (Kim et al. 2005). To confirm the assignment of this peak, we collected the 1H NMR spectrum of DPP-IVH740Q. Unexpectedly, the resonance at 16.1 ppm remained present (Fig. 6A), indicating that this resonance does not arise from the active-site histidine. This result led us to conduct a more detailed investigation of the origin of the downfield resonances.
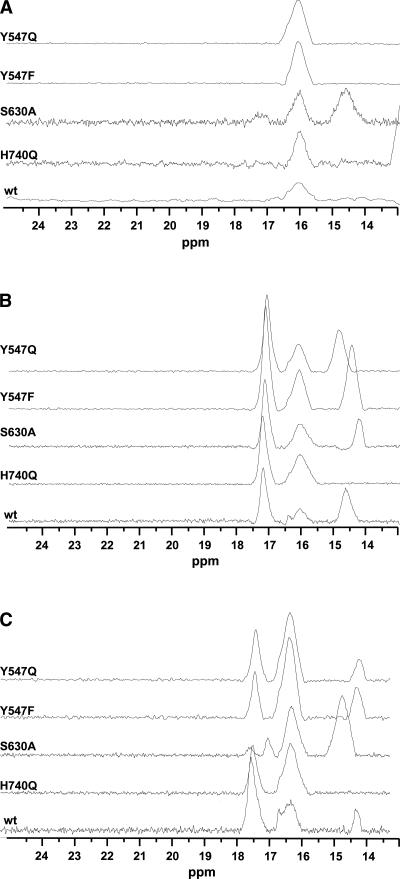
Figure 6.
Expansion of the downfield 1H NMR spectra of wild type and the four DPP-IV mutant proteins in the absence of any inhibitor (A) and in the presence of 1.2-molar excess BMS-538305 (B) or saxagliptin (C).
We collected the 1H NMR spectrum of all four mutant proteins, DPP-IVH740Q, DPP-IVS630A, DPP-IVY547F, and DPP-IVY5470Q, both in the absence (Fig. 6A) and presence of BMS-538305 (Fig. 6B) or saxagliptin (Fig. 6C). Examination of the NMR spectra for the mutant proteins indicates the resonance near 16.1 ppm is present in each mutant protein (Fig. 6A). Moreover, observation of this resonance is independent of the presence or absence of the ligands.
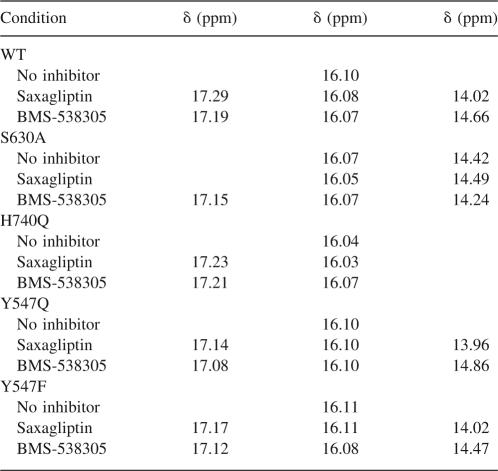
The addition of BMS-538305 to the DPP-IV proteins resulted in the appearance of new resonances near 17.2 and 14.7 ppm in all spectra, except that of H740Q (Fig. 6B). These data suggest BMS-538305 binds similarly to each of the mutant and wild-type proteins. The absence of a 14.7-ppm peak in all DPP-IVH740Q spectra allows us to assign this resonance to the H740 imidazole proton. Similarly, the addition of saxagliptin to the DPP-IV proteins resulted in the appearance of new resonances near 17.2 and 14.1 ppm in the 1H NMR spectrum. As seen for the complex with BMS-538305, the resonance near 14.1 ppm is absent in the DPP-IVH740Q–saxagliptin complex, again consistent with this resonance arising from the H740 imidazole proton.
A strong resonance is observed at 17.2 ppm in the 1H NMR spectra of all complexes except for DPP-IVS530A-saxagliptin. In the DPP-IVS530A-saxagliptin complex, this resonance is significantly reduced in intensity, is shifted, and appears to be split into two resonances. (Note that spectra for DPP-IVS530A– and DPP-IVS530A–saxagliptin complexes collected on two different protein preparations months apart yielded the same reproducible spectrum.) Comparison of the 1H NMR spectra of DPP-IVS530A in the presence and absence of saxagliptin indicates very little difference between the spectra; resonances are observed near 16.1 and 14.4 ppm in each spectrum. Because the resonance at 14.4 ppm is present in the spectra of both free and complexed DPP-IVS530A, it is unlikely to be related to the observed resonance at 14.1 ppm (or 14.7 ppm for the BMS-538305 complex). Instead, the 1H NMR spectrum suggests a structural reorganization of residues in the active site of DPP-IVS530A. It is clear from the spectrum of DPP-IVS530A–BMS-538305 and the corresponding ITC data, however, that this mutant protein remains competent for binding inhibitors. Finally, the NMR spectra for DPP-IVY547F and DPP-IVY547Q indicate both mutants bind saxagliptin and BMS-538305 similarly to the wild-type protein.
Discussion
The details of the catalytic and inhibition mechanisms of serine proteases remain of great interest due both to the historical importance of serine proteases in studies on enzyme catalysis and to the continuing medical interest in their inhibition. Considerable excitement surrounding new pharmacological agents based on DPP-IV inhibition has emerged recently with several molecules undergoing clinical evaluation (Ahren et al. 2005). All currently known DPP-IV inhibitors occupy the S1–S2 pocket in the enzyme active site. Although interaction with the catalytic residue(s) is not a necessary prerequisite for inhibitor potency and efficacy (Kim et al. 2005), an intriguing feature of cyanopyrrolidine-containing inhibitors is their potential for transient covalent bonding with S630 (Oefner et al. 2003). Inhibition of DPP-IV by these compounds should logically involve the presence of the catalytic triad. To test this hypothesis, we mutated three residues of human DPP-IV (S630, H740, and Y547; Fig. 3). These amino acids were proposed to facilitate substrate proteolysis, and they project into the S1/S2 pockets.
