Abstract
The Baltic Sea is one of the largest brackish environments on Earth. Despite extensive knowledge about food web interactions and pelagic ecosystem functioning, information about the bacterial community composition in the Baltic Sea is scarce. We hypothesized that due to the eutrophic low-salinity environment and the long water residence time (>5 years), the bacterioplankton community from the Baltic proper shows a native “brackish” composition influenced by both freshwater and marine phylotypes. The bacterial community composition in surface water (3-m depth) was examined at a single station throughout a full year. Denaturing gradient gel electrophoresis (DGGE) showed that the community composition changed over the year. Further, it indicated that at the four extensive samplings (16S rRNA gene clone libraries and bacterial isolates from low- and high-nutrient agar plates and seawater cultures), different bacterial assemblages associated with different environmental conditions were present. Overall, the sequencing of 26 DGGE bands, 160 clones, 209 plate isolates, and 9 dilution culture isolates showed that the bacterial assemblage in surface waters of the central Baltic Sea was dominated by Bacteroidetes but exhibited a pronounced influence of typical freshwater phylogenetic groups within Actinobacteria, Verrucomicrobia, and Betaproteobacteria and a lack of typical marine taxa. This first comprehensive analysis of bacterial community composition in the central Baltic Sea points to the existence of an autochthonous estuarine community uniquely adapted to the environmental conditions prevailing in this brackish environment.
The Baltic Sea covers an area of 377,000 km2 and is the second largest brackish sea on Earth, next to the Black Sea. Due to its regional importance for the fisheries industry and as a drainage area for ∼90 million people in 14 different countries (65), the Baltic Sea has been studied extensively. A comprehensive body of literature on bacterioplankton abundance and activity in the Baltic Sea is available (reviewed in reference 30); however, only a few studies of bacterial community composition and dynamics exist.
The surface water of the Baltic Sea is characterized by a salinity gradient from north to south of ∼2 to 8, and the water column in the Baltic proper is stratified due to a strong pycnocline (75). With a salinity too high for freshwater organisms and too low for marine organisms, the species diversity of eukaryotes in the Baltic Sea is low (49). Further, with its short history (<10,000 years), specialized, long-lived brackish-water species have had only a limited time to develop. In contrast, bacteria multiply quickly, making evolutionary adaptations feasible within this time frame (50). Also, with a water residence time of >5 years (81), bacterioplankton species do not risk being flushed from the system. Hence, it is conceivable that the Baltic Sea contains bacteria specifically adapted to the brackish conditions. In addition, selection among freshwater and marine bacterial populations advected to the Baltic Sea via terrestrial runoff and salt water inflows from the North Sea, respectively, may contribute to the formation of a unique brackish community (17).
The few studies available on bacterial community composition and dynamics in the Baltic Sea are mainly on the northern part (Bothnian Bay/Sea). These concern isolation-based analyses of single bacterial phylotypes (31, 40, 52, 55) or bacterial dynamics in experimental manipulations (see, e.g., references 41, 42, and 45), rather than bacterial community composition per se. Hagström et al. (31) noted a conspicuous presence of members of the alphaproteobacterial genus Sphingomonas and the gammaproteobacterial genera Pseudomonas and Shewanella but a lack of typical marine genera of Gammaproteobacteria (Vibrio, Pseudoalteromonas, and Alteromonas) and Alphaproteobacteria (Roseobacter) among 38 plate isolates. This suggests an avid selection against marine bacterial taxa in the northern Baltic Sea (salinity, ∼2 to 4); however, due to the gradient in salinity and productivity from north to south and the presence of multiple subbasins and sills, the Baltic Sea cannot be regarded as a uniform water mass (65). Some bacterial populations are sensitive to or require salt (18, 40, 45), while others respond to the supply of allochthonous and autochthonous material (37, 41, 42, 61). Therefore, pronounced regional differences in bacterial community structure are to be expected, and information about bacterial community composition in the northern part may not be valid for the central Baltic Sea.
In the present study, we examined the bacterial community composition at the surface (3-m depth) of the Baltic proper throughout a 1-year cycle in relation to environmental variables. Previous microbiological studies from this Baltic Sea region have focused mainly on the depth distribution of bacterial taxa (34, 35, 43). In the Baltic Sea, as in strictly marine waters, a large proportion of the bacterioplankton are non-colony forming, growing in seawater dilution cultures, or not yet cultured at all (72). However, it also appears that bacteria forming colonies on agar plates constitute a significant proportion of the bacterial community in the Baltic Sea (41, 52, 55, 72). Therefore, in addition to molecular methods (clone libraries, denaturing gradient gel electrophoresis [DGGE] profiles, and band identification), we applied cultivation (agar plates and seawater dilution cultures) to maximize resolution and detail in the analysis of bacterial community composition. Our study shows that bacterial assemblages in surface waters of the central Baltic Sea harbor members of typical freshwater bacterial groups and lack several typical marine taxa. Conceivably, the atypical estuarine/brackish local conditions have shaped a uniquely adapted bacterioplankton community.
MATERIALS AND METHODS
Sampling.
Samples were collected from 3-m depth at a single station every 1 to 3 weeks from March 2003 to May 2004 using a 5-liter Niskin bottle. The sampled Landsort Deep station (BY31) (58°35.90′N 18°14.21′E; depth, 459 m) is part of the Swedish Marine Monitoring Program funded by the Swedish Environmental Protection Agency. Data on salinity, temperature, nutrients, cyanobacterial abundance, and chlorophyll a (Chl a) were extracted from this program (http://www.smhi.se) (for methods, see references 36 and 46). All samples were analyzed for bacterial and flagellate abundance. The bacterial community compositions in 13 samples covering March to October 2003 and in one sample from May 2004 were examined by DGGE. On four occasions (14 April, 8 October, and 2 July 2003 and 17 May 2004), samples were obtained for bacterial production and cultivation and for the construction of 16S rRNA gene clone libraries. No samples were obtained during winter (November to February).
Bacterial and flagellate abundances.
Fifty-milliliter aliquots were fixed with 0.2-μm-filtered formalin (2% final concentration) and stored at 4°C. For bacterial counts, 0.5- to 1-ml aliquots were stained with 4′,6-diamidino-2-phenylindole (DAPI) (1-μg ml−1 final concentration) (57) and filtered onto black 0.2-μm-pore-size polycarbonate filters (Poretics), and >200 bacteria (or >15 fields filter−1) were counted at a magnification of ×1,250 using epifluorescence microscopy (Zeiss Axioplan). To estimate variability among filters, three sets of duplicate filters were counted. For flagellate abundance, 10- to 40-ml aliquots were stained with DAPI (1 μg ml−1 final) and filtered onto 1.0-μm black polycarbonate filters (Poretics). One filter diameter on each filter was counted at a magnification of ×788, and heterotrophic flagellates were defined as flagellate-sized cells without autofluorescence.
Bacterial production.
Bacterial production was measured by [methyl-3H]thymidine incorporation (23) as modified for microcentrifugation (73). Triplicate 1.7-ml aliquots were incubated with [methyl-3H]-thymidine (10 nM final concentration; Amersham Pharmacia Biotech) in sterile 2.0-ml-capacity polypropylene tubes for ca. 1 h on deck in a flowing surface water bath. Samples with 5% trichloroacetic acid added prior to the addition of [3H]thymidine served as blanks.
Bacterial isolation from agar plates.
