Abstract
Association of specific antimicrobial resistance patterns with unrelated selective traits has long been implicated in the maintenance of antimicrobial resistance in a population. Previously we demonstrated that Escherichia coli strains with a specific resistance pattern (resistant to streptomycin, sulfadiazine, and tetracycline [SSuT]) have a selective advantage in dairy calf intestinal environments and in the presence of a milk supplement commonly fed to the calves. In the present study we identified the sequence of the genetic element that confers the SSuT phenotype and show that this element is present in a genetically diverse group of E. coli isolates, as assessed by macrorestriction digestion and pulsed-field gel electrophoresis. This element was also found in E. coli isolates from 18 different cattle farms in Washington State. Using in vitro competition experiments we further demonstrated that SSuT strains from 17 of 18 farms were able to outcompete pansusceptible strains. In a separate set of experiments, we were able to transfer the antimicrobial resistance phenotype by electroporation to a laboratory strain of E. coli (DH10B), making that new strain more competitive during in vitro competition with the parental DH10B strain. These data indicate that a relatively large genetic element conferring the SSuT phenotype is widely distributed in E. coli from cattle in Washington State. Furthermore, our results indicate that this element is responsible for maintenance of these traits owing to linkage to genetic traits that confer a selective advantage in the intestinal lumens of dairy calves.
Antimicrobial resistance is found frequently among enteric flora, and multidrug resistance is of immediate importance to public health. Thus, the mechanisms by which multiple antimicrobial drug resistance genes are maintained in constant constellation is of interest, particularly when these genes are maintained within a population in the absence of specific antimicrobial selection pressure. One mechanism for maintaining these genes requires close genetic linkage of the resistance genes with other selectively advantageous genes, whereby antimicrobial resistance genes “hitchhike” with the closely linked genes (1, 6, 10, 11). Alternatively, if the resistance genes and associated genetic elements do not convey a selective disadvantage to the host (2), then the loss of the antimicrobial resistance genes from a population is a matter of chance rather than selection. In both of these cases we might expect a very long period of time without drug selection pressure before genetic disassociation and drift remove these genes from a population. Gene expression can also be closely regulated according to the presence of the drug of interest (e.g., tetracycline resistance efflux pump genes tetA and tetB and others) (17, 19, 22), and this probably limits the metabolic burden associated with carriage of these genes. In other cases the antimicrobial resistance genes might convey secondary and selectively advantageous functions that would result in their long-term maintenance in a population (9, 15, 18).
We have previously demonstrated that Escherichia coli isolates with the resistance pattern SSuT (resistant to streptomycin, sulfadiazine, tetracycline) and susceptible to ampicillin, chloramphenicol, and nalidixic acid have the most abundant resistance pattern in the intestinal tracts of neonatal calves at a small dairy farm (Pullman, WA) and that the high prevalence of this pattern is closely correlated with the use of milk supplement (regardless of the presence or absence of the antimicrobial drug oxytetracycline in the supplement) (12, 14). In a separate study we have shown that the antimicrobial drug resistance genes themselves do not convey obvious selective advantage in the absence of antimicrobial use (13). The aim of the present study was to identify the nucleotide sequence of the DNA of the SSuT antimicrobial resistance genes and to examine the distribution of this genetic element among strains of E. coli and among cattle farms in Washington State. For the isolates examined we found that the SSuT element (14,581 bp) is never associated with other antimicrobial resistance genes, and that, according to our pulsed-field gel electrophoresis (PFGE) data, this element is distributed among a genetically diverse population of strains. Furthermore, we found that the SSuT strains from different farms are able to outcompete non-SSuT strains in vitro.
MATERIALS AND METHODS
PFGE.