As expected, all mutants were catalytically inactive at cleavage of the standard pseudosubstrate. To ensure that the observed lack of proteolytic activity was the result of impaired catalytic potential rather than from misfolding or dimer disruption (Chien et al. 2004), we characterized the mutants using several biophysical methods. Taken together with our CD and 1H NMR spectral data, the SEC/MALS and binding data strongly suggested that all mutant proteins bear similar folding and dimerization patterns to the DPP-IVWT (Table 1). However, the inhibitor binding abilities of the proteins for two inhibitors (measured by ITC) were significantly altered in the mutants (Table 3). Whereas we did not observe any binding of saxagliptin to S630A either by ITC or TSE (Figs. 4A,B, 5), binding of BMS-538305 was only minimally affected (K d of 31 ± 11 nM for DPP-IVS630A vs. 14 ± 3 nM for DPP-IVWT; Table 3).
The only structural difference between the saxagliptin and BMS-538305 is the presence (saxagliptin) or absence (BMS-538305) of a nitrile group (Fig. 1); our X-ray crystal structure shows this group covalently bound to S630 in DPP-IVWT, accounting for the ∼20-fold enhanced potency of saxagliptin relative to BMS-538305 (K i = 500 pM and 10 nM for saxagliptin and BMS-538305, respectively, data not shown). By eliminating the nucleophilic hydroxyl in DPP-IVS630A, we envisioned that the covalent complex formation would be obviated while maintaining the rest of the affinity determinants. We thus expected that DPP-IVS630A would bind saxagliptin and BMS-538305 with similar affinities. Unexpectedly, our results show that binding of saxagliptin to this mutant was totally obliterated, whereas binding of BMS-538305 was only mildly perturbed (Table 3). To better understand these findings, we modeled the DPP-IVS630A–saxagliptin complex using the X-ray structure of the DPP-IVWT–saxagliptin complex. Our modeling studies support that saxagliptin cannot bind to the DPP-IVS630A. In the complex with DPP-IVWT, the nitrile carbon of saxagliptin adopts sp 2 geometry as it forms a covalent bond with the serine hydroxyl. However, unable to form a covalent interaction in DPP-IVS630A, the nitrile carbon retains its sp character. In this hybridization geometry, the nitrile of saxagliptin causes a large steric clash with the Ala630 side chain of DPP-IVS630A, which is reflected in the K i value.
Study of the DPP-IVH740Q mutant confirmed the hypothesis that enhanced nucleophilicity of Ser630 by the basicity of H740 is an essential component of the mechanism by which saxagliptin covalently binds to DPP-IV. Despite the presence of the Ser630 hydroxyl in DPP-IVH740Q for potential covalent interaction, its ability to function as a nucleophile was compromised by a dysfunctional catalytic triad as a result of the H740Q mutation. The K d for saxagliptin decreased by three orders of magnitude (Table 3), whereas DPP-IVH740Q bound the nitrile-deficient inhibitor BMS-538305 with similar affinity as DPP-IVWT. As with DPP-IVS630A, this change in saxagliptin K d is larger than expected; similar to DPP-IVS630A, the inability of the enzyme to prepare the inhibitor for covalent interaction translates into a suboptimal geometric configuration and steric hindrance.
Examination of the X-ray co-crystal structure of saxagliptin bound to DPP-IVWT reveals that a saxagliptin-derived imidate nitrogen is within hydrogen-bonding distance to the side-chain hydroxyl of Y547 (Fig. 3). This observation is consistent with reports that Y547 plays a major role in catalysis, presumably by stabilizing the oxyanion hole through hydrogen bonding (Aertgeerts et al. 2004). Loss of proteolytic activity has been observed upon mutation of Y547 (Bjelke et al. 2004) and in our present studies. To determine whether Y547 plays a role in stabilizing the enzyme–inhibitor complex, the binding of saxagliptin was measured for both DPP-IVY547Q and DPP-IVY547F. Ideally an increased K d of saxagliptin in DPP-IVY547F, but not in DPP-IVY547Q, would indicate the importance of this residue in stabilization of the enzyme–inhibitor complex. We found both mutant proteins bound saxagliptin very tightly (<5 nM), agreeing well with its potent K i of 0.5 nM for wild-type protein (Kim et al. 2006). Thus, replacing this residue either by phenylalanine or glutamine did not alter the binding potency significantly. While this result initially seems to disagree with the activity data, we cannot rule out potential changes in K d beyond the detection limit of our ITC assay. For example, even if a 10-fold reduction in the K d could occur, it would be undistinguishable in our assay, and thus the role of Y547 cannot be assessed unambiguously from our data. Alternatively, mutation of tyrosine to phenylalanine could allow for an additional water molecule to fill the space previously occupied by the hydroxyl, thereby potentially compensating for some of the lost binding energy resulting from the disrupted hydrogen-binding patterns (Bjelke et al. 2004) with the inhibitor, but not the oxyanion of intermediate during the catalysis.