Seawater dilutions (1, 10−1, and 10−2) were spread on ZoBell (5 g peptone, 1 g yeast extract, 15 g agar, 800 ml filtered seawater, 200 ml Milli-Q water; autoclaved at 121°C; 20 min) and R2A (Difco) agar plates (five replicates) and incubated in the dark at 20°C. Triplicate colonies from as many different colony morphotypes as possible were isolated throughout 4 weeks, clean streaked three times, and frozen in glycerol at −80°C. Isolates, from each season separately, were dereplicated by means of restriction fragment length polymorphism (RFLP) of the PCR-amplified 16S rRNA gene. A colony was dissolved in 10 μl TE (10 mM Tris, 1 mM EDTA, pH 8.0), heated for 5 min at 95°C, and centrifuged, and 1 μl of the supernatant was used as the PCR template. In a few cases DNA extraction by means of a kit (described below) or a more extensive DNA extraction protocol (described below) was required for successful PCR amplification. Further, for a few isolates the use of a high-yield polymerase (Phusion DNA polymerase; Finnzymes, Espoo, Finland) was necessary. PCR mixtures (50 μl) contained 0.8 mM deoxynucleoside triphosphates, 0.2 μM of primers 27F and 1492R (25), 100 ng bovine serum albumin, 1× PCR buffer with MgCl2, and 1 U Taq DNA polymerase (Roche). The PCR thermal cycling program was as follows: 95°C for 2 min; 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 45 s; and 72°C for 7 min. PCR products were cleaved using HaeIII and NdeII (Roche) and analyzed on 2% agarose gels. Two isolates having identical colony morphology and cleavage patterns were defined as identical; hence, one isolate would be discarded. Despite different RFLP patterns (based on ∼1,400 bp), partial 16S rRNA gene sequences (∼500 bp) were identical for some isolates. Colony morphology and enzyme profiles (ApiZym; BioMérieux) were then compared, and if they were identical, one isolate was discarded. The criterion for distinguishing two ApiZym profiles was set arbitrarily at two activity units for a single enzyme. This was based on our experience from replicate cultures.
Bacterial isolation from seawater dilution cultures.
The dilution-to-extinction method using 96-well culture plates (Falcon) was performed essentially as described by Simu et al. (72). Onboard the ship, medium water was obtained by a concomitant virus concentration procedure. Water was filtered through a 0.2-μm capsule filter (Advantec MFS Inc.) and a tangential flow filtration system (100,000-molecular-weight-cutoff prep/scale, TFF cartridge; Millipore), filtered again through a 0.2-μm filter, and then used as medium. Two dilutions were made of the inoculum (1:100 and 1:500 or 1:5,000), and 200 μl of these was added to the first row of 96-well plates. Twofold dilutions were made in 12 successive rows, and seawater medium was added to a final volume of 200 μl well−1. Ten replicate plates from each inoculum dilution were incubated for >3 weeks at 15°C in the dark. Dilution culture plates were screened for growth as described in detail elsewhere (14). Briefly, all wells were screened for growth and cell morphology uniformity by DAPI microscopy, and growth on ZoBell plates was tested in order to exclude colony-forming bacteria. Potentially pure non-colony-forming cultures were transferred to 5 or 100 ml 0.2-μm-filtered, autoclaved seawater and grown for an additional 3 weeks. In a few cases, mixed cultures were detected when analyzing sequencing electropherograms from these cultures (below). These cultures were then subjected to a further dilution to extinction to ensure purity of the final culture. Dilution culture bacteria forming colonies on ZoBell plates were screened by RFLP (as described above), and unique isolates were sequenced. The dilution cultivation failed for the sampling on 14 April 2003.
Sequencing of unique isolates.
For plate isolates, DNA was extracted from ZoBell liquid cultures using the DNeasy tissue kit (Qiagen) and quantified fluorometrically (PicoGreen; Molecular Probes), and 10 to 50 ng was PCR amplified as described above. For dilution culture isolates, 25 ml culture was filtered on a 25-mm, 0.2-μm-pore-size Supor-200 membrane filter (Pall Corporation). DNA was obtained by enzymatic lysis and phenol-chloroform extraction as described by Boström et al. (8) and PCR amplified using the Phusion DNA polymerase. Bidirectional sequencing was performed with the DYEnamic ET terminator cycle sequencing kit (Amersham Biosciences) using primers 27F and 519R (44) and an ABI PRISM 377 sequencer (Applied Biosystems) as described by the manufacturer.
Extraction, PCR, and DGGE analysis of community DNA.
For DNA extraction, 2 to 3 liters of water was prefiltered through 47-mm, 3.0-μm pore-size Isopore membrane filters (Millipore) (to prevent filter clogging, <1 liter was filtered per filter) and then through a 0.22-μm Sterivex capsule filter (Millipore) via peristaltic pump (∼100 ml min−1). Sterivex filters were frozen at −20°C until extraction. DNA was extracted from the filters using an enzyme/phenol-chloroform protocol as described by Riemann et al. (61) but with a 30-min lysozyme digestion (5-mg ml−1 final concentration) at 37°C and an overnight proteinase K digestion (100-μg ml−1 final concentration) at 55°C (8). DNA was resuspended in TE and quantified using PicoGreen.
Bacterial 16S rRNA genes were PCR amplified using 0.04 g DNA μl−1 and primers GC341F and 907R as described by Riemann et al. (63). Products were purified using the QIAquick nucleotide removal kit (Qiagen) and quantified using PicoGreen. Sixty nanograms of PCR product was analyzed by DGGE using the D Gene system (Bio-Rad) at 60°C for 6 h at 150 V. Gels were stained with SYBR green I nucleic acid stain (Molecular Probes). DGGE bands were excised, eluted, reamplified, cloned, and realigned as described previously (64). Inserts were bidirectionally sequenced using M13 primers. A dendrogram was constructed from DGGE banding patterns by the software Quantity One 4.6.2 (Bio-Rad) using the Dice coefficient and cluster analysis by the unweighted-pair group method using arithmetic average. DGGE bands were visually detected, and the presence and position of bands were employed in the similarity analyses.
16S rRNA gene clone libraries and phylogenetic analyses.
Bacterial 16S rRNA genes were PCR amplified using 0.05 g DNA μl−1 and primers 27f and 1492r (phosphorylated at their 5′ ends), and products were cloned using the CloneSmart vector system (Lucigen) as described by Pommier et al. (56). The 16S rRNA gene inserts were partially sequenced with primer 27f using an ABI 3100 sequencer (Applied Biosystems) at the Swegene Centre of Genomic Ecology in Lund, Sweden. The quality of the sequences was controlled by removing traces of the sequencing primers and vector contamination (for the clones) and by using PHRED (22) with a base-calling score of n ≥ 20. Ambiguous base calls (i.e., “N”s) at the ends of the sequences were also trimmed away. All sequences were analyzed using the programs Mallard (5) and CHECK_CHIMERA from the Ribosomal Database Project (48). Neither program detected any chimeras. Sequences were aligned using the multiple-alignment program ClustalW (13). Phylogenetic relationships inferred by the maximum-parsimony and neighbor-joining analyses using PAUP*4.0b10 (77) gave congruent results. Neighbor-joining analyses and associated bootstrapping results are presented here. Sequence taxonomic identities were assessed by performing local BLAST searches (4) against the nucleotide database available from GenBank on 14 February 2006 (http://www.ncbi.nlm.nih.gov/).
Nucleotide sequence accession numbers.
Sequences have been deposited in GenBank under the following accession numbers: for plate isolates (BAL, Baltic Sea), spring 2003, BAL62 to -100, DQ063002 to DQ063040; summer 2003, BAL101 to -133, DQ063041 to DQ063073; fall 2003, BAL134 to -202, DQ063101 to DQ063141; spring 2004, BAL204 to -207 and BAL210 to -271, DQ063142 to DQ063207, and BAL203 and BAL208 to -209, AY962019 to AY962021; for dilution cultures, BAL271 to -280, DQ063208 to DQ063216; for DGGE bands (BP, Baltic proper), BP1 to BP26, DQ270271 to DQ270296; and for clone libraries, EF627821 to EF627971.
RESULTS
Environmental data.