PFGE was used to ascertain the clonality of E. coli strains isolated from calves at the Washington State University (WSU) dairy. A convenience sample of SSuT (n = 113) and non-SSuT (n = 91) isolates of E. coli from calves of different ages were subjected to single-enzyme PFGE (8), where plug slices were digested with an XbaI endonuclease overnight. PFGE was performed on a CHEF-DRII PFGE apparatus (Bio-Rad) using a 1% agarose gel (Seakem Gold-agarose; FMC BioProducts) in 0.5× Tris-borate-EDTA at 14°C and 6 V/cm, with an initial switch time of 2.2 s and a final switch time of 54.2 s. Gels were run for 18 h and were stained with ethidium bromide for UV illumination. Resulting band patterns were analyzed using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) with 1% optimization and 3% tolerance settings. The average similarity within the two groups was calculated by exporting Dice coefficient matrices from Bionumerics into Excel (Microsoft, Redmond, WA) and averaging the values for SSuT and non-SSuT strains.
Sequence of the SSuT fragment.
Three regions between the SSuT antimicrobial drug resistance genes were amplified with High Fidelity (HF) polymerase (Invitrogen) using the sul2_fwd and the PSTetB1 primers, the PSTetB3 and the strA_rvs primers, the strA_fwd and the strB_rvs primers, and the tet(B)_fwd and tet(B)_rvs primers (Table 1). The SSuT-25 strain of E. coli, isolated from a calf at the WSU dairy (Pullman, WA), served as a template for the above amplification. The PCR fragments were cloned into the pCR-XL-TOPO vector (Invitrogen). The sequences of the fragments were determined using a primer walking strategy by Amplicon Express (Pullman, WA), and the sequence data were analyzed using Vector NTI (Invitrogen, Carlsbad, CA).
TABLE 1.
PCR primers used in this study
| Primer | Sequence | Source or reference |
|---|---|---|
| PSTetB1 | GCAACCGGTGTTATTGGCCC | Present study |
| PSTetB3 | GCGATCTTTGTCGAACTATT | Present study |
| tet(B)_fwd | CAGTGCTGTTGTTGTCATTAA | 4 |
| tet(B)_rvs | GCTTGGAATACTGAGTGTAA | 4 |
| tet(A)_fwd | TTGGCATTCTGCATTCACTC | 4 |
| tet(A)_rvs | GTATAGCTTGCCGGAAGTCG | 4 |
| sul2_fwd | TCGTCAACATAACCTCGGACAG | 10 |
| sul2_ rvs | GTTGCGTTTGATACCGGCAC | 10 |
| strA_fwd | CAACTGGCAGGAGGAACA | 10 |
| strA_rvs | CGCAGATAGAAGGCAAGG | 10 |
| strB_fwd | TTCTCATTGCGGACACCT | 10 |
| strB_rvs | GGCATTGCTCATCATTTG | 10 |
Detection of the SSuT element in E. coli from WSU and other farms in Washington State.
The following sets of isolates were analyzed: (i) 30 SSuT and 30 susceptible isolates of E. coli from calves at the WSU dairy collected in a period of 6 months in 2003; (ii) 12 ASSuT (resistant to ampicillin, streptomycin, sulfadiazine, and tetracycline) and 12 ASSuTCh (resistant to ampicillin, streptomycin, sulfadiazine, tetracycline, and chloramphenicol) isolates of E. coli isolated from calves at the WSU dairy collected over period of 6 months in 2003, which were also tested specifically for the tetB and tetA genes; and (iii) SSuT E. coli isolates from 18 farms in eastern Washington (n = 5 isolates per farm) selected from a bank of previously collected isolates (Field Disease Investigation Unit, Pullman, WA). Farms included both feedlots and dairy operations. The presence of the SSuT genetic element was determined by methods described below.
Genomic DNA (DNAeasy kit; QIAGEN) was prepared from all isolates, and fragments between the sul2 and tetB genes (∼7 kbp) and between the tetB and strA genes (∼7 kbp) were amplified with HF polymerase using the corresponding primers (Table 1). PCR mixtures (50 μl) consisted of 38.6 μl of PCR water, 5 μl of 10× buffer, 2 μl of 5 mM MgSO4, 0.5 μl of each 20 μM primer, 0.4 μl of 10-U/ml HF polymerase, and 1 μl of genomic DNA (100 ng/ml). The thermocycler program included 1 cycle at 94°C for 1 min; 30 cycles, each consisting of a 20-s denaturation at 94°C, a 20-s annealing at 58°C, and a 7-min extension at 68°C; and final dwell at a temperature of 4°C. The amplified fragments were then purified by ethanol precipitation (20) and digested with SacI (Promega, Madison, WI) and HindIII (Promega, Madison, WI) restriction enzymes to generate four distinct fragments discriminated by gel electrophoresis (1% agarose, 90 V, 3 h, 0.5× Tris-borate-EDTA), which matched those predicted by the genetic sequence. Positive controls included genomic DNA (isolated with a DNAeasy kit; QIAGEN) of strain SSuT-25.
