Abstract
Research on aging in Drosophila continues to provide new insights into this complex process. Drosophila is highly amenable to study aging because of its short generation time, comprehensive resources for genetic manipulation, and functionally conserved physiology. Importantly, many of these physiological processes such as heart function, sleep, and metabolism functionally senescence in older flies. As the evolutionarily conserved insulin and TOR pathways are critical regulators of aging, the influence of insulin and TOR signaling on these processes is an important area for future research. An important emerging theme is determining the age-dependent alterations that occur at the organ level and how this functional senescence is regulated by different tissues.
Keywords: endocrine systems, senescence, insulin, TOR, KCNQ, dietary restriction, sleep, circadian rhythm, cardiac aging, arrhythmia, heart failure, heart disease
Sleep
The fruit fly, Drosophila melanogaster, can be used as an effective model organism to study sleep (Hendricks et al., 2000; Shaw et al., 2000). Sleep in the fly is identified using behavioral criteria that were established in the days before the electroencephalogram became the standard for identifying sleep in mammals and birds. The criteria include: 1) Prolonged periods of quiescence; 2) Reduced responsiveness to external stimuli; 3) Rapid reversibility, which distinguishes sleep from hibernation or coma; 4) Homeostatic regulation – the increased need for sleep that follows sleep deprivation (Tobler, 1983). These criteria continue to be used to evaluate sleep-like states in organisms in which electrophysiological recordings are not possible (Campbell and Tobler, 1984). They help to avoid interpretive problems resulting from comparisons of brain activity patterns between species having very different neuroanatomical organizations (Campbell and Tobler, 1984; Greenspan et al., 2001; Hartse, 1994).
Careful observations of fly behavior demonstrate that Drosophila exhibit consolidated periods of quiescence (Hendricks et al., 2000; Shaw et al., 2000). During periods of quiescence lasting 5 minutes or longer, flies are unresponsive to mild external stimuli but could be quickly aroused with stronger stimulation. Furthermore, when flies are kept awake they respond by exhibiting large compensatory increases in the amount of quiescence the next day (Shaw et al., 2000). Moreover, adenosine antagonists such as caffeine increase waking while antihistamines increase sleep and reduce its latency (Hendricks et al., 2000; Shaw et al., 2000). More importantly, several molecular markers that are modulated by sleep and waking in mammals are similarly modulated in Drosophila. Altogether, behavioral, ontogenetic, pharmacological, molecular and genetic studies indicate that quiescence in Drosophila shares many of the critical features of mammalian sleep.
Aging is associated with sleep disruptions in humans, monkeys, dogs, cats, rats, mice, and flies (Pandi-Perumal et al., 2002; Shaw et al., 2002; Shiromani et al., 2000). Because sleep deprivation, sleep fragmentation and insomnia activate catabolic pathways and induce hormonal profiles that are commonly found during aging, it has been suggested that decreased sleep quality increases the wear and tear of living and accelerates aging processes (Van Cauter et al., 2000). Several recent epidemiological studies have found that sleep duration and insomnia are associated with an increased risk of all-cause mortality (Kripke et al., 2002; Manabe et al., 2000; Tamakoshi and Ohno, 2004). More importantly, sleep deficits, particularly in older adults, are associated with chronic health problems, impaired quality of life, decreased psychomotor performance, reduced attention, and increased fatigue (Akerstedt and Nilsson, 2003; Buysse and Ganguli, 2002; Pandi-Perumal et al., 2002; Spiegel et al., 1999).
In humans, sleep duration declines with age and is associated with increased nighttime awakenings and reduced slow wave sleep (Cajochen et al., 2006). Aging is also associated with modification in circadian timing as measured by reduced period and amplitude of temperature, sleep and hormonal rhythms (Dijk and Lockley, 2002). Frequently the circadian sleep cycle is phase advanced such that older adults both go to sleep earlier and wake up earlier than younger subjects. Not surprisingly, the prevalence of sleep disorders such as sleep apnea, periodic limb movement disorder, restless legs syndrome, and insomnia increase with age and pose an increased risk for health problems and reduced quality of life (Cooke and Ancoli-Israel, 2006).