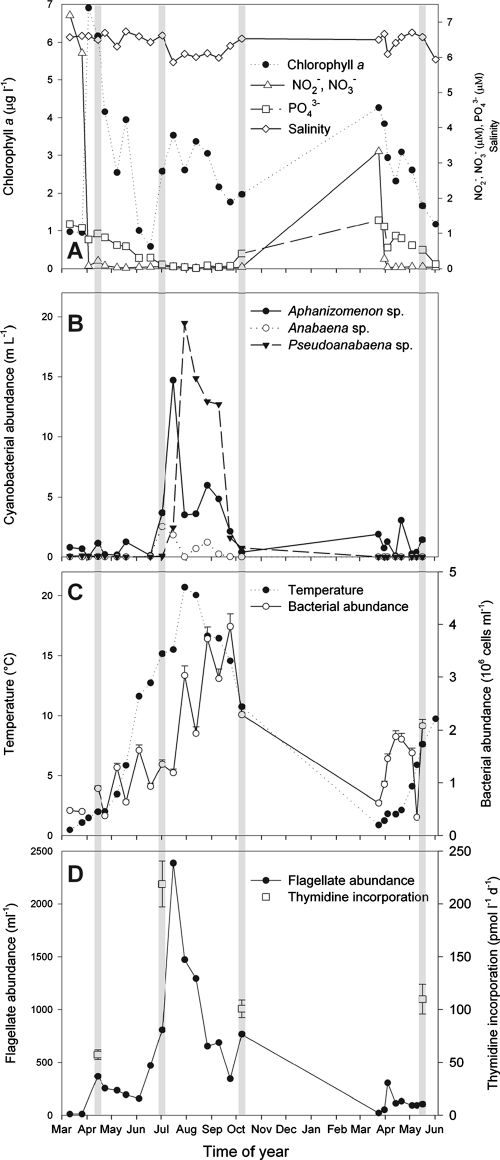
In 2003, the phytoplankton biomass (proxied by Chl a concentration) was low in early spring (1 μg l−1) but increased to 6.8 μg l−1 as a diatom-dominated spring bloom developed (Fig. 1A). This caused a pronounced decrease in nitrite and nitrate, from >7 to ∼0 μM (Fig. 1A). Phosphate concentrations decreased from 1.3 μM in March to <0.1 μM in mid-summer (July to September). In this period the phytoplankton biomass (2 to 3 μg Chl a l−1) consisted predominantly of the cyanobacterium Aphanizomenon sp. (1 to 15 July), followed by Pseudoanabaena sp. (late July to September) (Fig. 1A and B). The temperature increased from ∼1°C in March to 15 to 20°C in June and July, respectively (Fig. 1C), and then decreased again, while the salinity was 5.9 to 6.7 throughout the sampling period. Bacterial abundance fluctuated but was low in early spring (0.5 × 106 ml−1) and increased steadily to a maximal abundance of 3 × 106 to 4 × 106 ml−1 in late July to September (Fig. 1C). Flagellate abundance and bacterial production roughly mirrored the dynamics in bacterial abundance (Fig. 1D). Parameters for spring 2004 were similar to those for spring 2003, except that the spring bloom was less pronounced (up to 4.3 μg Chl a l−1).
FIG. 1.
Dynamics of biotic and abiotic parameters in the Baltic proper in 2003 and 2004. Salinity and concentrations of Chl a and inorganic nutrients (A), cyanobacterial abundance (B), bacterial abundance and temperature (C), and flagellate abundance and bacterial production (D) are shown. Winter time data are not presented. Gray lines indicate dates when isolates and clone libraries were obtained. Values for salinity, Chl a, and nutrients are calculated as averages for samples from 0- and 5-m depths. Cyanobacterial abundance is from integrated samples (0 to 20 m). All other data are from 3-m depth. Error bars indicate standard deviations.
DNA fingerprint of bacterial community dynamics.
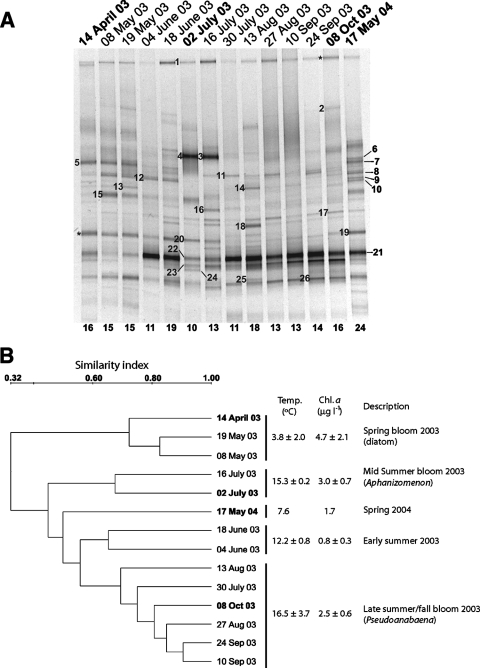
DGGE was used to get an impression of variability in bacterial community composition (Fig. 2). Except for the sample from May 2004 (24 bands), sample profiles consisted of 10 to 19 discernible bands. The 26 sequenced bands (Fig. 2A; Table 1) were related to heterotrophic and photosynthetic groups commonly found in aquatic systems. Except for four bands related to Bacteroidetes (bands 3, 6, 9, and 14; range, 87.6 to 96.5%), all band sequences were closely related to known sequences (range, 96.1 to 100%; average, 97.9%). These sequences were predominantly from uncultivated bacteria (14 of 26) and originated from freshwater (15 bands), estuarine (2 bands), and marine (9 bands) environments. No obvious heteroduplexes or chimeric sequences were detected. For a few vertical positions (e.g., bands 1 and 20), it was confirmed that vertically aligned bands from different samples had identical sequences. However, bands 3 and 4 (97.7% similarity) seemingly had the same vertical position but were identical only on the first 491 bp of the total length (576 bp).
FIG. 2.
(A) Bacterial community composition profiles analyzed by DGGE. A denaturing gradient of 29 to 52% (top to bottom) was applied. Dates when isolates and clone libraries were obtained are indicated in bold. Excised and sequenced bands are numbered to the left of the lanes. Asterisks denote vertical positions for which duplicate bands were sequenced. The number of discernible bands used in the construction of the dendrogram (B) is indicated below each lane. Relationships of excised band sequences to sequences in GenBank are shown in Table 1. (B) Dendrogram obtained by the unweighted-pair group method using arithmetic average, based on presence/absence of DGGE bands. Selected environmental data are shown to the right of the dendrogram.
TABLE 1.
Closest relatives of the sequenced DGGE bands
| Band | % Similarity | Closest relative | Environment | Taxonomic affiliation as reported in GenBank | Accession no.a |
|---|---|---|---|---|---|
| 1 | 99.1 | Clone PI_4z10f | Estuary | Alphaproteobacteria | AY580535 |
| 2 | 98.8 | Clone PLY-P3-42 | Marine | Bacteroidetes | AY354752 |
| 3 | 96.5 | Clone SW146 | Marine | Bacteroidetes | AF493676 |
| 4 | 97.0 | Clone 131677 | Marine | Bacteroidetes | AY922211 |
| 5 | 99.7 | Isolate ARK10048 | Marine (ice) | Bacteroidetes | AF468409 |
| 6 | 98.6 | Clone NorSea47 | Marine | Bacteroidetes | AM279194 |
| 7 | 98.3 | Isolate DG1021 | Marine | Bacteroidetes | AY258128 |
| 8 | 99.1 | DGGE Cytophagales ESR 4 | Lake | Bacteroidetes | AF268288 |
| 9 | 95.6 | Isolate GPl-11, Cytophaga sp. | Marine | Bacteroidetes | AJ456975 |
| 10 | 97.5 | Koliella spiculiformis, chloroplast gene | Lake | Plastid gene | AF278746 |
| 11 | 98.8 | Isolate WL5-12, CFB group | River | Bacteroidetes | AF497890 |
| 12 | 97.5 | DGGE band Cytophagales ESR 4 | Lake | Bacteroidetes | AF268288 |
| 13 | 97.6 | Clone SF54, Sphingobacteriales | Lake | Bacteroidetes | AJ697701 |
| 14 | 87.6 | Isolate CNJ537, Saprospira sp. | Marine | Bacteroidetes | AY527411 |
| 15 | 95.5 | Isolate DG945 | Marine | Bacteroidetes | AY258123 |
| 16 | 99.8 | Koliella spiculiformis, chloroplast gene | Lake | Plastid gene | AF278746 |
| 17 | 99.7 | Clone SF85, Sphingobacteriales | Lake | Bacteroidetes | AJ697706 |
| 18 | 99.5 | Sphingomonas subarctica, strain NKF1 | Freshwater biofilm | Alphaproteobacteria | X94104 |
| 19 | 97.3 | Chlorella mirabilis plastid DNA | Lake | Plastid gene | X65100 |
| 20 | 97.1 | Isolate BAL58 | Estuary | Betaproteobacteria | AY317112 |
| 21 | 99.6 | Synechococcus sp. strain MW73B4 | Lake | Cyanobacteria | AY151250 |
| 22 | 99.8 | Clone S9JA-48 | Lake | Actinobacteria | AB154320 |
| 23 | 99.6 | DGGE band betaproteobacterium zo33 | Lake | Betaproteobacteria | AF531004 |
| 24 | 99.8 | Synechococcus sp. strain MW99B6 | Lake | Cyanobacteria | AY151247 |
| 25 | 99.6 | Clone S9D-06 | Lake | Actinobacteria | AB154303 |
| 26 | 100 | Clone N5 | Lake | Actinobacteria | AJ575532 |
Nucleotide sequences can be accessed via http://www.ncbi.nlm.nih.gov/Entrez/.
