Abstract
Airborne microorganisms have been studied for centuries, but the majority of this research has relied on cultivation-dependent surveys that may not capture all of the microbial diversity in the atmosphere. As a result, our understanding of airborne microbial ecology is limited despite the relevance of airborne microbes to human health, various ecosystem functions, and environmental quality. Cultivation-independent surveys of small-subunit rRNA genes were conducted in order to identify the types of airborne bacteria and fungi found at a single site (Boulder, CO) and the temporal variability in the microbial assemblages over an 8-day period. We found that the air samples were dominated by ascomycete fungi of the Hypocreales order and a diverse array of bacteria, including members of the proteobacterial and Cytophaga-Flavobacterium-Bacteroides groups that are commonly found in comparable culture-independent surveys of airborne bacteria. Bacterium/fungus ratios varied by 2 orders of magnitude over the sampling period, and we observed large shifts in the phylogenetic diversity of bacteria present in the air samples collected on different dates, shifts that were not likely to be related to local meteorological conditions. We observed more phylogenetic similarity between bacteria collected from geographically distant sites than between bacteria collected from the same site on different days. These results suggest that outdoor air may harbor similar types of bacteria regardless of location and that the short-term temporal variability in airborne bacterial assemblages can be very large.
Bacteria and fungi are ubiquitous in the atmosphere, and microbial biomass can represent a significant proportion of the organic carbon fraction of airborne particulate matter (36, 53). Many microbes can remain viable even after extended periods of time aloft despite the challenges associated with surviving in the atmosphere, including extended UV exposure, low moisture levels, and extremely oligotrophic conditions (21). Atmospheric transport is a key mode of microbial dispersal (49), and the transmission of airborne plant and animal pathogens can have significant impacts on ecosystems, human health, and agricultural productivity. For example, unidentified microbes transported across the Atlantic by desert dust storms have been implicated as the causative agents of coral diseases, contributing to the decline of reef ecosystems in the Caribbean basin (48). In addition, many airborne bacteria and fungi can cause human diseases (45), particularly in individuals that are immunocompromised or otherwise sensitive to a broad range of allergenic and toxigenic biological material (11, 44). With the prevalence of asthma increasing worldwide in recent decades (4, 20), there is a growing need to better understand the diversity and spatiotemporal dynamics of airborne microbes.
Few studies have used molecular techniques to describe the extent of bacterial and fungal diversity suspended in air, since most literature in this arena has relied on cultivation techniques to isolate and identify the types of microbes present in air samples collected from either outdoor or indoor air. Since only a small percentage of microbes can be readily cultivated in the laboratory (39, 42), cultivation-dependent approaches are likely to underestimate airborne microbial diversity. This factor is likely to be exacerbated in the atmosphere, where large disparities between colony counts of airborne microbes and direct counts are typically observed (40).
The development of molecular methods for the identification of bacteria in environmental samples gives us the ability to survey all, or nearly all, of the bacteria present in a given volume of air without introducing a cultivation bias (40). Using such molecular survey techniques, a handful of recent studies have shown that the taxonomic diversity of airborne bacterial communities is far higher than we would have expected from traditional, culture-based surveys (3, 8, 35, 41, 52). Since similar techniques are less commonly used to characterize airborne fungal diversity (40), we do not know the extent to which cultivation-dependent surveys underestimate airborne fungal diversity. However, research on airborne fungi (5, 40) and fungi in other habitats (7, 38) has shown that molecular surveys are capable of identifying many fungi that are likely to be overlooked by cultivation-dependent or phenotypic surveys. While molecular methods of microbial community analysis are no panacea, such methods can be used to greatly advance our understanding of the identity, distribution, and abundance of airborne microbes.
As with any microbial habitat, the atmosphere is a heterogeneous environment, and the diversity of airborne microbes may vary spatially and temporally. It is well documented that air samples collected from different locations may differ with respect to the relative abundances of specific bacterial and fungal groups (8, 27, 46). Even when samples are collected from one location, the types of microorganisms present in the atmosphere can change significantly across a given day, month, or year (6, 24, 27). These temporal and spatial shifts are likely to be driven by changes in local and regional meteorological conditions (e.g., wind speed, solar radiation, and humidity) (21, 24, 29) and/or inoculum source (e.g., soils, plant surfaces, and sporulation release cycles) (26, 28).
For this project, we used a cultivation-independent molecular approach to identify the bacteria and fungi present in air samples collected from a single site in Boulder, CO. We documented shifts in the types of bacteria and fungi found in air samples collected over an 8-day period in order to assess the temporal variability associated with the diversity of airborne microbes.
MATERIALS AND METHODS
Sample collection.