Given its short lifespan, ≈ 60 days, Drosophila is an ideal model system to study the relationship between sleep and aging. As with humans, sleep time starts to decline in middle aged Drosophila and continues to fall in older flies (Figure 1) (Shaw et al., 2000). In addition to changes in sleep time, older Drosophila display other alterations in sleep architecture that resemble those found in older human adults. Thus, sleep becomes fragmented, longer sleep episodes become rare and the animals display an increase in nighttime awakenings. Although the field of Drosophila sleep research is only a few years old, several studies have evaluated lifespan in mutant flies that display reduced and/or disrupted sleep (Bushey et al., 2007; Cirelli et al., 2005; Hendricks et al., 2003; Kume et al., 2005; Seugnet et al., 2004; Shaw et al., 2002). The focus of these studies has not been to evaluate the effects of sleep on aging per se. Rather, these studies have used lifespan to determine whether the changes in sleep that are observed in the identified mutant are beneficial or detrimental to the organism. For example, a mutation that slows the cost of waking may reduce the need for sleep. While these flies might sleep substantially less than wild-type animals they should have a normal lifespan. On the other hand, mutations that disrupt sleep regulatory pathways may interfere with the animal's ability to obtain needed sleep thereby resulting in both reduced sleep time and a shorter lifespan. Not surprisingly then, short sleeping flies have been reported to have both a reduced (Cirelli et al., 2005; Hendricks et al., 2003; Seugnet et al., 2004) and a normal lifespan (Bushey et al., 2007; Kume et al., 2005; Pitman et al., 2006). While these studies were not designed to assess the relationship between sleep and aging, they have identified specific genetic tools that may ultimately be useful in elucidating common mechanisms between these two important processes.
Figure 1. Sleep is disrupted in older flies.
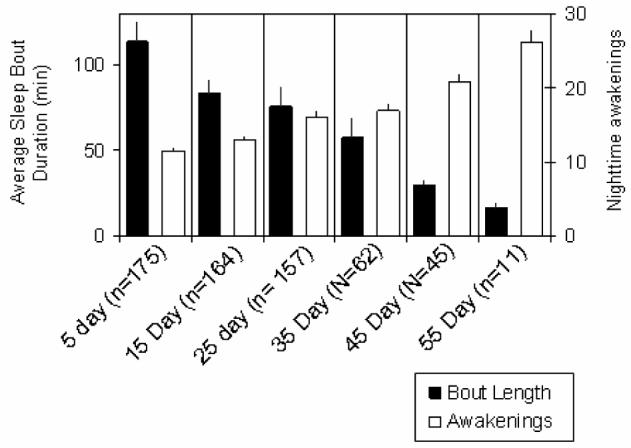
Average sleep bout duration declines with age (black), while the number of nighttime awakenings increases.
In contrast to studies that have focused on lifespan in sleep mutants, Sehgal and colleagues (2006) have evaluated sleep and aging. In particular, they have shown that sleep fragmentation is associated with a reduction in the strength of sleep-wake cycles as measured by fast Fourier transform (Koh et al., 2006). As flies become very old, daytime sleep increases, presumably in an effort to compensate for poor sleep at night such that total sleep duration is not reduced further. Aging in the fly can be easily manipulated by simply rearing the animals at different ambient temperatures. Flies reared at warm temperatures (29°C) have a reduced lifespan compared to flies reared at cooler temperatures (18°C). Interestingly, when flies were housed at cooler temperatures sleep impairments were slowed substantially over time compared with their accelerated-aging siblings. Thus, Sehgal and colleagues (2006) demonstrated that sleep fragmentation is associated with physiological not chronological age. Such a conclusion would not be possible using mammalian protocols and emphasize the utility of the fly to address problems that have been intractable using traditional approaches. It should be noted that intensive research efforts have begun to separately elucidate biological principles of aging and sleep (Cirelli et al., 2005; Hendricks et al., 2001; Hwangbo et al., 2004; Shaw et al., 2002). By combining these tools it should be possible to determine the precise relationship between sleep and aging.