Banding patterns varied between samples but with a few bands existing throughout the sampling period (band 1) or for prolonged periods of time (e.g., bands 5, 20, 21, 22, and 26). Several bands were dominant during and after the diatom spring bloom in 2003. Band 20, visible until mid-summer, showed high similarity (97.1%) to a betaproteobacterial dilution culture isolate previously obtained from the Baltic Sea (71). Band 5, which was seen as a bright band until 19 May, was almost identical (99.7%) to a Bacteroidetes isolate from arctic sea ice (12). During late spring and early summer, other Bacteroidetes-related bands appeared (bands 12, 13, 15, and 17), followed by two dense bands on 2 and 16 July (bands 3 and 4). Interestingly, their appearance coincided with an Aphanizomenon sp. bloom. Also, other photosynthesizing organisms appeared and disappeared (e.g., band 16 [Koliella, Chlorophyta] and band 24 [Synechococcus, Cyanobacteria]) along with members of heterotrophic groups (bands 11, 22 and 23). Later, as a Pseudoanabaena sp. bloom developed, another Synechococcus strain appeared (band 21), followed by a Prochlorophyte strain (band 19, Cyanobacteria). At this time, bands related to Bacteroidetes (band 14) and Alphaproteobacteria (band 18) also appeared, along with bands 25 and 26, which were related to clones within the actinobacterial lineages acI and aciV, respectively (80).
It was assumed that DGGE fingerprinting yields a qualitative representation of at least major variations in bacterial community composition (62). A dendrogram based on the presence/absence of bands showed that the four extensive samplings of bacterial community composition (cultivation and clone libraries [see below]) were affiliated with four of the five clusters (Fig. 2B). This suggested that the extensive samplings represent examinations of different bacterial assemblages. Further, branching patterns suggested that the bacterial community composition changed with seasons, possibly affected by the seasonal variation in temperature, nutrient availability, grazing, and phytoplankton biomass and community composition. Since examination of seasonal variability in bacterial community composition was outside the scope of this study, no extensive correlation analysis between community composition and environmental data was attempted. Consequently, the suggested connections are tentative.
The alignment of sequences from DGGE bands, isolates, and clones yielded several 100% matches (see “Analysis of the bacterial components” below). However, it should be noted that the overlapping 16S rRNA gene region is only ∼155 bp.
Bacterial isolates from nutrient agar plates and dilution cultures.
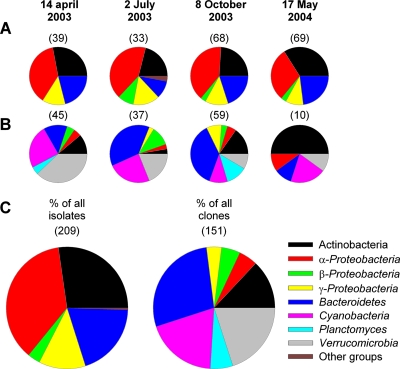
Dereplication of ∼550 colony-forming strains yielded a total of 209 unique isolates, which were sequenced. Isolates were predominantly Alphaproteobacteria (37%), Actinobacteria (27%), and Bacteroidetes (20%) while fewer Gammaproteobacteria (12%), Betaproteobacteria (3%), and Firmicutes (one isolate) were found. The distribution among phylogenetic groups was relatively stable between samplings (Fig. 3A). A number of unique isolates (according to RFLP of ∼1,400 bp) had identical 16S rRNA gene sequences (∼500 bp); however, based on additional pairwise comparisons of colony morphologies and ApiZym enzyme profiles, only two instances of identical isolates were observed between seasons: BAL126 and BAL186 (Afipia, Alphaproteobacteria), isolated in summer and fall 2003, respectively, and BAL110, BAL162, and BAL253 (Achromobacter, Betaproteobacteria), isolated in summer and fall 2003 and in spring 2004.
FIG. 3.
Compositions of plate isolates (A) and clone libraries (B) obtained from the four individual samplings, as well as the composition of all plate isolates/clones (C). The total number of analyzed isolates/clones is given above each pie chart.
The seawater dilution-to-extinction method was used to isolate cultures of possible dominant bacterioplankton species (Table 2). Inherent to this method is that the strains isolated from the wells with the highest dilution represent dominant types in the inoculum, assuming that the medium can support growth of all relevant bacteria (72). Thirty-two pure cultures of non-colony-forming isolates were obtained from a total of 2,688 inoculated wells. These cultures and even cultures with identical isolates originated from a range of inoculum dilutions (rows 2 to 10; see Materials and Methods), preventing a qualified estimate of in situ concentrations. Excluding replicate cultures, four, three, and two isolates were obtained from summer and fall 2003 and spring 2004, respectively. These consisted of three Alpha-, three Beta-, and two Gammaproteobacteria and one member of Bacteroidetes. Interestingly, BAL273 (Polaribacter, Bacteroidetes), obtained on 2 July, was identical to DGGE band 3 (dense band in July sample [Fig. 2A]) and to a cluster of clones (see below). The three betaproteobacterial isolates (BAL274 and BAL275 from fall 2003 and BAL280 from spring 2004) were almost identical (>99.5% similarity) to a dilution culture isolate previously obtained from the Baltic Sea (BAL59) (71). Further, isolate BAL277 (alphaproteobacterium, fall 2003) was similar (98.9%) to a dilution culture isolate obtained from the Skagerrak (71). Another alphaproteobacterial isolate from fall 2003 (BAL278, Sphingomonas) was almost identical (99.7%) to an isolate from the northern Baltic Sea (41). BAL272 was only distantly related (<96% identity) to various uncultured Alphaproteobacteria. Two gammaproteobacterial isolates (BAL276 and BAL279, Oceanobacter) obtained in fall 2003 and spring 2004, respectively, showed a single base pair difference from each other. BAL276 was identical to a plate isolate from this study (BAL235, spring 2004).
TABLE 2.
Non-colony-forming isolates obtained by the seawater dilution-to-extinction technique
| Season | No. of:
|
Isolate names | ||||
|---|---|---|---|---|---|---|
| Total wells inoculated | Wells with growth | Wells showing growth on agar plates | Oligotrophic bacterioplankton culture candidates | Sequenced cultures (different culturesa) | ||
| Summer 2003 | 864 | 294 | 157 | 137 | 22 (4) | BAL272-BAL275 |
| Fall 2003 | 864 | 545 | 391 | 154 | 8 (3) | BAL276-BAL278 |
| Spring 2004 | 960 | 136 | 85 | 51 | 2 (2) | BAL279-BAL280 |
| Total | 2,688 | 975 | 633 | 342 | 32 (9) | |
Based on 100% 16S rRNA gene similarity.
Environmental DNA clone libraries.