In vitro competition experiments.
SSuT isolates from 18 different farms were used for in vitro competition experiments with susceptible E. coli (susceptible to streptomycin, sulfadiazine, tetracycline, ampicillin, chloramphenicol, and nalidixic acid). The latter included a mixture of four susceptible strains (Pans 66, Pans 68, Pans 69, and Pans 71) that were coincubated with the resistant isolates to determine competitive ability in vitro over 8 days in Luria-Bertani (LB) broth as previously described (14). E. coli isolates with the resistance patterns ASSuT and ASSuTCh were also used for in vitro competition experiments against the same mixture of susceptible E. coli isolates. These experiments included a mixture of three ASSuT isolates or three ASSuTCh isolates for each experiment. Competition experiments were repeated three times for each comparison. Each in vitro competition experiment included (i) a positive control competition between SSuT (mixture of SSuT-22 and SSuT-25) and susceptible E. coli (mixture of Pans 66, Pans 68, Pans 69, and Pans 71) strains, where SSuT strains dominate the culture (competition index [CI] > 0.5), and (ii) a negative control competition between SSuT (SSuT-23 and SSuT-24) and susceptible E. coli (mixture of Pans 66, Pans 68, Pans 69, Pans 71) strains, where susceptible strains dominate the culture (CI < −0.5) (14).
Laboratory strain DH10B with the SSuT element.
Guided by a preliminary hypothesis that the SSuT element was plasmid borne (14), we isolated plasmid DNA from an SSuT E. coli strain using a Midi plasmid kit (QIAGEN). Plasmid preparation eluate (2 μl at 300 ng/μl) was then electroporated into DH10B electrocompetent cells (50 μl) (Invitrogen) and plated on an LB agar plate containing tetracycline (10 μg/ml).
In vitro growth and competitive ability of the DH10B strain with and without the SSuT genetic element.
Growth curves were generated for DH10B and SA1 (DH10B with the SSuT element) E. coli strains in LB broth and LB broth supplemented with milk supplement as described previously (12). The milk supplement was prepared by mixing 8.3 kg of dried skim milk, 156 g of vitamin D premix, and 241 g of vitamin A30 premix (all components were from Thomas Products Inc., Hubbard, OR). Each well of a 96-well microtiter plate contained 185 μl of appropriate media and was inoculated with 1.2 μl of culture (prepared in LB at 37°C for 24 h; ≈ 109 CFU). Each culture was inoculated into 6 or 12 replicate wells to calculate an average value for each time point, and each experiment was replicated three times. Some wells (between 16 and 24) were left uninoculated as controls for contamination during the experiments (no contamination was observed). The plate was incubated (37°C) as a stationary culture in a SPECTRAmax 384 Plus (Molecular Devices) plate reader. The culture was agitated before collecting absorbance values (A600), and optical densities (OD) were compared at 24 h using Student's t test.