Cardiomyopathies and Arrhythmias
Novel methods for investigating fly heart function have recently become available that promise to dramatically expand the possibilities for screening mutants and for analysis of cardiac contractility and stress response in Drosophila (Wessells et al., 2004; Wolf et al., 2006; Ocorr et al., 2007a,b). One method uses a non-contact echography, optical coherence tomography (OCT), discussed last year in Exp. Gerontol. (Lim et al., 2006), to image the contractions of the heart (Wolf et al., 2006). This non-invasive ultrasound-like method seems to be applicable for high throughput screening to identify mutants with ‘dilated heart’ phenotypes. It was found that cardiac overexpression of a mutant cardiomyopathic form of δ-sarcoglycan, a component of the Dystrophin Glycoprotein Complex (DGC), exhibit systolic dysfunction phenotypes reminiscent of ‘dilated cardiomyopathy’ in humans (Wolf et al., 2006). Recently, loss-of-function mutants of δ-sarcoglycan, which in addition to a dilated heart phenotype, also exhibit reduced motor function and a shortened lifespan (Allikian et al., 2007). A reduced life expectancy was also observed with reduced function of DGC genes, namely dystrophin and dystroglycan (Shcherbata et al., 2007). This suggests that compromised muscle function significantly truncates a fly's life expectancy similar to that of dystrophic mdx mice (Chamberlain et al., 2007).
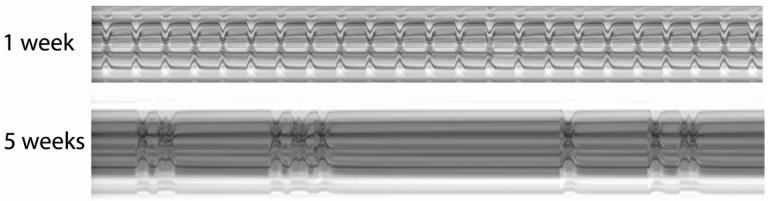
Another method for detailed analysis of heart contractions uses surgically exposed fly's hearts that are accessible for high-speed digital video microscopy (Ocorr et al., 2007a). This semi-intact fly preparation is well-suited for the study of cardiac arrhythmias and other dynamic measures of heart function. To monitor the movement of the edges of the heart, ‘M-modes’ from movies of exposed heart tube preparations display the dynamics of the heart wall contractions over time (Figure 2, top panel). These images are remarkably similar to the M-mode of human cardiac ultrasound measurements. The high speed of the movies (up to 200 frames per second) permits the racking of very rapid changes in and deviations of the regular heart wall movements. Significantly, as the fly ages, Ocorr et al. (2007a) find that the regularity of the heartbeat progressively deteriorates, displaying various forms of arrhythmia (Figure 2, bottom panel). These age-dependent changes in heart rhythm bear a resemblance to the increased incidence of atrial fibrillation in the elderly (see Lakatta and Levy, 2003).These findings suggest that the Drosophila heart may provide a powerful genetic model of cardiac aging.
Figure 2. M-mode traces prepared from high speed movies of dissected flies.
A 1 pixel-wide region with both edges of the heart tube is defined in a single movie frame. Same regions are electronically cut from all of the frames in the movie and aligned horizontally to produce the trace. A regular heart beat is seen in young flies (at 1 week of age; top trace). Arrhythmic heart beats are evident in old flies (at 5 weeks of age; bottom trace). For quantitative analysis, see Ocorr et al 2007a.
Ocorr et al. (2007a) tested this hypothesis by examining mutations in the single fly homolog of the human KCNQ genes, which encode the alpha subunits of the IKs current. KCNQ mutations in humans cause type 1 long QT syndrome (LQT1) and are associated with an increased risk for torsades des pointes (TdP) arrhythmias and sudden death (Jentsch, 2000; Towbin, 2004; Sanguinetti and Tristani-Firouzi, 2006). Deletion mutants of the Drosophila KCNQ are viable but show an elevated risk to pacing-induced heart failure (Ocorr et al., 2007a), a method that assesses age-dependent cardiac performance under stress (Wessells et al., 2004). Further investigation of the heart rhythm of KCNQ mutants with high-speed video recording revealed episodes of prolonged heart contraction and fibrillation in KCNQ mutants, and importantly these phenotypes get progressively more severe with age (Ocorr et al., 2007a). The observed arrhythmias in mutant flies apparently experience a broadening of the cardiac action potential due delayed relaxation of the cardiac myocytes. Interestingly, in contrast to the fly and human heart, these channels seem to be of lesser importance in the much faster beating mouse heart. Therefore, flies develop congenital and age-dependent arrhythmias as do humans, suggesting that at least some of the causes and underlying mechanisms are similar and can be genetically examined in Drosophila to further elucidate the molecular basis for functional aging in an organ system.