All air samples were collected from a single location in the middle of the University of Colorado campus in Boulder, CO (1660 meters above sea level, 40.01°N, 105.27°W). The sampling location is bordered by buildings, paved lots, and a 0.5-ha maintained grass field. Air samples were collected from the site for a 4-h period at mid-day on each of 5 days between 17 and 25 May 2006. Meteorological data were collected with a Davis Instruments Vantage Pro 2 weather station (Hayward, CA) maintained by the University of Colorado and located approximately 250 m from the sampling site (http://foehn.colorado.edu/weather/paos1/PAOSstation.html). Samples were collected by using two swirling aerosol collectors (BioSampler; SKC, Inc., Eight Four, PA) located 2 m apart and 1.5 m above the ground surface, with the inlets oriented to the east. We sampled using liquid impingers because they are well-characterized sampling devices that have a high collection efficiency in the aerodynamic diameter range between 0.3 and 10 μm, and these glass devices are easy to sterilize and render DNA-free (40). We did not prefilter any of the samples because we wanted to survey the largest extent of microbiological diversity afforded by capture in these types of liquid impingers. Prior to sampling, the impingers were washed with ethanol, autoclaved, and filled with 20 ml of diethyl pyrocarbonate-treated 0.1 M phosphate-buffered saline (PBS) solution (pH 7). The flow rate through each aerosol collector was maintained at 12.5 liters min−1 over the entire 4-h collection period. We added sterile diethyl pyrocarbonate-treated H2O to the collectors at 20-min intervals to maintain a constant volume and maximize the trapping efficiency. The H2O and PBS solutions were filtered through a 0.2-μm-pore-size filter and autoclaved to ensure sterility immediately prior to use.
Clone library construction.
DNA was extracted from the solution in the liquid impingers immediately after sampling. The impinger solutions were filtered onto sterile cellulose nitrate filters with a 0.45-μm pore size (Nalgene, Rochester, NY), and the filters were placed directly into an UltraClean plant DNA isolation kit (MoBio Laboratories, Carlsbad, CA) with DNA extracted according to the manufacturer's instructions. Since this kit uses a vigorous bead-beating method to lyse cells, it should effectively disrupt most types of cells (40). The DNA samples extracted from the two replicated liquid impingers were pooled together prior to further analyses, and DNA concentrations were determined by PicoGreen fluorometry (Invitrogen, Carlsbad, CA). Parallel aliquots of sterile PBS collection solution were run through an identical DNA extraction procedure to check for sample contamination. These DNA extraction “blanks” were PCR amplified alongside the DNA samples extracted from the impingers.
DNA samples were PCR amplified using the universal primer pair 515f (5′-GTGCCAGCMGCCGCGGTAA-3′) and 1391r (5′-GACGGGCGGTGWGTRCA-3′). This primer pair amplifies small subunit rRNA genes from all three domains (Archaea, Bacteria, and Eukarya) (3, 23, 43), yielding a PCR product that is 850 to 1,100 bp in length. Although this primer set may not necessarily amplify all taxonomic groups equally, any amplification bias should be consistent across all of the collected samples. Each 50-μl PCR contained 1× PCR buffer, 2.5 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphate,1 mg of bovine serum albumin ml−1, 1 M betaine, 0.2 μM concentrations of each primer, 0.5 U of Taq polymerase, and 2 μl of DNA sample as described in the molecular aerobiology study by Angenent et al. (3). We ran three replicate PCR amplifications per DNA sample and pooled the amplicons from each sample prior to cloning. Amplicons were cleaned by using a QIAquick PCR purification kit (Qiagen, Valencia, CA), with the amplicon length confirmed by agarose electrophoresis.
Amplicons were cloned by using a TOPO TA for Sequencing kit (Invitrogen) according to the manufacturer's instructions. A total of 768 clones were bidirectionally sequenced per sample at the Broad Institute of Massachusetts Institute of Technology and Harvard. Bar-coded bioassay dishes were plated and incubated at 37°C overnight. White colonies were imaged and picked by using a QPix2 Benchtop colony picker (Genetix, Boston, MA). Each picked colony was inoculated into a single well of a 384-well growth plate. Plates were grown overnight at 37°C before plasmid amplification and sequencing. Capillary sequencing was performed on ABI 3730 DNA Analyzers (Applied Biosystems, Foster City, CA).
Sequence analyses.
Sequences were binned into major taxonomic groups (i.e., plant, metazoan, bacteria, and fungi) by using the BLAST algorithm (2) against the complete European rRNA database (http://www.psb.ugent.be/rRNA/index.html). Sequences with expect (E) values greater than 1e−100 to the nearest neighbors were not included in the analyses. Of the 768 clones sequenced per library, between 6 and 12% of the sequences were of low quality and/or chimeric and were not included in the analyses. The taxonomic classification of the fungal, metazoan, and plant sequences were based on the common lineage of closest matches to the European rRNA database. The bacterial sequences were aligned against the Greengenes database (16) by using the NAST aligner (15) and classified into taxonomic groups with the Greengenes “classify” utility (http://greengenes.lbl.gov/cgi-bin/nph-classify.cgi). Only sequences sharing >90% similarity to reference sequences over a 600-bp region were classified. The number of unique phylogenetic groups of bacteria (sequences with ≥97% sequence similarity) was estimated with the FastGroupII algorithm (55). We conducted more detailed phylogenetic analyses of specific bacterial groups that were common in the air samples, and these analyses are described in the caption of Fig. 3.
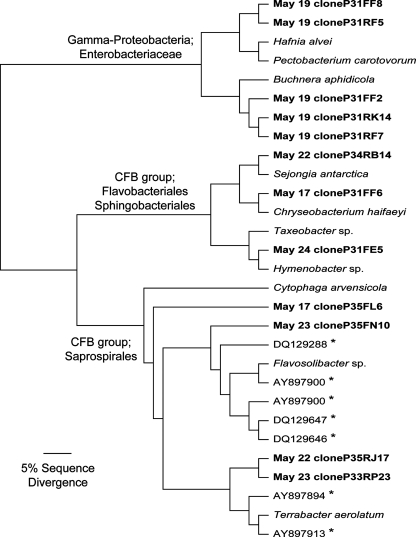
FIG. 3.