The thermodynamic parameters measured by ITC for inhibitor binding to DPP-IV variants are shown in Tables 3 and 4. Comparison of the differences in observed heats of binding (ΔH obs; Table 3) is somewhat compromised by the moderate (4.88 kcal/mol) heat of ionization of HEPES (Goldberg et al. 2002). However, we expect that, when the differences in the observed heats of binding between saxagliptin and BMS-538305 are relatively large, such a comparison may still be informative. We also note that the heats are uncorrected for potential release or uptake of protons during binding. The observed enthalpy for binding DPP-IVWT is nearly −10 kcal/mol greater for saxagliptin than for BMS-538305, reflecting the formation of the weak covalent bond between the cyano group of saxagliptin and Ser630 (see Fig. 3); this cyano group is absent in BMS-538305. When S630 was mutated to alanine, binding of saxagliptin was abolished, and no binding enthalpy was detected. In contrast, BMS-538305 bound DPP-IVWT and DPP-IVS630A with similar affinities and observed enthalpies, indicating that binding of BMS-538305 was unperturbed by the mutation. Binding of saxagliptin to DPP-IVH740Q was reduced 220-fold relative to DPP-IVWT; this reduction in binding is reflected in the reduction of observed binding heat for saxagliptin for DPP-IVH740Q (−11.3 kcal/mol) compared to DPP-IVWT (−17.6 kcal/mol). Moreover, the observed ΔH of binding of saxagliptin to DPP-IVH740Q was essentially the same as that for BMS-538305, consistent with the absence of covalent bond formation. For DPP-IVY547Q, the high favorable observed ΔΔH of binding (−7.9 kcal/mol) for saxagliptin relative to BMS-538305 is also consistent with covalent bond formation. Overall, the comparisons show that large changes in observed binding enthalpy are consistent with formation of the covalent complex with saxagliptin observed in the DPP-IVWT crystal structure.
Table 5.
Summary of the downfield NMR resonance shifts for WT and mutant DPP-IV proteins
Mechanism-based serine protease inhibitors when complexed with the enzyme often feature short strong hydrogen bonds readily detectable by 1H NMR (Cassidy et al. 1997). In our previous publication (Kim et al. 2005), we reported our observation of several resonances which could be related to the protons of the catalytic triad featuring SSHB in both free, as well as saxagliptin-complexed, DPP-IV. In addition to large downfield chemical shifts, these resonances had reduced fractionation factors (∼0.5) with respect to the fractionation factors of protons engaged in “normal” hydrogen bonds. By comparison to literature spectra, we postulated that the resonance at 16 ppm in the 1H NMR spectrum was the Nδ1 proton of H740. In an attempt to confirm our assignments, we examined the spectra of the mutant proteins. Surprisingly, the resonance at 16 ppm was present in H740Q mutant as well. We found the same resonance in all of our mutant proteins, free and complexed with either inhibitor, suggesting that this signal does not arise from an active-site residue. A similar observation has been reported for serine proteases belonging to the class of prolyl oligopeptidases: prolyl oligopeptidase (PO) and oligopeptidase B of Escherichia coli (OpB) (Kahayaoglu et al. 1997). Downfield chemical shifts (∼17 and ∼16 ppm) were observed for the wild type and an active-site H652A mutant of OpB. Nuclear Overhauser enhancement (NOE) experiments provided strong evidence that these two resonances belonged to two different noncatalytic histidines. Because DPP-IV contains 19 histidine residues, further mutagenesis studies to assign this resonance would be impractical to execute. Examination of the structure of DPP-IV reveals no His-Asp (or Glu) pairs that are sufficiently close to form a SSHB. A likely candidate for the resonance at 16.1 ppm is a hydrogen bond that is observed in the active site between the carboxyl groups of E206 and D663. Not only is the hydrogen-bonding distance short (2.5 Å), the hydrogen-bond configuration is observed in all structures of DPP-IV, both complexed and uncomplexed, determined to date. Our NMR data are consistent with this observation. The conserved configuration of this hydrogen bond may serve to preorganize the acid moieties of the active site (E205, E206) for binding substrate and inhibitor, and it may contribute an additional restraint for enzyme specificity (see below).
Upon either saxagliptin or BMS-538305 binding to DPP-IVWT and both Y547 mutants, resonances appeared at ∼17 ppm and at either ∼14 or ∼14.6 ppm. The resonances at ∼14 and ∼14.6 ppm were completely absent in H740Q mutant, and therefore we suggest that this resonance is reflective of an Hδ1 in a protonated dyad as a result of the enzyme–inhibitor complex formation. Although a similar resonance at 14.4 ppm appears in DPP-IVS630A, it cannot be that of the histidine because DPP-IVS630A did not bind saxagliptin. Additionally, this resonance was also present in the free (S630A) enzyme as well.
Upon BMS-538305 binding to DPP-IVS630A, a resonance appears at 14.2 ppm; the origin of this resonance is unclear. Because we have no evidence of catalytic residue involvement in BMS-538305 binding (its affinity for DPP-IVS630A is comparable to that for DPP-IVWT; Table 3), this resonance could be the same as that observed in the free enzyme, just slightly perturbed in chemical shift by being in spatial proximity of ring current effects from the inhibitor. Alternatively, the resonance could be a direct result of complex formation due to engagement with inhibitor, as a similar resonance appears in DPP-IVWT and both Y547 mutants upon binding BMS-538305. Even though mechanism of inhibition by BMS-538305 does not require serine nucleophilicity, binding compound might “tighten up” the active site, deshielding Nδ1 the same way that protonation of Nε2 does in the case of saxagliptin binding.