In vitro competition experiments were accomplished by inoculating LB broth with 10 μl of overnight culture of each DH10B and SA1 isolate and incubating this mixture overnight (37.0°C) on a shaker platform (200 rpm). We repeated the experiments involving competition between an isolate of DH10B that was selected to be nalidixic acid resistant (16) and an SA1 E. coli isolate susceptible to nalidixic acid and conversely repeated the experiments involving competition between an isolate of SA1 that was selected to be nalidixic acid resistant and a DH10B E. coli isolate susceptible to nalidixic acid. The latter experiments were completed to control for potential horizontal transmission of the SSuT element during the in vitro competition experiments. Overnight culture (10 μl) was transferred into fresh LB broth (3 ml) for eight consecutive days. On day 8, a competition index was calculated by estimating the CFU/ml for the SA1 (resistant) and DH10B (susceptible) strains. The SSuT colonies were counted on LB agar supplemented with streptomycin (12 μg/ml), whereas the number of susceptible colonies was determined by subtracting the number of resistant colonies from the same dilution grown on LB agar media without antimicrobial drugs. Three replicate counts were made at each time point and averaged. A CI was calculated as (X − Y)/(X + Y), where X was the number of SSuT colonies and Y was the number of susceptible colonies. CI values near +1 indicated dominance by resistant strains, whereas CI values near −1 indicated dominance by susceptible strains.
RESULTS
Relative diversity of SSuT strains in calves.
A convenience sample of SSuT (n = 113) and non-SSuT (n = 91) E. coli isolates were analyzed by PFGE with XbaI endonuclease. Genetic similarity was assessed for each strain-by-strain comparison using a Dice coefficient where a value of 1 means the isolates share all bands whereas a value of 0 means the isolates share no bands. The average Dice coefficient for all non-SSuT E. coli isolates was 78.5%, and the average Dice coefficient for all SSuT E. coli isolates was 86.8%. Between the two groups there were no identical fingerprints. Because the PFGE data indicated that SSuT strains have multiple, distinct genotypes, we surmised that the genes conferring the SSuT phenotype are harbored on a horizontally transmissible element.
Sequence of the SSuT element.
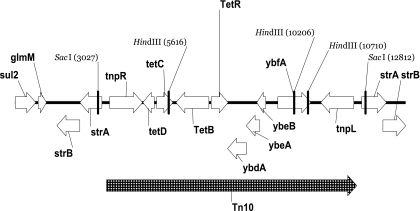
Using PCR we generated four DNA fragments that spanned the sequence between resistance genes. These included sul2 to tetB (∼7 kbp), tetB to strA (∼7 kbp), strA to strB (∼1 kbp), and the tetB gene itself. Fragment sequences formed a 14,581-bp contig that harbors the SSuT resistance genes (GenBank accession no. EF646764) (Fig. 1). The tetracycline resistance gene (tetB) is contained within a classic Tn10 transposon (7). The streptomycin resistance gene strA is followed by the streptomycin resistance gene strB as commonly described (21), and both genes are found flanking the Tn10 transposon. A sequence similar to that of the phosphoglucosamine mutase gene (glmM) (GenBank accession no. VSA289135) is present downstream from the sulfadiazine resistance gene (sul2).
FIG. 1.
The assembled sequence of the 14,284-bp SSuT element from E. coli strain SSuT-25. Arrows show predicted coding regions and direction of transcription. Genetic identity was assigned using BLASTx (e score < 10−30). The hatched arrow shows the portion of the sequence that comprises a Tn10 transposon. SacI and HindIII restriction sites are indicated.
Presence of the sequenced SSuT genetic element in isolates from 18 different farms in Washington State.
Initially we tested 30 SSuT isolates from the WSU dairy for the presence of the SSuT element and found restriction digestion profiles consistent with the sequenced portion of the SSuT genetic element. To determine the distribution of the sequenced SSuT element, 90 SSuT isolates from 18 farms (5 per farm) in the eastern part of Washington State were tested for the presence of the SSuT element by PCR and restriction digestion; 90% of the tested SSuT isolates had a restriction profile consistent with the sequenced segment.
The two other most common resistance patterns at the WSU dairy were ASSuT and ASSuTCh (13). None of the tested isolates with these two phenotypes harbored the SSuT element described herein. ASSuT isolates (n = 12) consistently harbored a tetB gene, while ASSuTCh isolates (n = 12) harbored a tetA gene.
In vitro competition studies.