Phylogenetic relationships between representative sequences of the dominant airborne bacteria. Boldface type indicates sequences from the present study. Nearest-neighbor bacterial isolates are indicated by italics. Asterisks indicate representative sequences from other studies of airborne bacteria; the GenBank accession numbers starting with AY and DQ indicate sequences obtained from outside air samples by Angenent et al. (3) and Brodie et al. (8), respectively. Sequences were aligned by using MUSCLE (17), and the neighbor-joining tree was constructed by using PAUP (50). The archaeon, Haloferax volcanii, was included as the outgroup (not shown).
Forward and reverse sequence reads were not assembled prior to conducting the phylogenetic analyses. We kept the sequences unassembled so we could use them as an internal control to compare the results from the forward and reverse sequence reads separately and assess the robustness of our taxonomic assignments and between-sample comparisons of bacterial and fungal diversity. In all cases, the forward and reverse reads gave us nearly identical results so the results reported here are based on the forward reads alone (as in Table 2 and Fig. 1 to 3) or an averaging of the distances from the forward and reverse reads together (as in Table 3 and Fig. 4).
TABLE 2.
Identity of the most abundant bacterial sequences and the proportional abundances of the phylogenetic groups in each librarya
| Sampling date | Putative identity | Taxonomic group | % of bacterial clones in the sample |
|---|---|---|---|
| May 17 | Flavobacterium/Chryseobacterium | CFB group | 40 |
| Flexibacter | CFB group | 24 | |
| Acidovorax | β-Proteobacteria | 5 | |
| Bradyrhizobium | α-Proteobacteria | 4 | |
| Caulobacter | α-Proteobacteria | 2 | |
| May 19 | Enterobacteriaceae | γ-Proteobacteria | 97 |
| Acinetobacter | γ-Proteobacteria | 1 | |
| May 22 | Pseudomonas | γ-Proteobacteria | 20 |
| Rhizobium | α-Proteobacteria | 20 | |
| Sphingomonas | α-Proteobacteria | 13 | |
| Flavobacterium/Chryseobacterium | CFB group | 9 | |
| Flexibacter | CFB group | 9 | |
| May 23 | Flavobacterium/Chryseobacterium | CFB group | 13 |
| Methylobacterium | α-Proteobacteria | 10 | |
| Flexibacter | CFB group | 9 | |
| Comamonadaceae | β-Proteobacteria | 9 | |
| Burkholderia | β-Proteobacteria | 8 | |
| Rhodobacter | α-Proteobacteria | 5 | |
| May 25 | Arthrobacter | Actinobacteria | 14 |
| Hymenobacter | CFB group | 13 | |
| Planococcus | Firmicutes | 7 | |
| Pseudomonas | γ-Proteobacteria | 7 | |
| Comamonadaceae | β-Proteobacteria | 6 | |
| Nitrosovibrio | β-Proteobacteria | 5 | |
| Sphingomonas | α-Proteobacteria | 5 |
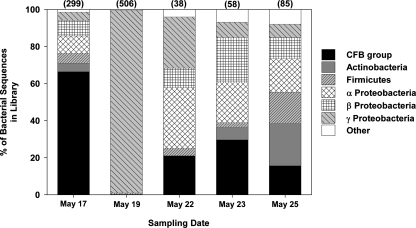
Figure 2 details the total number of bacterial clones in each library. In all cases the sequences were >95% identical to the closest match in the database. In many cases, taxonomic identities could not be determined with a high degree of certainty, so sequences were only classified to the genus or family level. The phylogenetic placements of representative bacterial sequences assigned to the CFB group (Flavobacterium/Chryseobacterium and Flexibacter genera) and the Enterobacteriaceae are further detailed in Fig. 3.
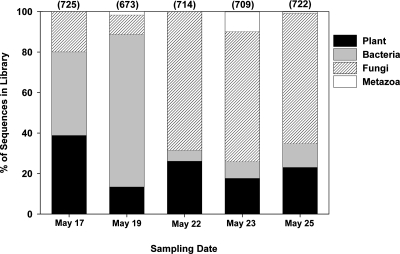
FIG. 1.
Taxonomic identities of the clones sequenced from each library. The numbers in parentheses indicate the total number of sequenced clones (out of 768 per library) that were nonchimeric and could be assigned to one of the four taxonomic groups (E < 1e−100).
TABLE 3.
Correlations between the phylogenetic distance (weighted UniFrac distance) of airborne bacteria and fungi identified from the 5 sampling events and measured meteorological conditions during each 4-h sampling event (Euclidean distances)a
| Meteorological variable | Correlation (Spearman's r value)
|
|
|---|---|---|
| Bacteria | Fungi | |
| Humidity | 0.41* | -0.29 |
| Air temp | 0.29 | -0.37 |
| Solar irradiance | 0.08 | -0.06 |
| Barometric pressure | -0.15 | -0.07 |
| Avg wind speed | -0.30 | -0.47 |
| Peak wind speed | -0.30 | -0.47 |
| Wind direction | -0.66 | -0.59 |
UniFrac distances between samples are identical to those in Fig. 4, except that we have only included samples collected from Boulder, CO, in the analyses. Combining variables did not significantly improve any of the correlations, so we have only shown correlations between individual meteorological variables and the UniFrac distances. A negative correlation indicates that the samples that were more similar with respect to the measured meteorological variable were less similar with respect to their bacterial or fungal assemblages. The asterisk indicates a significant correlation (P = 0.05, corrected for multiple comparisons).