A new resonance also appeared at ∼17.2 ppm when the wild type and all mutant proteins were complexed with either inhibitor. This resonance is clearly related to ligand binding but is unrelated to either of the catalytic residues. A prominent and a very distinct feature of DPP-IV relative to other proteases is the requirement of the substrate N-terminal free amine for binding and proteolysis. Hydrogen-bond formation between E205/E206 and the N terminus of the substrate is the most important interaction for aligning the peptide before cleavage (Rasmussen et al. 2003). This same interaction is maintained in all DPP-IV inhibitors known to date: an amine anchored by the same glutamates (Fig. 3). In the X-ray co-crystal structure of saxagliptin bound to DPP-IVWT, a network of hydrogen bonds is observed between the basic amine of the inhibitor, Y662, and the carboxyl groups of E205 and E206. Although the intermolecular hydrogen bonds are too long (2.8–2.9 Å) to fall into the category of a SSHB, the unusually downfield chemical shift of the resonance ascribed to this proton can be readily attributed to the electrostatic effect on the hydrogen-bonding proton from the negatively charged carboxyl groups of E205 and E206. At neutral pH, the amine will be positively charged. Thus, the interaction between the inhibitor basic amine and the enzyme carboxyl groups may be better categorized as a tight ionic hydrogen bond. Increasing the basicity of the amine causes the resonance at 17.2 ppm to shift to 15.7 ppm upon complexation with DPP-IV (data not shown), supporting the notion that it is the basic amine of saxagliptin giving rise to the resonance at 17.2 ppm. Definitive assignment of the resonance to this hydrogen bond needs to be confirmed by further mutagenesis studies.
We have confirmed that saxagliptin is a mechanism-based inhibitor and that H740 is a critical residue that is required to enhance S630 nucleophilicity for covalent adduct formation. Despite H740 making no physical contact with the inhibitor, mutating H740 to Q reduces saxagliptin binding by approximately three orders of magnitude (from 0.5 nM to 1.1 μM) without altering the interaction of BMS-538305 (K d of 14 vs. 6 nM for DPP-IVWT and DPP-IVH740Q, respectively), which lacks the electrophilic serine trap of a nitrile group. Protonation of the catalytic triad (at Nε1 of H740) during the covalent complex formation is accompanied by a new resonance at ∼14 ppm in the downfield 1H NMR spectrum that is characteristic of a SSHB. By stabilizing the first step of the acylation reaction, this hydrogen bond can facilitate both deprotonation of S630 by S630 and the subsequent nucleophilic attack on the nitrile carbon of saxagliptin.
Materials and Methods
Cloning and expression of DPP-IV proteins
Baculovirus
The wild-type DPP-IV (S39-P766) gene was amplified from the template pD439BacHis plasmid DNA by PCR and then cloned into baculovirus transfer vector pAcGP67-B (BD Biosciences Pharmagen) at BamHI/EcoRI sites to generate the C-terminal His-tagged DPP-IV (S39-P766) construct. The specific PCR primers designed for baculovirus expression plasmid were forward, 5′-GCGCGGATCCAGTCGCAAAACTTACACTCT-3′; and reverse, 5′-GCGCGAATTCTCAATGATGATGATGATGATGAGGTAAAGAGAAACATTG-3′. Mutants were generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, cat. no. 200521-5) to mutate the residues S630A, H740Q, Y547F, and Y547Q. With the above wild-type DPP-IV clone in pAcGP67-B as the template, the four mutants were generated with the following primer sets: Ser630A-forward (GCAATTTGGGGCTGGGCATATGGAGGGTACGTAACC) and S630A-reverse (GGTTACGTACCCTCCATATGCCCAGCCCCAAATTGC); H740Q-forward (GGTATACTGATGAAGACCAGGGAATAGCTAGCAGCACAGC) and H740Q-reverse (GCTGTGCTGCTAGCTATTCCCTGGTCTTCATCAGTATACC); Y547F-forward (CCTCTACTATTAGATGTGTTTGCAGGCCCATGTAGTC) and Y547F-reverse (GACTACATGGGCCTGCAAACACATCTAATAGTAGAGG); and Y547Q-forward (CCTCTACTATTAGATGTGCAGGCAGGCCCATGTAGTCAAAAAGC) and Y547Q-reverse (GCTTTTTGACTACATGGGCCTGCCTGCACATCTAATAGTAGAGG).
Recombinant baculovirus was generated via co-transfection with BaculoGold (BD Biosciences Pharmingen), and amplified once. The optimized culture condition was determined by screening DPP-IV expression in Sf9 or Hi5 cells, varying the multiplicity of infection (MOI) and time of infection in a 24-well block. The second-passage virus was used to infect Hi5 cells at a density of 2 × 106/mL with MOI of 1 at 27°C in a 25/50 wave reactor (Wave Biotech) for a 20-L cell culture. The supernatant was harvested 48–72 h post-infection at which time the cell viability was ∼70%. Expression levels for the wild-type S39-P766 were determined by activity using Gly–Pro–pNA pseudosubstrate in 25 mM Tris-HCl (pH 7.5), 0.15 M NaCl, 0.1% Triton X-100 (v/v). The rate of the absorbance at 405 nm was measured using a SpectraMax microtiter plate reader. The expression levels of the four mutants were measured by ELISA using purified wild-type DPP-IV as reference standard. The 96-well microtiter plates (Nunc) were coated with 100 μL of 3 μg/mL adenosine deaminase from calf intestine (Roche Diagnostics) in PBS and incubated overnight at 4°C. Wells were washed with 20 mM sodium phosphate, 0.15 M NaCl, pH 7.4 (PBS), containing 0.05% Tween-20 and incubated for 1 h in blocking buffer (PBS containing 10 mg/mL BSA and 0.05% Tween-20). Following a 1-h incubation with mutant DPP-IV samples, the wells were incubated with 100 μL of 1 μg/mL of an anti-DPP-IV monoclonal antibody MAB1180 (R&D Systems) in blocking buffer for 1 h. The plate was washed and then incubated with goat anti-rat IgG-HRP (Southern Biotechnology) in blocking buffer for 30 min. The color was developed with TMB peroxidase substrate (Pierce), and the reaction was stopped with 100 μL of 1 M sulfuric acid. The absorbance was read at 450 nm. Expression levels were typically around 10–14 mg/L.