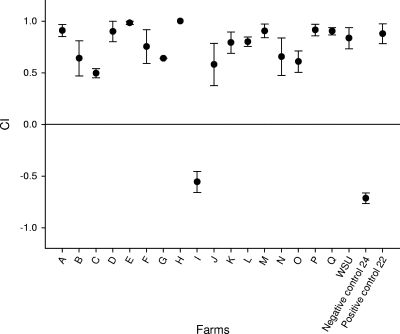
SSuT isolates from 18 farms were tested for their ability to compete with susceptible strains in vitro (14), and, with the exception of those from one farm, the SSuT strains were more competitive than the susceptible strains (Fig. 2). ASSuT and ASSuTCh E. coli strains from the WSU dairy were separately competed against mixture of susceptible strains. Both ASSuT (CI = −0.9) and ASSuTCh (CI = −0.95) E. coli strains were less competitive than the susceptible strains after 8-day passage in LB broth.
FIG. 2.
In vitro competition experiments between SSuT E. coli isolates from 18 farms (indicated by letters A to Q) and susceptible E. coli isolates. Each circle represents average of three replicate experiments. Error bars, standard errors. CI is given by (X − Y)/(X + Y), where X is the number of SSuT colonies and Y is the number of susceptible colonies. When CI is 1, only SSuT strains remain in the broth culture. WSU, Washington State University dairy SSuT isolates.
Laboratory strain DH10B with the SSuT element.
We have shown previously that the majority of SSuT strains harbor a large plasmid (14). To determine whether the plasmid is responsible for the carriage of the resistance genes, we attempted to transfer the plasmid to E. coli laboratory strain DH10B by broth and filter mating, but we were unable demonstrate transfer of the plasmid (data not shown). We then electroporated the plasmid into DH10B, and a single colony grew on an LB agar plate supplemented with 10 mg/liter of tetracycline after 48 h at 37°C. Further testing with HF PCR and restriction digestion as described above showed that DH10B acquired the SSuT resistance gene element. We confirmed the identity of the DH10B strain containing the SSuT sequence, which we designated strain SA1, with PFGE using the XbaI enzyme. DH10B and SA1 were identical. Numerous attempts to identify an electroporated plasmid in SA1 were unsuccessful. The SSuT element appears to have integrated into the chromosome of SA1, but our PFGE assay lacked the sensitivity to detect the integration.
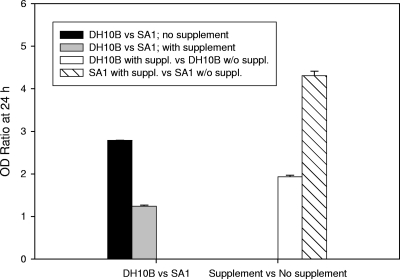
The growth curve of the SA1 strain compared to the DH10B strain showed a significant reduction in OD at 24 h; the final OD for the DH10B strain was 2.8-fold greater than that for the SA1 strain (Fig. 3). When these strains were grown in LB with 1% milk supplement, however, the growth of SA1 improved significantly relative to DH10B, with the final OD ratio at 24 h being 1.2, significantly lower than that determined in the absence of milk supplement (Student's t test; P < 0.001) (Fig. 3). Furthermore, when the SA1 strain was grown in milk-supplemented media, it was much more responsive, with a 24-h OD ratio of 4.3 compared to growth in nonsupplemented media (Student's t test; P < 0.001). This level of responsiveness was significantly greater than the response seen for the DH10B strain (OD ratio = 1.9, Student's t test; P < 0.001; Fig. 3). In vitro competition experiments between the DH10B and SA1 strains showed that the SA1 strain consistently outcompetes the DH10B strain (CI = 1). Using nalidixic acid resistance markers, we confirmed that the competition results were due to a competitive advantage for the SA1 strain rather than horizontal transfer of the SSuT element to DH10B (data not shown).
FIG. 3.
OD (at 600 nm) ratios at 24 h growth for DH10B and SA1 strains grown in LB broth with and without milk supplement. Error bars, standard errors.