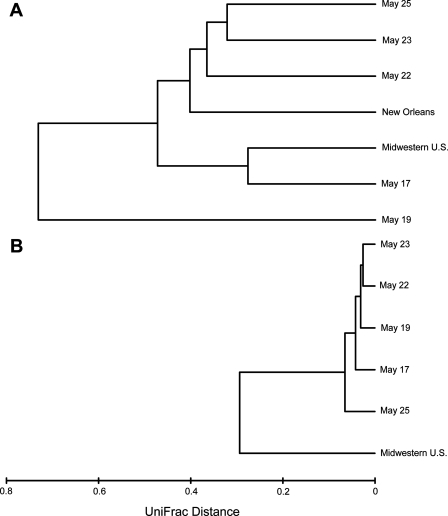
FIG. 4.
Weighted UniFrac distance between the bacterial (A) and fungal (B) sequences identified from outside air samples collected in Boulder, CO (the present study, identified by dates in May); New Orleans, LA (Rodríguez-Hernández, unpublished data); and an undisclosed location in the midwestern United States (3). Since very few fungal sequences were identified in the libraries constructed from the New Orleans air, these samples were not included in the fungal UniFrac analyses. For the Boulder sequences, we averaged the calculated distances from the forward and reverse reads from each sample since the two directions yielded very similar results.
We used the weighted UniFrac algorithm (31, 32) to compare the diversity of bacterial and fungal sequences across the five collected samples. UniFrac provides an overall estimate of the phylogenetic distance between each pair of communities by examining the fraction of the total branch length within a single phylogenetic tree that is unique to either of the two communities (as opposed to being shared by both) (30-32). Prior to conducting the UniFrac analyses, bacterial sequences were aligned by using the Greengenes NAST aligner (minimum length, 400 bp; minimum identity, 75%), and fungal sequences were aligned by using MUSCLE (17) against 814 high-quality aligned fungal sequences. The phylogenetic tree was then built by inserting these alignments into an index tree. We performed Mantel tests (the RELATE routine [14]) using PRIMER version 5 (Primer-E, Ltd., Plymouth, United Kingdom) to examine correlations between the phylogenetic distance (UniFrac distance) of airborne bacteria and fungi identified from the five sampling events and the measured differences in meteorological conditions at the sampling times (Euclidean distances). All Mantel tests were conducted with 1,000 randomized runs to determine significance with a Pearson correction for multiple comparisons. To compare the temporal variability in bacterial and fungal communities at our study site to the spatial variability in airborne microbial communities from geographically distinct sites, we compared the bacterial and fungal sequences from the five libraries constructed for the present study with sequences from clone libraries of outdoor air samples collected by using identical methods in New Orleans, LA, on 25 September 2005 (M. Rodríguez-Hernández, unpublished data) and an undisclosed location in the midwestern United States in August 2001 (3).
Nucleotide sequence accession numbers.
The nonredundant sequences from the present study have been deposited in the GenBank database. The bacterial sequences have accession numbers EU170658 through EU172623, and the fungal sequences have accession numbers EU172624 through EU175853.
RESULTS
Meteorological characteristics.
It rained 4 mm on May 9; otherwise, meteorological conditions were relatively stable prior to the start of the 8-day sampling period (May 17 to 25). The only measurable precipitation during the sampling period itself was a <2-mm rain event on May 22 from h 2200 to 2300. Wind speed and direction were variable during the sampling period, with recorded wind speeds as high as 40 km h−1 (Table 1). During the individual 4-h sampling times air temperatures varied from 24 to 29°C, with relative humidities between 18 and 26%, conditions typical for late spring at this location (Table 1).
TABLE 1.
Sampling dates (in 2006) and meteorological conditions during the 4-h sampling period on each datea
| Parameter | Sampling date
|
||||
|---|---|---|---|---|---|
| May 17 | May 19 | May 22 | May 23 | May 25 | |
| Avg temp (°C) | 25.5 | 29.1 | 26.5 | 23.5 | 26.6 |
| Avg wind speed (km h−1) | 6.1 | 1.8 | 3.1 | 15.3 | 2.6 |
| Peak wind gust (km h−1) | 35.2 | 16.5 | 19.2 | 40.7 | 9.6 |
| Predominant wind direction | NNW | NNE | ESE | WSW | NNE |
| Humidity (%) | 20.2 | 17.6 | 25.5 | 23.3 | 22.2 |
| Barometric pressure (hPa) | 836 | 833 | 829 | 831 | 833 |
| Solar irradiance (W m−2) | 394 | 628 | 588 | 903 | 873 |
Data were collected from a weather station located approximately 250 m from the sampling site (see Materials and Methods).
Phylogenetic characteristics of the airborne microorganisms.