Pichia pastoris
The P. pastoris strains KM71H, GS115, and X-33, and the pPICZα-A vector were purchased from Invitrogen. The gene encoding DPP-IV (39–766) was amplified from the template pD439BacHis plasmid and was subcloned into the pPICZα-A P. pastoris expression vector at the EcoRI/XbaI sites. The specific PCR primers designed for the P. pastoris expression plasmid pPICZα/DPP-IV (39–766) were forward, 5′-GCGCGAATTCAGTCGCAAAACTTACACTCT-3′; and reverse, 5′-GCGCTCTAGAGGAGGTAAAGAGAAACATTG-3′. The recombinant expression vector pPICZα/DPP-IV (39–766) was amplified in E. coli DH5α, and positive colonies were selected. The pPICZα/DPPIV (39–766) plasmid was purified and confirmed by DNA sequencing. The construct was linearized with PmeI and the DNA recovered using the QIAquick PCR Purification Kit (QIAGEN). The fragment was resuspended in water and used to transform P. pastoris KM71H, GS115, and X-33 cells which were made competent in accordance with the manufacturer's instructions (Invitrogen). Transformation was performed using a 2-mm gap cuvette at 50 μF, 1500 V, with a BTX Electro Cell Manipulator. After pulsing, cells were suspended in 1 mL of cold 1 M sorbitol and plated onto YPD plates (2% peptone, 1% yeast extract, and 2% dextrose) containing 100 μg/mL Zeocin and incubated at 30°C for 3 d. Colonies were re-streaked onto fresh YPD plates containing 50 μg/mL Zeocin, and individual clones were screened for expression of DPP-IV. Colonies were placed into 24-well plates (Whatman) containing 2 mL of BMGY (2% peptone, 1% yeast extract, 1% glycerol, 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, and 100 mM potassium phosphate buffered to pH 6.0) and 25 μg/mL Zeocin and shaken at 220 rpm for 24 h at 30°C. The plates were centrifuged at 2000g for 5 min, and the supernatant removed. Cells were resuspended in 2 mL of BMMY (BMGY with 1% methanol in place of glycerol) and 25 μg/mL Zeocin and shaken at 220 rpm for 24 h. An additional 20 μL of methanol was added to each well, and after a further 24-h incubation the plates were centrifuged and supernatants removed for DPP-IV analysis. The DPP-IV activity was determined using the same activity assay described above for baculovirus-expressed wild-type DPP-IV (39–766). Selected clones possessing DPP-IV activity from the expression screening were confirmed for DPP-IV expression when performed in shake flasks (0.4 L of BMGY in 2-L flasks) using the same media, induction, and culture conditions. One clone, clone 44 in KM71H cells, was chosen to be scaled up for purification. A colony of clone 44 was inoculated into a shake flask containing 200 mL of BMGY with 25 μg/mL Zeocin and shaken for 48 h at 250 rpm. This inoculum was used to inoculated five 2-L shake flasks, each containing 0.45 L of BMGY with 25 μg/mL Zeocin. After 48 h at 30°C and 250 rpm, the cells were centrifuged at 2000g for 10 min, and the supernatant was removed. The cells were resuspended with 0.45 L of BMMY with 1% methanol and 25 μg/mL Zeocin, placed into new flasks, and returned to shaking at 250 rpm for 70 h, with methanol added to 1% at 24 and 44 h. Cells were harvested by centrifuging at 3000g for 20 min, and the supernatant was passed through a 0.45 μm filter.
Protein purification
DPP-IV expressed in baculovirus
The supernatant was concentrated 15-fold by tangential flow filtration using a 0.5 m2 30 kDa MW cut-off membrane (Millipore) and then buffer-exchanged with five volumes of Buffer A (20 mM Tris-HCl [pH 8], 0.1 mM EDTA). DPP-IV was captured on a 5 × 24 cm column of Q-Sepharose Fast Flow (Amersham Pharmacia) and then washed with four column volumes of Buffer A. DPP-IV was eluted with a linear gradient from Buffer A to Buffer B (Buffer A with 1 M NaCl) over 2.5 column volumes. DPP-IV eluted between 100 and 200 mM NaCl. Fractions were pooled, the NaCl concentration was adjusted to 0.5 M, and imidazole was added to 10 mM. The samples were loaded on a 2.6 × 5 cm Ni-NTA column (QIAGEN). The column was washed with 10 column volumes of Buffer C (50 mM sodium phosphate [pH 7.9], 0.5 M NaCl). Weak binding impurities were eluted with a gradient from Buffer C to 10% Buffer D (Buffer C with 0.3 M imidazole) over five column volumes. DPP-IV was eluted with 100% Buffer D. Lastly, the concentrated DPP-IV fractions were run on a 2.6 × 60 cm column of Superdex-200. Unlike DPP-IVWT, DPP-IVS630A, DPP-IVH740Q, and DPP-IVY547F, the DPP-IVY547Q mutant contained about 50% monomer which was removed on the Superdex-200 column. Concentrations were determined by the absorbance at 280 nm using the extinction coefficient of ε = 0.46 (mg/mL)−1 calculated from the amino acid composition.
DPP-IV expressed in P. pastoris
About 15 L of supernatant from P. pastoris was concentrated 45-fold by tangential flow filtration using a 0.1-m2 30-kDa MW cut-off membrane (Millipore). The solution was buffer-exchanged into 20 mM Tris-HCl, 0.1 mM EDTA, pH 8, as previously described. The EDTA concentration was brought to 5 mM, and solid ammonium sulfate was added slowly to a final concentration of 1.6 M while stirring with a magnetic stir bar. The solution was loaded on a 5 × 9.5 cm column of Butyl Sepharose. The column was washed with 25 mM Tris-HCl (pH 8), 5 mM EDTA, 1.6 M ammonium sulfate (Buffer E) until the absorbance reached baseline. The DPP-IV was eluted using a descending gradient from 100% to 30% Buffer E over 3.5 column volumes by mixing the buffer with Buffer E without the ammonium sulfate. The peak fractions that eluted between 1.4 and 1.1 M ammonium sulfate were pooled, concentrated, and dialyzed in 20 mM Tris-HCl (pH 8), 0.1 mM EDTA. The sample was run on a 2.6 × 10 cm column of Resource Q (Amersham Pharmacia). The column was washed with Buffer A followed by a gradient from 100% Buffer A to 25% Buffer B over 10 column volumes. DPP-IV eluted between 80 and 150 mM NaCl. The fractions were concentrated to 5 mg/mL and treated with 3 units of EndoH/μg protein (New England BioLabs) for 18 h at 37°C. The sample was run on a 2.6 × 60 cm column of Superdex-200 in 20 mM Tris-HCl (pH 7.8), 0.1 M NaCl, 0.05 mM EDTA.