DISCUSSION
In this paper we determined the sequence and organization of the SSuT resistance genes and intervening sequence by amplifying and sequencing the regions lying between the known antimicrobial drug resistance genes tetB, sul2, strA, and strB. We determined that this organization of the SSuT genes is uniformly present in all of the 30 SSuT E. coli isolates from the WSU dairy. Despite the fact that the SSuT sequence is conserved, a PFGE analysis of SSuT and non-SSuT strains revealed that the SSuT strains are not clonal but that, compared to the non-SSuT E. coli isolates, they are less genetically diverse. If the SSuT genetic element (and associated selectively advantageous genes) is horizontally transmissible, such transmission may be rare or restricted to a smaller pool of receptive strains in the population, or, if horizontal transmission does not occur, then the observed genetic variation represents expansion and subsequent diversification from a single ancestral clone some time in the past. Poor transmissibility is suggested by our inability to conjugate the SSuT element to a wide variety of hosts in vitro and a lack of identical SSuT and non-SSuT E. coli isolates according to PFGE fingerprinting analysis of field isolates.
To determine whether the SSuT element is present in other farms in Washington State, we examined SSuT isolates from 18 cattle operations (five isolates per farm) and found that the majority of the SSuT isolates from different farms had a sequence organization similar to that of the SSuT isolates from the WSU dairy. Such widespread distribution of a single element suggests a significant selective advantage for these strains in a farm environment. One of the ultimate goals of identifying the sequence of an “advantageous” element is to identify the specific gene(s) responsible for the advantageous trait(s). Our previous experiments have alluded to a few factors that can be advantageous to the SSuT strains; these include presence of vitamin D in a milk supplement (12) and the calf intestinal niche itself (14).
In our previous work we showed through in vitro competition experiments that the SSuT E. coli isolates are more competitive than the susceptible E. coli isolates at the WSU dairy (14). In the present study we used similar in vitro experiments and found that the laboratory strain of E. coli (DH10B) that acquired the SSuT resistance element (SA1) became uniformly more competitive that its parent susceptible strain. This occurred despite the fact that, when not in direct competition, in vitro growth of the SA1 strain is severely retarded compared to its parent strain. This result suggests the presence of a bacteriocin or similar colicin, although we have found no phenotypic evidence of such growth inhibitors (our unpublished data). We have preliminary data suggesting that the SSuT strains may grow better in the presence of volatile fatty acids, and that may be an advantage in vitro if cultures become partially or completely anaerobic. Previously, we demonstrated that the SSuT E. coli isolates grow to a significantly higher cell density in the presence of vitamin D (12) and that the SA1 strain also acquired the ability to grow to a higher density after acquiring the SSuT element. This observation further supports our previous evidence of a selectively advantageous linkage between the ability to take direct advantage of a dietary milk supplement and the SSuT antimicrobial resistance genes. We were unable to ascertain if the SSuT element acquired by SA1 was transferred as a large integrating plasmid or transposon or if this was a simple insertion by homologous exchange. Further work is needed to identify regions flanking the SSuT genes. An XbaI macrorestriction digestion of SA1 and DH10B strains showed no obvious band differences consistent with a large insertion. We have since confirmed that the plasmid harbored by SSuT-25 does not harbor the tetB gene based on microarray hybridization experiments (5). We also discovered that SSuT-25 and SA1 both harbor a cdiA contact-dependent growth inhibition gene (3), which might explain differential survival in culture (D. R. Call et al., unpublished data), but the presence of the cdiA gene is not closely correlated with the SSuT phenotype on the basis of tests of many SSuT strains (r = 0.5).
Results from past in vitro experiments have been consistent with results from in vivo competition experiments. For example, two strains of E. coli (SSuT-23 and SSuT-24) that were less competitive than the susceptible E. coli in vitro were also less competitive in vivo (in calves); conversely two strains (SSuT-22 and SSuT-25) that were shown to be more competitive in vitro were more competitive in vivo (14). In the present study we used the in vitro competition experiments to test the competitive ability of SSuT isolates from 18 farms in Washington State. SSuT isolates from most farms (17/18) were more competitive than the mixture of four susceptible strains from the WSU dairy. When we examined other E. coli isolates that have the SSuT resistance phenotype, with additional resistance to ampicillin or chloramphenicol, we found that they do not have a sequence organization similar to that of the sequenced SSuT fragment. Furthermore, these strains were uniformly less competitive in vitro when competed with susceptible strains. Consequently, we have now shown that the genetic factors attributable to the SSuT phenotype are widely distributed, probably owing to the environmental characteristics found in a dairy production environment, but the exact gene(s) conferring a selective advantage and thus responsible for maintaining the SSuT element in these E. coli populations remains to be identified.