Of the 160 16S rRNA gene clones obtained from the four extensive samplings, 9 clones did not show significant similarity to any known sequences. The remaining 151 clones consisted of Bacteroidetes (28%), Verrucomicrobia (20%), Cyanobacteria (19%), Actinobacteria (13%), Planctomyces (6%), Alphaproteobacteria (5%), Gammaproteobacteria (5%), and Betaproteobacteria (4%) (Fig. 3B). Large differences in composition were observed between libraries from different seasons. For instance, the proportion of clones related to Verrucomicrobia varied from 38% (spring 2003) to 9% (fall 2004). Actinobacteria accounted for 11 to 15% of the clones in spring and fall 2003 but for only 3% in summer 2003. In contrast, the proportion of Betaproteobacteria was highest in summer.
Analysis of the bacterial components.
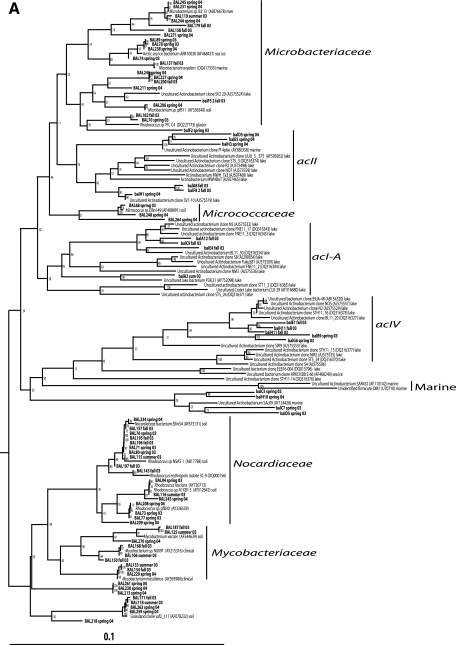
Forty-five of the 57 actinobacterial plate isolates clustered within the Nocardiaceae, Mycobacteriaceae, and Microbacteriaceae families (Fig. 4A), which are mostly known from soil (19), although members of Microbacteriaceae have also been isolated from lakes (33). Most (75%) of the actinobacterial clones and three DGGE bands (bands 22, 25, and 26) were affiliated with the acI, acII, and acIV clusters (33, 80). The majority of sequences within these clusters originate from freshwater habitats (85%, 93%, and 75%, respectively), while a minority originate from estuaries (13%, 7%, and 15%, respectively) (80). None of our sequences were affiliated with the marine Actinobacteria clade (58).
FIG. 4.
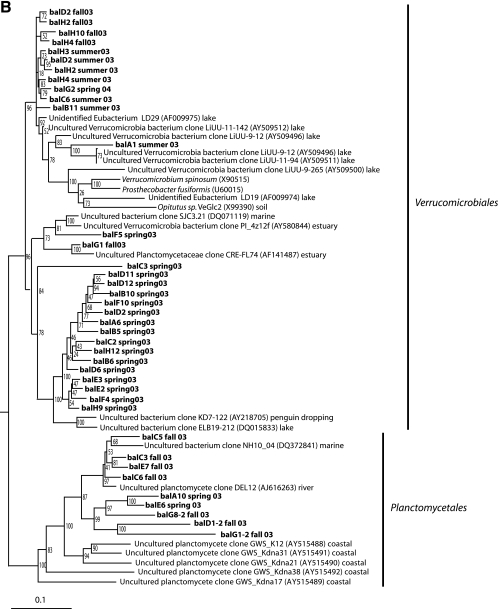
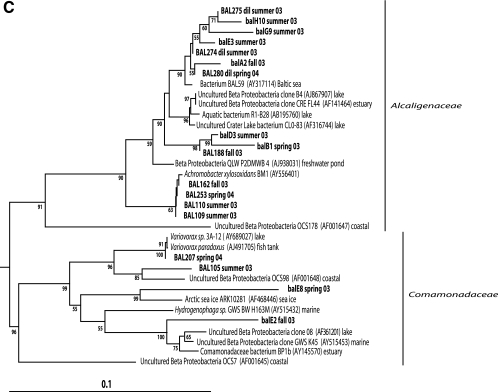
Neighbor-joining phylogenetic trees of 16S rRNA gene sequences (∼460 bp) obtained from clones (lowercase letters) or isolates (uppercase letters) of Actinobacteria (A), Verrucomicrobia and Planctomyces (B), and Betaproteobacteria (C). Sequences with the prefix dil originate from dilution culture isolates. The environment of origin is given for reference sequences if information was available in GenBank.
No cultured representatives were obtained within Verrucomicrobiales and Planctomycetales despite the fact that sequences within these orders accounted for 20% and 6% of the clones obtained, respectively. Nine clones were affiliated with Planctomycetales. Except for three clones, all clones within Verrucomicrobiales branched in two distinct clusters (Fig. 4B). One cluster of 16 clones from spring 2003 was only distantly related (<93% similarity) to known sequences. The other cluster consisted of clones from summer/fall 2003 and spring 2004 and was closely related (∼98% similarity) to a clone from a Dutch lake (84).
The so-called beta III freshwater cluster (27) within the Alcaligenaceae family was represented by one plate isolate (BAL188), three dilution culture isolates, and six of eight betaproteobacterial clones (Fig. 4C). In addition, four plate isolates closely related to Achromobacter xylosoxidans were found within this family. Two plate isolates and a clone belonged to the beta I cluster, which contains numerous cultured bacteria (27). Interestingly, no representatives of the freshwater cosmopolitan Polynucleobacter necessaries cluster were found (32).
Most of the Bacteroidetes isolates and clones obtained were affiliated with the Flavobacteriaceae, Sphingobacteriaceae, and Flexibacteriaceae families (see Fig. S1a in the supplemental material). Two-thirds of the clones were affiliated with the Flavobacteriaceae family. Except for three clones, all isolates were found within the Sphingobacteriaceae and Flexibacteriaceae families. Interestingly, five clones (bal A9, B10, C10, G9, and E12) from fall 2003 formed a distinct cluster with several environmental sequences of uncertain phylogenetic affiliation. These clones showed a 100% match with DGGE band 2, which was observed as a distinct band on this same date (Fig. 2; see Fig. S1a in the supplemental material). Two Polaribacter clones (bal E4 and F11, from spring 2003) were identical to band 5, which was seen as a distinct band also during spring 2003. Further, seven Polaribacter clones obtained in summer 2003 were almost identical (>99% similarity) to the dilution culture isolate BAL273 and to DGGE band 3 obtained from the same water sample. Polaribacter is a prominent genus in polar sea ice and water (6, 12). Overall, Bacteroidetes plate isolates and clones were segregated phylogenetically.
One dilution culture isolate and more than a third of the 77 alphaproteobacterial plate isolates were found within the Sphingomonadales order, predominantly within the Sphingomonas and Sphingopyxis genera (see Fig. S1b in the supplemental material). Bacteria related to Sphingomonas have commonly been isolated from the Baltic Sea (31). Other predominant groups were Brevundimonas/Caulobacter within Caulobacterales and the Rhizobiaceae and Bradyrhizobiaceae families. One plate isolate branched with the SAR11 clade but showed only 85% identity to SAR11. Most of the nine alphaproteobacterial clones were found in a cluster only distantly related to known sequences (<95% similarity). No isolates or clones clustered with the marine Roseobacter clade.
Eleven and four of the 24 gammaproteobacterial plate isolates were found within Pseudomonas and Shewanella, respectively (see Fig. S1c in the supplemental material). These genera have previously been found in the Baltic Sea (31, 35). A number of the clones/isolates obtained were related to sequences from soil or freshwater; however, other sequences were closely related to clones/isolates obtained from marine environments. Interestingly, two clones were affiliated with the SAR86 cluster. Bacteria within this cluster may be able to supplement chemoheterotrophic growth with energy from light (7). No sequences were affiliated with common marine gammaproteobacterial genera such as Alteromonas, Pseudoalteromonas, or Vibrio.