Analytical size-exclusion chromatography coupled to multiple-angle light scattering (SEC/MALS)
Isocratic separations were carried out using an Agilent 1100 series HPLC on a Phenomenex BioSep-SEC-S 3000 column, in buffer containing 200 mM K2HPO4 (pH 6.8 with HCl), 150 mM NaCl, and 0.02% sodium azide and run at 0.5 mL/min. The HPLC was outfitted with a diode-array absorbance detector. A Wyatt Technology MiniDawn Tristar laser light scattering instrument was plumbed downstream from the HPLC, followed by a Wyatt Optilab differential refractometer, both of which were calibrated according to the manufacturer's guidelines. BSA (Sigma) was run along with samples for analysis to verify system performance. GPC protein standards (Bio-Rad) were also run routinely to check chromatographic performance and as reference for retention time. Particulates were removed from protein samples with a centrifugal filtration device (Nanosep MF, 0.2 μm, Pall Corporation) prior to injection. Data were analyzed with Origin (MicroCal), Astra (Wyatt), and ChemStation (Agilent).
Crystal growth conditions and treatment
Crystals were grown of both the apo protein and protein complexed with inhibitor. The apo protein stock solution consisted of 10.0 mg/mL DPP-IV (expressed in P. pastoris) in 20 mM Tris-HCl, pH 7.8. Crystallization trials were prepared by the hanging-drop vapor diffusion method. Initial crystallization screens were prepared using a CyberLab (Gilson) C-240 on NeuroProbe hanging-drop trays using Hampton Research crystallization screening solutions Crystal Screen 1 and 2, Natrix, MembFac, Index, and SaltRx. Initial crystals were observed and conditions were successfully optimized for harvesting and data collection.
The optimized condition was prepared by the hanging-drop vapor diffusion on siliconized cover slips in 24-well Linbro plates. The 1-mL reservoir solution consisted of 15%–20% w/v PEG 3350, 0.2 M Mg2SO4, 15% v/v glycerol, 0.1 M Tris-HCl at pH 8.5. Drops were formed from 1 μL of the protein solution and 1 μL of the reservoir solution (total initial volume of 2 μL), mixed, and equilibrated at room temperature. Crystals appeared within 1 wk. Single crystals were removed and prepared for data collection at 100 K using the crystallization drop as the cryosolution. The diffraction patterns from crystals gave unit-cell parameters of a = 65.3 Å, b = 68.0 Å, c = 423.8 Å, and α = β = γ = 90°. The symmetry was consistent with space group P212121. With this unit cell and space group, the asymmetric unit was estimated to contain two molecules per asymmetric unit based on a Matthew's number of 2.8 and 55.9% solvent fraction.
Apo crystals were placed in a soaking solution consisting of 20%–25% w/v PEG 3350, 0.200 M Mg2SO4, 15% v/v glycerol, 0.1 M Tris-HCl at pH 8.5, and 3 mM ligand for a minimum of 1 h. Crystals remained stable for weeks in soaking solution. Crystals were prepared for data collection as previously described.
Co-crystallization with ligand used the same protein stock solution and crystallization procedure described above with the addition of 0.60 mM ligand (≥5× molar excess).
Apo crystals soaked with inhibitor tended to yield better-diffracting crystals that those co-crystallized with inhibitor.
Structural determination and refinement
Data collection was performed at beamline X29 at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY), with the wavelength tuned to 1.1 Å, nitrogen cryostream set to 100 K, and ADSC Quantum 315 detector positioned at 325 mm. The diffraction images were processed with DENZO and SCALEPACK from the HKL suite (Otwinowski and Minor 1997). The intensities were converted to structure-factor amplitudes and placed on an absolute scale with TRUNCATE (French and Wilson 1978) from the CCP4 suite (Bailey 1994). The initial phases were calculated by molecular replacement by AMoRe (Navaza 1994) with the dimer from 1FPU used as the search model. The structure was refined to 2.35 Å resolution with CNX (Brünger et al. 1998) and modeled with QUANTA (Accelrys, Inc.). The data collection and refinement statistics are summarized in Table 2.
Coordinates
The refined coordinates and structure factors were deposited in the Protein Data Bank (Berman et al. 2000) as ID 3BJM.
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry experiments were carried out at 25°C in a VP-ITC microcalorimeter (Microcal Inc.). DPP-IV proteins were dialyzed against 50 mM HEPES, pH 7.5, and concentrations were estimated after dialysis by absorbance using a molar extinction coefficient of 187,000 M−1 cm−1. Inhibitor was dissolved in dimethyl sulfoxide (DMSO) immediately prior to use. All solutions for titration contained dialysis buffer with 0.5% DMSO (v/v). Data were analyzed using the single-site binding model in Origin 7 (Microcal) software. In a typical experiment, the calorimetry cell containing 2.5 μM DPP-IV (as monomer) was titrated with a syringe solution containing 50 μM inhibitor.