Acknowledgments
This project received financial support from the Agricultural Animal Health Program (Animal Health Formula Funds), College of Veterinary Medicine, Washington State University, Pullman, and from USDA NRI 2004-35201-14112.
We specially thank J. Swain, S. LaFrentz, Autumn Ramsrud, Tremon Bell, M. Oatley, R. McClanahan, the WSU Field Disease Investigation Unit, and the WSU Dairy, Pullman, WA.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Aarestrup, F. M. 2000. Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 38:2774-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245-1248. [DOI] [PubMed] [Google Scholar]
- 4.Call, D. R., M. K. Bakko, M. J. Krug, and M. C. Roberts. 2003. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob. Agents Chemother. 47:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Call, D. R., M. S. Kang, J. Daniels, and T. E. Besser. 2006. Assessing genetic diversity in plasmids from Escherichia coli and Salmonella enterica using a mixed-plasmid microarray. J. Appl. Microbiol. 100:15-28. [DOI] [PubMed] [Google Scholar]
- 6.Calomiris, J. J., J. L. Armstrong, and R. J. Seidler. 1984. Association of metal tolerance with multiple antibiotic resistance of bacteria isolated from drinking water. Appl. Environ. Microbiol. 47:1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalmers, R., S. Sewitz, K. Lipkow, and P. Crellin. 2000. Complete nucleotide sequence of Tn10. J. Bacteriol. 182:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, M. A., D. D. Hancock, T. E. Besser, and D. R. Call. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enne, V. I., P. M. Bennett, D. M. Livermore, and L. M. Hall. 2004. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 53:958-963. [DOI] [PubMed] [Google Scholar]
- 10.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. [DOI] [PubMed] [Google Scholar]
- 11.Kehrenberg, C., and S. Schwarz. 2001. Occurrence and linkage of genes coding for resistance to sulfonamides, streptomycin and chloramphenicol in bacteria of the genera Pasteurella and Mannheimia. FEMS Microbiol. Lett. 205:283-290. [DOI] [PubMed] [Google Scholar]
- 12.Khachatryan, A. R., T. E. Besser, D. D. Hancock, and D. R. Call. 2006. Use of a nonmedicated dietary supplement correlates with increased prevalence of streptomycin-sulfa-tetracycline-resistant Escherichia coli on a dairy farm. Appl. Environ. Microbiol. 72:4583-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2006. Antimicrobial drug resistance genes do not convey secondary fitness advantages to calf-adapted Escherichia coli. Appl. Environ. Microbiol. 72:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo, N., S. Pereira, O. Sahin, J. Lin, S. Huang, L. Michel, and Q. Zhang. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 102:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall, B., D. Petrowski, and S. B. Levy. 1990. Inter- and intraspecies spread of Escherichia coli in a farm environment in the absence of antibiotic usage. Proc. Natl. Acad. Sci. USA 87:6609-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen, T. N., Q. G. Phan, L. P. Duong, K. P. Bertrand, and R. E. Lenski. 1989. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 6:213-225. [DOI] [PubMed] [Google Scholar]
- 18.Pruss, G. J., and K. Drlica. 1986. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc. Natl. Acad. Sci. USA 83:8952-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Sundin, G. W. 2000. Examination of base pair variants of the strA-strB streptomycin resistance genes from bacterial pathogens of humans, animals and plants. J. Antimicrob. Chemother. 46:848-849. [DOI] [PubMed] [Google Scholar]
- 22.Van Bambeke, F., Y. Glupczynski, P. Plesiat, J. C. Pechere, and P. M. Tulkens. 2003. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J. Antimicrob. Chemother. 51:1055-1065. [DOI] [PubMed] [Google Scholar]