Twenty-eight clones were related to Cyanobacteria. Except for two clones related to sequences from soil and a salt marsh, respectively, all clones were related to Synechococcus clones obtained from lakes (see Fig. S1d in the supplemental material). These were found during all seasons examined. Interestingly, the same nearest relatives were found for the three cyanobacterial DGGE bands (bands 19, 21, and 24) as for a large number of the clones. This is consistent with Synechococcus spp. (cellular size, <1 μm) making up a considerable part of the autotrophic biomass during summer in the Baltic Sea (74).
DISCUSSION
Temporal variations in bacterial community composition.
Bacterioplankton community composition usually displays a pronounced seasonal variability (24, 52, 67), and therefore we sampled during all seasons except winter. Here we used a common DNA fingerprinting method (DGGE) to depict temporal variability in bacterial community composition and to show that our four extensive samplings represent examinations of different bacterial assemblages. Although the purpose was not to examine seasonal dynamics per se, we observed some interesting differences between seasons. For instance, the proportion of Verrucomicrobia decreased from 38% in spring to 19% and 9% in summer and fall 2003, respectively, while Bacteroidetes increased from 13% to 38%. Also, Gammaproteobacteria and Planctomyces increased in fall 2003. While the composition of plate isolates did not differ significantly with seasons, pronounced differences between clone libraries or DGGE profiles were observed. This could represent a difference in the dynamics of various assemblages within the bacterial community (51). However, this comparison may be misleading, since the collection of colony-forming bacteria reflected species richness, due to the nonrandom sampling, rather than the relative abundance resulting from the clone libraries.
The clustering of DGGE banding patterns corresponded roughly with major changes in environmental conditions. For instance, spring, summer, and fall banding patterns formed discrete groupings, presumably driven by changes in, e.g., temperature, biomass, and identity of primary producers (Fig. 3) as well as by other factors related to seasonal chemical and physical forcing. Similarly, the formation of discrete spring and summer communities has been observed in the northern Baltic Sea (52). This indicates that the bacterial community composition in the Baltic proper is driven by temporal variations in environmental conditions, which is consistent with the pronounced seasonal variation and predictable seasonal patterns recently documented in several other aquatic environments (16, 24, 38).
A “native” brackish bacterioplankton community.
The Baltic Sea may be seen as a large estuary with a huge drainage area and a net outflow through the Danish straits, corresponding to the Mississippi River (30). A number of studies have examined bacterial community compositions along estuarine gradients (see, e.g., references 15, 17, 69, 78, and 82). These generally report a limited overlap in composition between freshwater and marine communities (70, 78), where estuaries typically appear as mixing zones. For instance, Crump et al. (15) found that about half of their clones from the Columbia River estuary were similar to clones from the river or the coastal ocean. However, the contemporary finding of specific fast-growing bacterial phylotypes associated with particles suggested that the formation of a uniquely adapted estuarine community was dependent on residence time and bacterial growth rate (15). Similarly, retention time appears as a steering factor for the formation of “native” bacterial communities in lakes (47). Recently, Crump et al. (17) showed that a shift from a mixture of allochthonous communities to a native estuarine community requires bacterial doubling times much shorter than the local water residence time (<10 days). In comparison, the water residence time in the Baltic Sea is orders of magnitudes longer (>5 years) (81), and it is therefore conceivable that niche differentiation and competitive exclusion have shaped a uniquely adapted bacterioplankton community in the Baltic proper.
Indeed, the surface (3-m depth) bacterioplankton community at a single station (BY31) in the northwestern Baltic proper shows estuarine characteristics with a pronounced influence of typical freshwater phylogenetic groups and a lack of typical marine groups. Since this station is situated far from any river mouth (>60 km) and our four extensive sampling campaigns were at times of average salinity (Fig. 1A), we consider a potential direct influence of advected freshwater bacteria on the indigenous bacterioplankton community to be minimal. Hence, we find it conceivable that the “freshwater” phylotypes we found are salt tolerant and have the capacity to proliferate at the salinity of ∼6 found in large parts of the central Baltic proper. In contrast, this salinity may constrain “marine” bacteria. Interestingly, a minimum eukaryote species richness is found at salinities of 5 to 8, which makes this region the most infected by invasive species (49). If this is also valid for prokaryotes, then the Baltic proper may be particularly prone to invasion resulting in a rapid formation of a distinguishable “brackish” bacterioplankton community. However, parameters other than salinity may also add to the proliferation of freshwater phylotypes in the Baltic proper. For instance, blooming of heterocystous potentially toxic cyanobacteria (in our study mainly Aphanizomenon and Pseudoanabaena) is a phenomenon generally associated with lakes rather than marine waters (reviewed in reference 74). Further, the high level of dissolved organic carbon (>300 μM C) (30) of freshwater origin may favor certain properties of the local bacterial assemblage (72).
Bacteroidetes was the overall dominant group in our samples, constituting large fractions of DGGE bands (54%), clones (28%), and dilution (11%) and plate (20%) isolates. Members of this group are abundant in both marine and freshwater environments (26, 39, 83) and tolerate a wide range in salinity (17, 39, 82), and successful immigration from freshwater to the brackish northern Baltic Sea has been observed (40). Hence, it is not surprising that Bacteroidetes dominate bacterioplankton assemblages in the brackish Baltic proper. Interestingly, members of the Flavobacteriaceae accounted for two-thirds of the Bacteriodetes clones. This family is commonly found in coastal seawater and is believed to be particularly important for processing of organic matter during algal blooms (53, 61).
More than a third of our analyzed clones belonged to phylogenetic groups which are usually prominent in freshwater (Verrucomicrobia [20%], Actinobacteria [13%], and Betaproteobacteria [5%]). Also, Actinobacteria and Betaproteobacteria accounted for about a third of the isolates obtained, and the majority of the sequenced DGGE bands showed highest resemblance to sequences from freshwater environments. Members of Verrucomicrobiales are common in freshwater and soil; e.g., the verrucomicrobial cluster CL0-14 was found in 90% of 81 lakes examined (85). However, they are scarce in marine environments. For example, we found only six Verrucomicrobia sequences among 7,195 clones obtained from nine coastal stations distributed worldwide (56). Actinobacteria are mostly known as soil bacteria (19). They constitute only a minor fraction of bacterioplankton communities in coastal and oceanic samples (1, 39, 56) but are of major importance in freshwater environments (2, 27, 80). Betaproteobacteria are predominant among freshwater bacteria (27, 83) but are also found in estuaries (70) and even in marine waters (59). Bouvier and del Giorgio (9) examined bacterial community compositions along salinity gradients in two estuaries and found that Betaproteobacteria were important in the freshwater parts but constituted less than ∼5% of the bacterial community at a salinity of 5 to 10. Interestingly, they found a positive relationship between dissolved organic carbon and the proportion of Betaproteobacteria, which broke down at a salinity of ∼5. This suggests salinity as an additional controlling factor for Betaproteobacteria (9).
Our findings suggest that at least some members of these typical freshwater groups (Verrucomicrobia, Actinobacteria, and Betaproteobacteria) are able to survive and proliferate in the brackish central Baltic Sea. In addition, similar to findings by Hagström et al. (31), we found a conspicuous presence of Sphingomonas (Alphaproteobacteria) and Pseudomonas and Shewanella (Gammaproteobacteria) among our plate isolates. Hence, these genera may consistently be part of the indigenous bacterial assemblage. No representatives of typical marine bacterioplankton taxa such as Vibrio, Alteromonas, or Pseudoalteromonas (Gammaproteobacteria) or SAR11 or Roseobacter (Alphaproteobacteria) were found. Recently, various Vibrio populations were detected in surface waters of the Baltic Sea by a sensitive PCR-based approach (20). To our knowledge, there is only one case where a typical marine bacterium (an Alteromonas species) was isolated from surface waters (10-m depth) of the Baltic proper (11). In contrast, members of the SAR11 clade and the Vibrionaceae, Alteromonadaceae, and Pseudoalteromonadaceae families have been found in and below the halocline (10, 11, 43). We speculate that such marine taxa thrive in the more saline deep water, which originates from the North Sea/Skagerrak where these taxa are present (54), but are constrained to this stratum by the low salinity found above the halocline. Following this reasoning, the brackish Black Sea, which has a higher salinity than the Baltic proper (salinity of 14 to 19 [http://maps.grida.no]), harbors typical marine bacteria in its surface layer (79). Consequently, the limit for their survival is below a salinity of 14 but higher than the salinity of ∼6 found in the Baltic proper.