Thermal stability enhancement (TSE)
The TSE effects of inhibitors on DPP-IV proteins were measured with a ThermoFluor instrument (3-Dimensional Pharmaceuticals, Inc.) (Pantoliano et al. 2001; Carver et al. 2005). Thermal denaturation curves were measured by monitoring the fluorescence enhancement of an extrinsic probe (1-anilino-8-napthalene sulfonate [ANS]) as it preferentially bound to the unfolded protein. Reactions contained 1.3 μM DPP-IV in the presence or absence of varying concentrations of inhibitor in final conditions of 50 mM HEPES, pH 7.5, 100 μM ANS, and 0.5% (v/v) DMSO. Reactions were monitored in 384-well plates (Abgene) by increasing temperature in 1°C increments with 60 sec equilibration at each temperature, and measuring four 10 sec exposure digital images (plus one dark-field image) per point from 25°C to 95°C. Thermal denaturation curves were analyzed with ThermoFluor analysis software (3DP, Inc.) to determine T m values (temperature at which the unfolding reaction is half complete).
Proton NMR experiments
NMR spectra were recorded on a Varian Inova 600 spectrometer equipped with a triple-resonance 5-mm probe with triax gradients using the 1-1 sequence to suppress the water signal with minimal saturation (Hore 1983). Spectra were acquired over 16,384 scans with a sweep width of 30 kHz, 8192 time domain points, and a relaxation delay of 4 sec, at 20°C. All DPP-IV solutions were 50 μM protein in 25 mM Tris, pH 7.5, 140 mM NaCl; 1.2 equivalents of saxagliptin or BMS-538305 (25 mM stock in d 6-DMSO) were added to the DPP-IV solution, yielding a final ligand concentration of 100 μM. DMSO concentration did not exceed 0.4%. Chemical shifts are reported relative to the water resonance referenced at 4.76 ppm.
Acknowledgments
We thank Howard Robinson and Stu Myers (PX operator) for assistance with data collection and beamline preparation and Andy Pudzianowski for insightful discussions on quantum mechanics and SSHBs.
Footnotes
Reprint requests to: William J. Metzler, Mail Stop H23-02, P.O. Box 4000, Princeton, NJ 08543-4000, USA; e-mail: william.metzler@bms.com; fax: (609) 252-6012.
Abbreviations: ANS, 1-anilino-8-naphthalene sulfonate; DPP-IV, dipeptidyl peptidase IV; GLP-1, glucagon-like peptide-1; ITC, isothermal titration calorimetry; pNA, p-nitroaniline; Pn–Pn ′, amino acid residues of the substrate which numerically indicate the position relatively to the scissile bond, n being the residues toward the N terminus and n′ being the residues toward the C terminus (this nomenclature was first defined by Schechter and Berger [1967]); SEC-MALS, size-exclusion chromatography–multiple angle light scattering; SKIE, solvent kinetic isotope effect; SSHB, short strong hydrogen bond; SIHB, short ionic hydrogen bond; TSE, thermal stability enhancement.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073253208.
References
- Aertgeerts, K., Ye, S., Tennant, M.G., Kraus, M.L., Rogers, J., Sang, B.-C., Skene, R.J., Webb, D.R., Prasad, G.S. Crystal structure of human dipeptidyl peptidase IV in complex with the decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Sci. 2004;13:412–421. doi: 10.1110/ps.03460604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahren, B., Simonsson, E., Larsson, H., Landin-Olson, M., Torgeirsson, H., Jansson, P.A., Sandquist, M., Bavenholm, P., Efendic, S., Eriksson, J.W. Inhibition of dipeptidyl peptidase IV improves metabolic control over a 4-week study period in type 2 diabetes. Diabetes Care. 2002;25:869–875. doi: 10.2337/diacare.25.5.869. [DOI] [PubMed] [Google Scholar]
- Ahren, B., Gomis, R., Standl, E., Mills, D., Schweizer, A. Twelve- and 52-week efficacy of the dipeptidyl peptidase IV inhibitor LAF237 in metformin-treated patients with Type 2 diabetes. Diabetes Care. 2004;27:2874–2888. doi: 10.2337/diacare.27.12.2874. [DOI] [PubMed] [Google Scholar]
- Ahren, B., Pacini, G., Foley, J.E., Schweizer, A. Improved meal-related β-cell function and insulin sensitivity by the dipeptidyl peptidase-IV inhibitor vildagliptin in metformin-treated patients with Type 2 diabetes over 1 year. Diabetes Care. 2005;28:1936–1940. doi: 10.2337/diacare.28.8.1936. [DOI] [PubMed] [Google Scholar]
- Augeri, D.J., Betebenner, D.A., Magnin, D.R., Robl, J.A., Khanna, A., Robertson, J.G., Wang, A., Simpkins, L.M., Taunk, P., Huang, Q., et al. Discovery and preclinical profile of saxagliptin (BMS-477118): A highly potent, long-acting, orally active dipeptidyl peptidase IV inhibitor for the treatment of Type 2 diabetes. J. Med. Chem. 2005;48:5025–5037. doi: 10.1021/jm050261p. [DOI] [PubMed] [Google Scholar]