More than 99% of the sequences could be identified as belonging to one of four taxonomic groups (plants, bacteria, fungi, and metazoa) (Fig. 1). No archaeal sequences were identified in any of the samples. Between 13 and 38% of the sequences in each library were from plants (Fig. 1), with 95% of the plant sequences assigned to the genus Pinus and the remaining 5% assigned to the genera Poa, Abies, Alnus, and Equisetum. Three of the five samples contained metazoan sequences (Fig. 1). All of the metazoan sequences from the samples collected on 19 May and 25 May were identified as belonging to dipteran flies (most likely of the genus Ornithoica or Ceratitis), while the metazoan sequences from the sample collected on May 23 were exclusively from a mite (genus Chortoglyphus).
The airborne bacteria collected at the site were phylogenetically diverse. Across all samples (985 valid bacterial sequences), we observed 367 unique operational taxonomic units (defined as sequences with ≥97% sequence similarity). Bacteria assigned to the CFB group (also known as the Cytophaga-Flavobacterium-Bacteroides group or the Bacteroidetes group) (22) and proteobacterial groups were the most abundant across the five sampling dates (Table 2 and Fig. 2). The air sample collected on 19 May was dominated by γ-proteobacteria of the Enterobacteriaceae group (Table 2). These sequences from 19 May were closely related to an aphid endosymbiont (Buchnera aphidicola) and bacteria frequently found on plants (Pectobacterium carotovorum and Hafnia alvei); however, the phylogenetic identity of these γ-proteobacterial sequences could not be determined with a high degree of certainty since even the closest matches had sequence similarities between 92 and 96% (Fig. 3). Bacteria affiliated with the CFB group were common in many of the air samples (Table 2 and Fig. 2), particularly bacteria assigned to the Flavobacteriales, Sphingobacteriales, and Saprospirales groups (Fig. 3). The taxonomy of these CFB groups is not well resolved, but representative sequences were similar to sequences identified from other air samples (Fig. 3), as well as bacterial isolates obtained from air samples (Terrabacter aerolatum and Hymenobacter sp.) (9) and low temperature environments (Chryseobacterium haifaeyi, Sejongia antarctica, and Taxeobacter sp.) (37, 54).
FIG. 2.
Major groups of bacteria identified from each clone library. Firmicutes refers to the Bacillus-Clostridium group and CFB refers to the Cytophaga-Flavobacterium-Bacteroides group. The numbers in parentheses indicate the total number of bacterial clones in each library.
Compared to airborne bacteria, the airborne fungi collected at the site were far less diverse. Approximately 97% of the fungal sequences were classified as Ascomycota, only 3% of the fungal sequences (out of 1,590 in all) were Basidiomycota (mainly Hymenomycetes). In particular, ascomycetes within the Hypocreales order (class Sordariomycetes) were the dominant fungi in all of the air samples, accounting for more than 90% of the fungal sequences in each of the five libraries. The sequences within the Hypocreales order were the closest matches to sequences from the Paecilomyces, Fusarium, Acremonium, Trichoderma, and Cordyceps genera.
Temporal variability in the airborne microorganisms.
There was significant variability in the relative abundances of bacteria and fungi across the five air samples (Fig. 1). In the first two samples collected, between 41 and 75% of the sequences in each library were bacterial (Fig. 1). In contrast, the relative abundance of bacteria was far lower in the latter three air samples collected, where 5 to 12% of the sequences in the each library were bacterial (Fig. 1). As with the airborne bacteria, we observed significant variability in the relative abundance of fungi across the five sampling times. The relative abundance of fungi ranged from 10 to 68% across all air samples collected, and fungi were particularly abundant in the air samples collected on the last three dates (Fig. 1).
We observed significant shifts in the phylogenetic diversity of airborne bacteria across the five sampling dates. The UniFrac results (Fig. 4) summarize shifts in the types of airborne bacteria identified across the sampling dates, shifts that are qualitatively evident in Fig. 2 and Table 2. The bacteria identified from the air sample collected on 19 May were distinct from those found in the other samples (Fig. 4), a result of the dominance of γ-proteobacteria in that sample. Most strikingly, the temporal variability in the types of bacteria identified from one site (Boulder) exceeded the large-scale spatial variability in airborne bacterial assemblages. In other words, the phylogenetic distance between bacterial assemblages identified in air samples collected from geographically distant locations (Boulder, New Orleans, and an undisclosed location in the midwestern United States) was less than the phylogenetic distance between bacteria identified from air samples collected on different dates at the same location (Fig. 4).
Compared to bacteria, airborne fungi exhibited much less temporal variability in their phylogenetic structure (Fig. 4). This result is to be expected considering that the air samples collected on all five sampling dates were dominated by a single taxonomic group of fungi. Relative to the five air samples collected in Boulder, the air sample collected in the midwestern United States harbored a phylogenetically distinct assemblage of fungi (Fig. 4). In particular, more than 65% of the fungal sequences in the outdoor air samples collected by Angenent et al. (3) were identified as Basidiomycota, and all of the identified ascomycetes were Dothideomycetes, a subclass of ascomycetes accounting for <3% of the fungal sequences collected in the present study.
With only five individual samples, we lack the statistical power to determine correlations between meteorological conditions and the observed shifts in bacterial and fungal assemblages. However, neither sampling time nor any of the measured meteorological variables listed in Table 3 appear to be correlated with the bacterial or fungal phylogenetic distance between samples and, in many cases, sampling times with more similar environmental conditions had less similar microbial assemblages (Table 3). Differences in humidity could partially predict phylogenetic distances between the bacterial assemblages, but the correlation was not particularly strong (Table 3). For the fungi, none of the measured meteorological variables were significantly correlated with the phylogenetic distance between samples. There was also no apparent correlation between bacterial and fungal phylogenetic distances across the five sampling points (Spearman's r = 0.1), nor did we find significant correlations between clone library sizes and the phylogenetic distances between the airborne bacterial (Spearman's r = 0.2) or fungal (Spearman's r = −0.3) assemblages.