Methodology and overlap between methods.
Only partial sequences of the 16S rRNA gene were used in this study, presumably limiting the taxonomic resolution relative to full-length sequences. However, since phylogenetic analyses based on partial sequences are largely congruent with those calculated using most of the 16S rRNA gene (31, 60, 68) this limitation should not affect the main conclusions drawn here. Overall, we found pronounced differences between the compositions of clones and isolates (Fig. 3C). Alpha- and Gammaproteobacteria were enriched on plates relative to clone libraries (Fig. 3C). Such enrichment is commonly observed (21, 28) but is not always the case (69). No members of Cyanobacteria, Planctomyces, or Verrucomicrobia were cultivated. To date, only a few members of Verrucomicrobia have been cultivated (66). Surprisingly, no Verrucomicrobia sequences were present among the identified DGGE bands. Consistent with the clone libraries, Bacteroidetes constituted the predominant group among sequenced DGGE bands. Within Actinobacteria and Bacteroidetes, no phylogenetic overlap was seen between plate isolates and clones. Similarly, Bacteroidetes sequences in GenBank show no overlap (3). In contrast, betaproteobacterial clones and plate and dilution culture isolates were phylogenetically mixed. Hence, while many abundant bacterioplankton species are not readily cultivable by standard methods (76), this seems to differ between bacterial taxa as well as between marine environments. For instance, a large overlap between clone libraries and isolates has been found in sea ice (12).
In order to identify sequence matches between methods, the overlapping 16S rRNA gene regions of clones, isolates, and DGGE bands were aligned and a phylogenetic tree generated (data not shown). Similar to the study by Kisand and Wikner (42), we found only a poor match at the species level. Due to the short sequence overlap (∼155 bp), this method was not phylogenetically consistent even when a 100% match was found; however, some interesting identifications were made. For instance, DGGE band 2, seen on 8 October 2003, was identical to five Flavobacteriaceae clones obtained on the same date and was closely related (98.4% similarity) to the nearest relative of these clones. Another example was DGGE band 5 and two Polaribacter clones (see Results). Interestingly, the dilution culture isolate BAL273 (from summer 2003) was almost identical (>99% similarity) to seven Polaribacter clones also obtained in summer 2003. Though limited by the short sequence overlap, the finding of Polaribacter by several independent methods indicates that these bacteria were an important constituent of the bacterial assemblage in summer/fall 2003.
The present study represents the first extensive analysis of bacterioplankton community composition in the central region of the estuarine Baltic Sea. Estuaries are typically nutrient-rich, net-heterotrophic environments (see, e.g., reference 29) where intermediate salinities compromise microbial requirement for or sensitivity to salt. Our molecular and cultivation-based analyses showed that the bacterioplankton assemblage in the Baltic proper was dominated by Bacteroidetes but exhibited a pronounced influence of typical freshwater phylogenetic groups and a lack of typical marine groups. This is indicative of an autochthonous estuarine community uniquely adapted to the environmental conditions prevailing in this brackish environment.
Supplementary Material
Acknowledgments
We thank Leif Lundgren and Berndt Abrahamsson, Department of Systems Ecology, Stockholm University, for help with sampling and P. Griekspoor for technical assistance in PCR-DGGE-16S rRNA gene sequencing analyses. We also thank H.-P. Grossart for thoughtful comments on the manuscript.
This work was supported by the EU through contract EVK3-CT-2002-00078 “BASICS” to Å.H. and by the Swedish Environmental Protection Agency through the National Marine Monitoring Program.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Allgaier, M., and H.-P. Grossart. 2006. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl. Environ. Microbiol. 72:3489-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, C., F. Warnecke, R. Amann, and J. Pernthaler. 2007. High local and global diversity of Flavobacteria in marine plankton. Environ. Microbiol. 9:1253-1266. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Béja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 8.Boström, K. H., K. Simu, Å. Hagström, and L. Riemann. 2004. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol. Oceanogr. Methods 2:365-373. [Google Scholar]
- 9.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 10.Brettar, I., and M. G. Höfle. 1993. Nitrous oxide producing heterotrophic bacteria from the water column of the central Baltic: abundance and molecular identification. Mar. Ecol. Prog. Ser. 94:253-265. [Google Scholar]
- 11.Brettar, I., E. R. B. Moore, and M. G. Höfle. 2001. Phylogeny and abundance of novel denitrifying bacteria isolated from the water column of the central Baltic Sea. Microb. Ecol. 42:295-305. [DOI] [PubMed] [Google Scholar]
- 12.Brinkmeyer, R., K. Knittel, J. Jürgens, H. Weyland, R. Amann, and E. Helmke. 2003. Diversity and structure of bacterial communties in Arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 69:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connon, S. A., and S. J. Giovannoni. 2002. High-thoughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crump, B. C., and J. E. Hobbie. 2005. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol. Oceanogr. 50:1718-1729. [Google Scholar]
- 17.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influence of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.del Giorgio, P. A., and T. C. Bouvier. 2002. Linking the physiologic and phylogenetic successions in free-living bacterial communities along a estuarine salinity gradient. Limnol. Oceanogr. 47:471-486. [Google Scholar]
- 19.Dworkin, M., S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt. 2006. The prokaryotes, vol. 3B. Archaea. Bacteria: Firmicutes, Actinomycetes p. 297-1115. Springer-Verlag, New York, NY. [Google Scholar]
- 20.Eiler, A., M. Johansson, and S. Bertilsson. 2006. Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl. Environ. Microbiol. 72:6004-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eilers, H., J. Pernthaler, and R. Amann. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 23.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 24.Fuhrman, J. A., I. Hewson, M. S. Schwalbach, J. A. Steele, M. V. Brown, and S. Naeem. 2006. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. USA 103:13104-13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Inc., New York, NY.
- 26.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Dennissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goosen, N. K., P. van Rijswijk, J. Kromkamp, and J. Peene. 1997. Regulation of annual variation in heterotrophic bacterial production in the Schelde estuary (SW Netherlands). Aquat. Microb. Ecol. 12:223-232. [Google Scholar]
- 30.Hagström, Å., F. Azam, J. Kuparinen, and U. L. Zweifel. 2001. Pelagic plankton growth and resource limitations in the Baltic Sea, p. 177-210. In F. Wulff, L. Rahm, and P. Larsson (ed.), A systems analysis of the Baltic Sea. Springer, Heidelberg, Germany.