- Bailey, S. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelke, J.R., Christense, J., Branner, S., Wagtmann, N., Olsen, C., Kanstrup, A.B., Rasmussen, H.B. Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV. J. Biol. Chem. 2004;279:34691–34697. doi: 10.1074/jbc.M405400200. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carver, T.E., Bordeau, B., Cummings, M.D., Petrella, E.C., Pucci, M.J., Zawadzke, L.E., Dougherty, B.A., Tredup, J.A., Bryson, J.W., Yanchunas J., Jr, et al. Decrypting the biochemical function of an essential gene from Streptococcus pneumoniae using ThermoFluor technology. J. Biol. Chem. 2005;280:11704–11712. doi: 10.1074/jbc.M413278200. [DOI] [PubMed] [Google Scholar]
- Cassidy, C.S., Lin, J., Frey, P.A. A new concept for the mechanism of action of chymotrypsin: The role of the low-barrier hydrogen bond. Biochemistry. 1997;36:4576–4584. doi: 10.1021/bi962013o. [DOI] [PubMed] [Google Scholar]
- Chien, C.-H., Huang, L.-H., Chou, C.Y., Chen, Y.S., Han, Y.-S., Chang, G.-G., Liang, P.-H., Chen, X. One site mutation disrupts dimer formation in human DPP-IV proteins. J. Biol. Chem. 2004;279:52338–52345. doi: 10.1074/jbc.M406185200. [DOI] [PubMed] [Google Scholar]
- French, S., Wilson, K. On the treatment of negative intensity observations. Acta Crystallogr. 1978;A34:517–525. [Google Scholar]
- Goldberg, R.N., Kishore, N., Lennen, R.M. Thermodynamic quantities for the ionization reactions of buffers. J. Phys. Chem. Ref. Data. 2002;31:231–370. [Google Scholar]
- Hore, P.J. Solvent suppression in Fourier transform nuclear magnetic resonance. J. Magn. Reson. 1983;55:283–300. [Google Scholar]
- Hughes, T.E., Mone, M.D., Russell, M.E., Weldon, S.C., Villhauer, E.B. NVP-DPP728 (1-[[[2-[(5-cyanopyridin-2-yl)amino]ethyl]amino]acetyl]-2-cyano-(S)-pyrrolidine), a slow-binding inhibitor of dipeptidyl peptidase IV. Biochemistry. 1999;38:11597–11603. doi: 10.1021/bi990852f. [DOI] [PubMed] [Google Scholar]
- Kahayaoglu, A., Haghjoo, K., Guo, F., Jordan, F., Kettener, C., Felfoldi, F., Polgar, L. Low barrier hydrogen bond is absent in the catalytic triads in the ground state but present in a transition-state complex in the prolyl oligopeptidase family of serine proteases. J. Biol. Chem. 1997;272:25547–25554. doi: 10.1074/jbc.272.41.25547. [DOI] [PubMed] [Google Scholar]
- Kim, D., Wang, L., Beconi, M., Eiermann, G.J., Fisher, M.H., He, H., Hickey, G.J., Kowalchick, J.E., Leiting, B., Lyons, K., et al. (2R)-4-Oxo-4[trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: A potent, orally active dipeptidyl peptidase IV inhibitor for the treatment of Type 2 diabetes. J. Med. Chem. 2005;48:141–151. doi: 10.1021/jm0493156. [DOI] [PubMed] [Google Scholar]
- Kim, Y.B., Kopcho, L.M., Kirby, M.S., Hamann, L.G., Weigelt, C.A., Metzler, W.J., Marcinkeviciene, J. Mechanism of Gly–Pro–pNA cleavage catalyzed by dipeptidyl peptidase-IV and its inhibition by saxagliptin (BMS-477118) Arch. Biochem. Biophys. 2006;445:9–18. doi: 10.1016/j.abb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Lambeir, A.M., Proost, P., Durinx, C., Bal, G., Senten, K., Augustyns, K., Scharpe, S., Van Damme, J., De Meester, I. Kinetic investigation of chemokine truncation by CD26/dipeptidyl peptidase IV reveals a striking selectivity within the chemokine family. J. Biol. Chem. 2001;276:29839–29845. doi: 10.1074/jbc.M103106200. [DOI] [PubMed] [Google Scholar]
- Lin, J., Westler, W.M., Cleland, W.W., Markley, J.L., Frey, P.A. Fractionation factors and activation energies for the low barrier hydrogen bonding proton in peptidyl trifluoromethyl ketone complexes of chymotrypsin. Proc. Natl. Acad. Sci. 1998;95:14664–14668. doi: 10.1073/pnas.95.25.14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza, J. AMoRe: An automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- Oefner, C., D'Arcy, A., Mac Sweeney, A., Pierau, S., Gardiner, R., Dale, G.E. High-resolution structure of human apo dipeptidyl peptidase IV/CD26 and its complex with 1-[([2-[(5-iodopyridin-2-yl)amino]-ethyl]amino)-acetyl]-2-cyano-(S)-pyrrolidine. Acta Crystallogr. 2003;D59:1206–1211. doi: 10.1107/s0907444903010059. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pantoliano, M.W., Petrella, E.C., Kwasnoski, J.D., Lobanov, V.S., Myslik, J., Graf, E., Carver, T., Asel, E., Springer, B.A., Lane, P., et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- Rasmussen, H.B., Branner, S., Wiberg, F.C., Wagtmann, N. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat. Struct. Biol. 2003;10:19–25. doi: 10.1038/nsb882. [DOI] [PubMed] [Google Scholar]
- Robillard, G., Schulman, R.G. High resolution nuclear magnetic resonance study of the histidine–aspartate hydrogen bond in chymotrypsin and chymotrypsinogen. J. Mol. Biol. 1972;71:507–511. doi: 10.1016/0022-2836(72)90366-x. [DOI] [PubMed] [Google Scholar]
- Schechter, I., Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Villhauer, E.B., Brinkman, J.A., Naderi, G.B., Dunning, B.E., Mangold, B.L., Mone, M.D., Russell, M.E., Weldon, S.C., Hughes, T.E. 1-[2-[(5-Cyanopyridin-2-yl)amino]ethylamino]acetyl-2-(S)-pyrrolidinecarbonitrile: A potent, selective, and orally bioavailable dipeptidyl peptidase IV inhibitor with antihyperglycemic properties. J. Med. Chem. 2002;45:2362–2365. doi: 10.1021/jm025522z. [DOI] [PubMed] [Google Scholar]