DISCUSSION
As in many well-studied microbial habitats, there is a disparity between cultivation-dependent and -independent surveys of microbial diversity (39, 42). Although a comparison of cultivation-dependent and -independent surveys was not the objective of the present study, such a disparity is likely to explain the qualitative differences between the airborne bacteria identified from the present study and those identified from other, cultivation-based surveys. For example, gram-positive bacteria, particularly bacteria of the Micrococcus and Bacillus genera, often dominate the culturable fraction of airborne bacteria (34, 46). In contrast, our samples were dominated by gram-negative bacteria (members of the CFB and proteobacterial groups) (Table 2 and Fig. 2), and sequences with close matches to the Bacillus or Micrococcus genera were relatively rare (<2% of the bacterial sequences). The influence of this apparent cultivation bias on surveys of airborne microorganisms is particularly relevant from a public health perspective, considering that many of the bacteria that are under-represented in cultivation-based surveys could be important pathogens or allergens (40).
As evident in Fig. 4, there was a significant amount of overlap in the types of bacteria found in our samples and the samples from two very different locations collected by similar methods. Although differences in methodologies, particularly those related to sample collection, can have a large influence on results (40), other cultivation-independent surveys not included in the UniFrac analyses, including those conducted in Texas (8), Antarctica (19), and France (35), identified groups of bacteria similar to those identified from the present study. For example, many of the bacterial groups abundant in the air samples collected from Texas (8), including those identified as Enterobacteriaceae, Sphingomonas, Comamonadaceae, Pseudomonas, and Sphingobacteriales (see supplemental Table 8 in reference 8), were also abundant in many of the Boulder air samples (Table 2). Together, these results suggest that specific phylogenetic groups of bacteria may be commonly found in the atmosphere, regardless of location. We hypothesize that these groups of bacteria are abundant in the atmosphere because they have characteristics that enhance their ability to become aerosolized and remain intact in the relatively inhospitable environment of the atmosphere. This speculation is anecdotally supported by the observation that many of the specific bacterial groups that are abundant in our air samples are also common in other cold and highly oligotrophic environments such as glacial ice and Antarctic soils (1, 13, 37).
The air samples collected on 19 May contained a unique bacterial assemblage relative to the other sampling dates (Fig. 3 and 4). The air sample was dominated by members of the Enterobacteriaceae frequently found associated with leaf surfaces (Pectobacterium carotovorum and Hafnia alvei) or plant-feeding aphids (Buchnera aphidicola) (Fig. 3). Since only a few metazoan sequences (from a dipteran fly) were found in the 19 May clone library (Fig. 1), it is highly unlikely that the bacteria are aphid endosymbionts. Instead, we speculate that these γ-proteobacteria originate from leaf surfaces and may have become aerosolized from the mowing of the neighboring grass field on that date. Support for this speculation comes from studies showing that leaf surfaces are often an important source of airborne bacteria (26, 29), and crop harvesting can significantly increase the inputs of bacteria and fungi into the atmosphere (25).
The phylogenetic diversity of airborne fungi was not particularly high at our site over this 8-day period since one group of ascomycete fungi (the Hypocreales order) was, by far, the most common type of fungi found in all five air samples. However, there is likely to be a significant amount of seasonal variability in airborne fungi (33), variability that would not have been captured in the present study, and we did find different types of fungi in air samples collected from distinct locations (Fig. 4). Many of the Hypocreales sequences from the Boulder air samples were close matches to genera (e.g., Fusarium and Trichoderma) commonly found in outdoor air samples and potentially allergenic (10, 18, 47). However, fungi that are generally considered to be the most abundant types of fungi in the atmosphere (Alternaria, Aspergillus, Cladosporium, and Penicillium spp.) (12, 47) were not particularly abundant in the Boulder samples. Without a direct comparison of cultivation-dependent and -independent surveys of airborne fungal diversity, we do not know whether these results reflect a cultivation bias in surveys of airborne fungi. However, the present study and others (5, 51) do demonstrate the utility of using molecular techniques to identify airborne fungi and examine spatiotemporal patterns in fungal diversity.
We observed significant temporal variability in the types of microorganisms identified from the air samples at our sampling site. Averaging across the 8-day sampling period, bacteria and fungi were nearly equal in abundance, confirming the results of other studies (3, 52); however bacterial/fungal ratios ranged from 8 to 0.08 across the five sampling days (Fig. 1). These large shifts in bacterial/fungal ratios do not correspond to particular changes in meteorological conditions (Table 1), suggesting that the short-term changes in bacterial/fungal ratios may be driven by sporadic phenomena, such as lawn mowing (see above), soil disturbances, or sporulation events (27, 33). By using the “universal” PCR primers, we were able to capture these major shifts in the airborne microbial communities across the sampling dates, shifts that would not have been detected if we had focused only on one microbial group.