- 31.Hagström, Å., J. Pinhassi, and U. L. Zweifel. 2000. Biogeographical diversity among marine bacterioplankton. Aquat. Microb. Ecol. 21:231-244. [Google Scholar]
- 32.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn, M. W., H. Lünsdorf, Q. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Höfle, M. G., and I. Brettar. 1995. Taxonomic diversity and metabolic activity of microbial communities in the water column of the central Baltic Sea. Limnol. Oceanogr. 40:868-874. [Google Scholar]
- 35.Höfle, M. G., and I. Brettar. 1996. Genotyping of heterotrophic bacteria from the central Baltic Sea by use of low-molecular-weight RNA profiles. Appl. Environ. Microbiol. 62:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Höglander, H., U. Larsson, and S. Hajdu. 2004. Vertical distribution and settling of spring phytoplankton in the offshore NW Baltic Sea proper. Mar. Ecol. Prog. Ser. 283:15-27. [Google Scholar]
- 37.Judd, K. E., B. C. Crump, and G. W. Kling. 2006. Variation in dissolved organic matter controls bacterial production and community composition. Ecology 87:2068-2079. [DOI] [PubMed] [Google Scholar]
- 38.Kan, J., B. C. Crump, K. Wang, and F. Chen. 2006. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol. Oceanogr. 51:2157-2169. [Google Scholar]
- 39.Kirchman, D. L., A. I. Dittel, R. R. Malmstrom, and M. T. Cottrell. 2005. Biogeography of major bacterial groups in the Delaware Estuary. Limnol. Oceanogr. 50:1697-1706. [Google Scholar]
- 40.Kisand, V., N. Andersson, and J. Wikner. 2005. Bacterial freshwater species successfully immigrate to the brackish water environment in the northern Baltic. Limnol. Oceanogr. 50:945-956. [Google Scholar]
- 41.Kisand, V., R. Cuadros, and J. Wikner. 2002. Phylogeny of culturable estuarine bacteria catabolizing riverine organic matter in the northern Baltic Sea. Appl. Environ. Microbiol. 68:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kisand, V., and J. Wikner. 2003. Combining culture-dependent and -independent methodologies for estimation of richness of estuarine bacterioplankton consuming riverine dissolved organic matter. Appl. Environ. Microbiol. 69:3607-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labrenz, M., G. Jost, and K. Jürgens. 2007. Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquat. Microb. Ecol. 46:177-190. [Google Scholar]
- 44.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langenheder, S., V. Kisand, J. Wikner, and L. J. Tranvik. 2003. Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol. Ecol. 45:189-202. [DOI] [PubMed] [Google Scholar]
- 46.Larsson, U., S. Hajdu, J. Walve, and R. Elmgren. 2001. Baltic Sea nitrogen fixation estimated from the summer increase in upper mixed layer total nitrogen. Limnol. Oceanogr. 46:811-820. [Google Scholar]
- 47.Lindström, E. S., M. P. Kamst-van Agterveld, and G. Zwart. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl. Environ. Microbiol. 71:8201-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. J. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paavola, M., S. Olenin, and E. Leppäkoski. 2005. Are invasive species most succesful in habitats of low native species richnes across European brackish water seas? Est. Coast. Shelf Sci. 64:738-750. [Google Scholar]
- 50.Papadopoulos, D., D. Schneider, J. Meier-Eiss, W. Arber, R. E. Lenski, and M. Blot. 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. USA 96:3807-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedrós-Alio, C. 2006. Marine microbial diversity: can it be determined? Trends Microbiol. 14:257-263. [DOI] [PubMed] [Google Scholar]
- 52.Pinhassi, J., and Å. Hagström. 2000. Seasonal succession in marine bacterioplankton. Aquat. Microb. Ecol. 21:245-256. [Google Scholar]
- 53.Pinhassi, J., M. M. Sala, H. Havskum, F. Peters, Ò. Guadayol, A. Malits, and C. Marrasé. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinhassi, J., A. Winding, S. J. Binnerup, U. L. Zweifel, B. Riemann, and Å. Hagström. 2003. Spatial variability in bacterioplankton community composition at the Skagerrak-Kattegat front. Mar. Ecol. Prog. Ser. 255:1-13. [Google Scholar]
- 55.Pinhassi, J., U. L. Zweifel, and Å. Hagström. 1997. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl. Environ. Microbiol. 63:3359-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pommier, T., B. Canbäck, L. Riemann, K. H. Boström, K. Simu, P. Lundberg, A. Tunlid, and Å. Hagström. 2007. Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16:867-880. [DOI] [PubMed] [Google Scholar]
- 57.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 58.Rappé, M. S., D. A. Gordon, K. Vergin, and S. J. Giovannoni. 1999. Phylogeny of Actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst. Appl. Microbiol. 22:106-112. [Google Scholar]
- 59.Rappé, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 60.Rath, J., K. Y. Wu, G. J. Herndl, and E. F. DeLong. 1998. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat. Microb. Ecol. 14:261-269. [Google Scholar]
- 61.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riemann, L., G. F. Steward, L. B. Fandino, L. Campbell, M. R. Landry, and F. Azam. 1999. Bacterial community composition during two consecutive NE monsoon periods in the Arabian Sea studied by denaturing gradient gel electrophoresis (DGGE) of rRNA genes. Deep-Sea Res. II 46:1791-1811. [Google Scholar]
- 63.Riemann, L., J. Titelman, and U. Båmstedt. 2006. Links between jellyfish and microbes in a jellyfish dominated fjord. Mar. Ecol. Prog. Ser. 325:29-42. [Google Scholar]
- 64.Riemann, L., and A. Winding. 2001. Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol. 42:274-285. [DOI] [PubMed] [Google Scholar]
- 65.Rönnberg, C., and E. Bonsdorff. 2004. Baltic Sea eutrophication: area-specific ecological consequences. Hydrobiology 514:227-241. [Google Scholar]
- 66.Sangwan, P., S. Kovac, K. E. R. Davis, M. Sait, and P. H. Janssen. 2005. Detection and cultivation of soil Verrucomicrobia. Appl. Environ. Microbiol. 71:8402-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schauer, M., V. Balagúe, C. Pedrós-Alio, and R. Massana. 2003. Seasonal changes in the taxonomic composition of bacterioplankton in a coastal oligotrophic system. Aquat. Microb. Ecol. 31:163-174. [Google Scholar]
- 68.Schmidt, T. M., E. F. DeLong, and N. R. Pace. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Selje, N., T. Brinkhoff, and M. Simon. 2005. Detection of abundant bacteria in the Weser estuary using culture-dependent and culture-independent approaches. Aquat. Microb. Ecol. 39:17-34. [Google Scholar]
- 70.Selje, N., and M. Simon. 2003. Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat. Microb. Ecol. 30:221-237. [Google Scholar]
- 71.Simu, K., and Å. Hagström. 2004. Oligotrophic bacterioplankton with a novel single-cell life strategy. Appl. Environ. Microbiol. 70:2445-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simu, K., K. Holmfeldt, U. L. Zweifel, and Å. Hagström. 2005. Culturability and coexistence of colony-forming and single-cell marine bacterioplankton. Appl. Environ. Microbiol. 71:4793-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 74.Stal, L. J., P. Albertano, B. Bergman, K. von Bröckel, J. R. Gallon, P. K. Hayes, K. Sivonen, and A. E. Walsby. 2003. BASIC: Baltic Sea cyanobacteria. An investigation of the structure and dynamics of water blooms of cyanobacteria in the Baltic Sea—responses to a changing environment. Cont. Shelf Res. 23:1695-1714. [Google Scholar]
- 75.Stigebrandt, A. 2001. Physical oceanography of the Baltic Sea, p. 19-74. In F. Wulff, L. Rahm, and P. Larsson (ed.), A systems analysis of the Baltic Sea. Springer, Heidelberg, Germany.
- 76.Suzuki, M. T., M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Ströbel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swofford, D. L. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA.
- 78.Troussellier, M., H. Schäfer, N. Batailler, L. Bernard, C. Courties, P. Lebaron, G. Muyzer, P. Servais, and J. Vives-Rego. 2002. Bacterial activity and genetic richness along an estuarine gradient (Rhone River plume, France). Aquat. Microb. Ecol. 28:13-24. [Google Scholar]
- 79.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Warnecke, F., R. Amann, and J. Pernthaler. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 81.Wulff, F., and A. Stigebrandt. 1989. A time-dependent budget model for nutrients in the Baltic Sea. Global Biochem. Cycles 3:63-78. [Google Scholar]
- 82.Zhang, Y., N. Jiao, M. T. Cottrell, and D. L. Kirchman. 2006. Contribution of major bacterial groups to bacterial biomass production along salinity gradient in the South China Sea. Aquat. Microb. Ecol. 43:233-241. [Google Scholar]
- 83.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S.-K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 84.Zwart, G., R. Huismans, M. P. van Agterveld, Y. Van de Peer, P. De Rijk, H. Eenhoorn, G. Muyzer, E. J. van Hannen, H. J. Gons, and H. J. Laanbroek. 1998. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol. Ecol. 25:159-169. [Google Scholar]
- 85.Zwart, G., E. J. van Hannen, M. P. Kamst-van Agterveld, K. Van der Gucht, E. S. Lindström, J. Van Wichelen, T. Lauridsen, B. C. Crump, S.-K. Han, and S. Declerck. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.