We found a high degree of temporal variability in airborne bacterial diversity (Fig. 4), a pattern observed in a number of other studies (8, 27, 35). At this point, we cannot clearly identify the process(es) responsible for the temporal variability observed at our site since we measured only a few meteorological variables, and none of these variables correlated with shifts in bacterial population structure (Table 3). While this may be a result of our limited sampling campaign (only five sampling times within an 8-day period), many studies find that identifying the specific causes of observed shifts in airborne microbial assemblages is a challenging endeavor since a wide variety of biotic and abiotic processes can interact to move microbes into and out of the atmosphere (21, 27). A significantly more extensive sampling and monitoring effort is required if we want to accurately determine the processes responsible for observed shifts in airborne microbial assemblages.
We have documented that the temporal shifts in airborne bacterial diversity at one site across a relatively short time period can be greater in magnitude than the shifts in bacterial assemblages across geographically distant locations (Fig. 4). These results suggest that studies attempting to compare airborne microbial populations across sites must carefully consider sampling protocols since air samples collected a few days apart can harbor very distinct types of microorganisms. Since the identification of bacteria and fungi from an individual air sample may not provide a representative picture of the microbial populations typically found at that site, the quantification of spatial patterns in airborne bacterial diversity requires estimating the magnitude of temporal variability in airborne bacterial populations at individual sites.
Acknowledgments
We thank Alex Krahn and Heather Hamilton for help with sample collection and clone library construction. We also thank the Broad Institute for assistance with the sequencing and sequence analyses.
This study was supported in part by grants from the Andrew W. Mellon Foundation and the National Science Foundation to N.F. and a grant from the National Academies Keck Future Initiative to R.K. Some analyses were performed using the Keck RNA Bioinformatics Facility at Boulder.
Footnotes
Published ahead of print on 2 November 2007.
REFERENCES
- 1.Aislabie, J. M., K.-L. Chhour, D. J. Saul, S. Miyauchi, J. Ayton, R. F. Paetzold, and M. R. Balks. 2006. Dominant bacteria in soils of Marble Point and Wright Valley, Victoria Land, Antarctica. Soil Biol. Biochem. 38:3041-3056. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angenent, L. T., S. T. Kelley, A. St. Amand, N. R. Pace, and M. T. Hernandez. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. USA 102:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beasley, R. 2002. The burden of asthma with specific reference to the United States. J. Allergy Clin. Immunol. 109:S482-S489. [DOI] [PubMed] [Google Scholar]
- 5.Boreson, J., A. M. Dillner, and J. Peccia. 2004. Correlating bioaerosol load with PM2.5 and PM10cf concentrations: a comparison between natural desert and urban-fringe aerosols. Atmos. Environ. 38:6029-6041. [Google Scholar]
- 6.Bovallius, A., B. Bucht, R. Roffey, and P. Anas. 1978. Long-range air transmission of bacteria. Appl. Environ. Microbiol. 35:1231-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge, P., and B. Spooner. 2001. Soil fungi: diversity and detection. Plant Soil 232:147-154. [Google Scholar]
- 8.Brodie, E., T. Z. DeSantis, J. Parker, I. Zubietta, Y. Piceno, and G. Andersen. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. USA 104:299-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buczolits, S., E. B. M. Denner, D. Vybiral, M. Wieser, P. Kampfer, and H. J. Busse. 2002. Classification of three airborne bacteria and proposal of Hymenobacter aerophilus sp. nov. Int. J. Syst. Evol. Microbiol. 52:445-456. [DOI] [PubMed] [Google Scholar]
- 10.Burge, H. 1985. Fungus allergens. Clin. Rev. Allergy 3:319-329. [DOI] [PubMed] [Google Scholar]
- 11.Burge, H., and C. Rogers. 2000. Outdoor allergens. Environ. Health Perspect. 108:653-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burge, H. A. 2002. An update on pollen and fungal spore aerobiology. J. Allergy Clin. Immunol. 110:544-552. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, S. M., and J. M. Foght. 2007. Cultivation-independent and -dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol. Ecol. 59:318-330. [DOI] [PubMed] [Google Scholar]
- 14.Clarke, K., and R. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd ed. PRIMER-E, Ltd., Plymouth, United Kingdom.
- 15.DeSantis, T., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horner, W. E., A. Helbling, J. E. Salvaggio, and S. B. Lehrer. 1995. Fungal allergens. Clin. Microbiol. Rev. 8:161-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes, K., H. McCartney, T. Lachlan-Cope, and D. Pearce. 2004. A preliminary study of airborne microbial biodiversity over peninsular Antarctica. Cell. Mol. Biol. 50:537-542. [PubMed] [Google Scholar]
- 20.Isolauri, E., A. Huurre, S. Salminen, and O. Impivaara. 2004. The allergy epidemic extends beyond the past few decades. Clin. Exp. Allergy 34:1007-1010. [DOI] [PubMed] [Google Scholar]
- 21.Jones, A., and R. Harrison. 2004. The effects of meteorological factors on atmospheric bioaerosol concentrations: a review. Sci. Total Environ. 326:151-180. [DOI] [PubMed] [Google Scholar]
- 22.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 23.Lane, D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, West Sussex, United Kingdom.
- 24.Lighthart, B. 1997. The ecology of bacteria in the alfresco atmosphere. FEMS Microbiol. Ecol. 23:263-274. [Google Scholar]
- 25.Lighthart, B. 1984. Microbial aerosols: estimated contribution of combine harvesting to an airshed. Appl. Environ. Microbiol. 47:430-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lighthart, B., and B. Shaffer. 1995. Airborne bacteria in the atmospheric surface layer: temporal distribution above a grass seed field. Appl. Environ. Microbiol. 61:1492-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lighthart, B., and L. Stetzenbach. 1994. Distribution of microbial bioaerosol, p. 68-98. In B. Lighthart and A. Mohr (ed.), Atmospheric microbial aerosols: theory and applications. Chapman & Hall, New York, NY.
- 28.Lindemann, J., H. Constantinidou, W. Barchet, and C. Upper. 1982. Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Appl. Environ. Microbiol. 44:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindemann, J., and C. Upper. 1985. Aerial dispersal of epiphytic bacteria over bean plants. Appl. Environ. Microbiol. 50:1229-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozupone, C., M. Hamady, S. Kelley, and R. Knight. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madelin, T. 1994. Fungal aerosols: a review. J. Aerosol Sci. 25:1405-1412. [Google Scholar]
- 34.Mancinelli, R. L., and W. A. Shulls. 1978. Airborne bacteria in an urban environment. Appl. Environ. Microbiol. 35:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maron, P. A., D. P. H. Lejon, E. Carvalho, K. Bizet, P. Lemanceau, L. Ranjard, and C. Mougel. 2005. Assessing genetic structure and diversity of airborne bacterial communities by DNA fingerprinting and 16S rDNA clone library. Atmos. Environ. 39:3687-3695. [Google Scholar]
- 36.Matthias-Maser, S., V. Obolkin, T. Khodzer, and R. Jaenicke. 2000. Seasonal variation of primary biological aerosol particles in the remote continental region of Lake Baikal/Siberia. Atmos. Environ. 34:3805-3811. [Google Scholar]
- 37.Miteva, V. I., P. P. Sheridan, and J. E. Brenchley. 2004. Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl. Environ. Microbiol. 70:202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Brien, H., J. Parrent, J. Jackson, J. Moncalvo, and R. Vilgalys. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71:5544-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-739. [DOI] [PubMed] [Google Scholar]
- 40.Peccia, J., and M. Hernandez. 2006. Incorporating polymerase chain reaction-based identification, population characterization, and quantification of microorganisms into aerosol science: a review. Atmos. Environ. 40:3941-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radosevich, J. L., W. J. Wilson, J. H. Shinn, T. Z. DeSantis, and G. L. Andersen. 2002. Development of a high-volume aerosol collection system for the identification of airborne micro-organisms. Lett. Appl. Microbiol. 34:162-167. [DOI] [PubMed] [Google Scholar]
- 42.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 43.Reysenbach, A., and N. Pace. 1995. Reliable amplification of hyperthermophilic archaeal 16s rRNA genes by the polymerase chain reaction, p. 101-107. In F. Robb (ed.), Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 44.Ross, M. A., L. Curtis, P. A. Scheff, D. O. Hryhorczuk, V. Ramakrishnan, R. A. Wadden, and V. W. Persky. 2000. Association of asthma symptoms and severity with indoor bioaerosols. Allergy 55:705-711. [DOI] [PubMed] [Google Scholar]
- 45.Salem, H., and D. Gardner. 1994. Health aspects of bioaerosols, p. 304-330. In B. Lighthart and A. Mohr (ed.), Atmospheric microbial aerosols: theory and applications. Chapman & Hall, New York, NY.
- 46.Shaffer, B. T., and B. Lighthart. 1997. Survey of culturable airborne bacteria at four diverse locations in Oregon: urban, rural, forest, and coastal. Microb. Ecol. 34:167-177. [DOI] [PubMed] [Google Scholar]
- 47.Shelton, B. G., K. H. Kirkland, W. D. Flanders, and G. K. Morris. 2002. Profiles of airborne fungi in buildings and outdoor environments in the United States. Appl. Environ. Microbiol. 68:1743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinn, E. A., G. W. Smith, J. M. Prospero, P. Betzer, M. L. Hayes, V. Garrison, and R. T. Barber. 2000. African dust and the demise of Caribbean coral reefs. Geophys. Res. Lett. 27:3029-3032. [Google Scholar]
- 49.Stetzenbach, L., M. Buttner, and P. Cruz. 2004. Detection and enumeration of airborne biocontaminants. Curr. Opin. Biotechnol. 15:170-174. [DOI] [PubMed] [Google Scholar]
- 50.Swofford, D. 2006. PAUP 4.0: phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, MA.
- 51.Williams, R. H., E. Ward, and H. A. McCartney. 2001. Methods for integrated air sampling and DNA analysis for detection of airborne fungal spores. Appl. Environ. Microbiol. 67:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Womiloju, T. O., J. D. Miller, P. M. Mayer, and J. R. Brook. 2003. Methods to determine the biological composition of particulate matter collected from outdoor air. Atmos. Environ. 37:4335-4344. [Google Scholar]
- 54.Yi, H., H. I. Yoon, and J. Chun. 2005. Sejongia antarctica gen. nov., sp. nov. and Sejongia jeonii sp. nov., isolated from the Antarctic. Int. J. Syst. Evol. Microbiol. 55:409-416. [DOI] [PubMed] [Google Scholar]
- 55.Yu, Y., M. Breitbart, P. McNairnie, and F. Rohwer. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]