Coordination of Tissue Aging
Tissue and functional aging of organs is an important component of the regulation of organismal lifespan, yet the underlying regulation and coordination of these processes remains largely unknown. One mechanism influencing lifespan is genetic variation. For example, there are polygenic influences contributing to lifespan regulation (Wilson et al., 2006). Although many quantitative trait loci (QTLs) have been identified that contribute to the variation in longevity, how these QTLs functionally interact with established pathways that control aging remains to be determined.
A first foray toward an understanding of genetic interactions in longevity has been made in a screen of fifty ‘real’ wild-type lines, generated by inbreeding of individual wild-caught flies, for defects in heart function (Ocorr et al., 2007c). These ‘wild’ lines revealed a continuous spectrum of pacing-induced heart failure, including some extreme cases where young flies exhibit an incidence of failure that is as high as is normally found in old flies (Ocorr et al., 2007c; see also Wessells et al., 2004; Wessells et al., 2007). High-speed video analysis of the inbred lines with high rates of inducible heart failure shows that they also exhibit elevated occurrence of arrhythmias and contractile disorders. This suggests that flies in the wild exhibit naturally occurring ‘cardiomyopathies’. Although the inbred lines examined generally showed an increased frequency of heart failure with age, a few lines displayed an unusually low frequency of failure upon pacing-induced stress (Ocorr et al., 2007c). This finding is reminiscent of hearts with decreased insulin or TOR signaling (Wessells et al., 2004; Luong et al., 2006). Thus, is seems that in the wild there exists a remarkable spectrum of genetic variation in age-dependent cardiac performance. It remains to be seen what genes are affected in these natural variants. Using QTL and other quantitative genetic measures, the traits can now be mapped and nature of the genetic variation can be identified. Elucidating why these variations exist will be important for understanding the epidemiology of heart disease.
Changes in the endocrine system and metabolism are known to affect the aging process. Determining the age-dependent alterations in endocrine function and metabolism will be critical to develop an integrated understanding of the functional changes that occur in senescent organ systems (Tatar, 2004). Recent work has shown that components of the endocrine system, such as TOR, are involved in aging, possibly by influencing insulin signaling (Luong et al., 2006; Kapahi et al., 2004). For example, reduction of TOR function leads to elevated insulin levels in Drosophila. The nutrient-sensitive Tuberous Sclerosis Complex proteins (TSC 1-2)/ Target of Rapamycin (TOR) pathway interacts in many ways with the Insulin Receptor (InR) pathway in controlling metabolism and aging (Oldham and Hafen, 2003; Wullschleger et al., 2006). Akt/PKB activation by PI3K inhibits not only FOXO but also TSC2, thus interfering with the tumor suppressor TSC1-2 complex, which negatively regulates TOR signaling. Interestingly, TOR seems to influence Akt/PKB activity by Ser473 phosphorylation (Sarbassov et al., 2005). The TOR kinase, in turn, stimulates the translation factors S6K and eIF4E, the latter by inhibiting 4E-BP (Fingar and Blenis, 2004). S6K may also need the phosphoinositide-dependent kinase PDK1 and the TSC-dependent Rheb GTPase for full-activation, although the exact relationships are somewhat controversial. InR-TOR signaling may control these processes both autonomously and non-autonomously, as evidence in Drosophila suggests (Tatar, 2004). For example, FOXO signaling in the IPCs changes DILP levels (Luong et al., 2006), and increases stress resistance and longevity when expressed in the head fatbody (Hwangbo et al., 2004). Also, the temporal dynamics of FOXO overexpression during aging were examined to determine when FOXO action acts in the fatbody to extend lifespan (Giannakou et al., 2007). These results showed that loss of insulin signaling can lead to long-range effects on the aging trajectory. Tissue specificity and temporal requirement of aging and metabolism will be particularly important areas of future research.
TOR signaling is an integral sensor of nutritional status of the environment and may also be part of the dietary restriction (DR) response in various species. Interestingly, a recent study by Promislow and colleagues shows evidence that DR has different effects on the functional decline in different tissues and at different times (Burger et al., 2007). For example, starvation resistance is increased in young flies but decreased in old flies by DR. DR also has little effect on oxidative stress response at young ages, while having a negative effect in older flies. Furthermore, DR had differential effects on responses to different pathogens, which suggests that the functional interaction of DR with the innate immune responses is complex. Thus, although DR increases lifespan, it has differential effects on functional changes at the organ level. As an important goal of aging research is to prevent the age-dependent decline in organ function, more work is needed to understand how DR affects these organ-specific traits. Remarkably, the sense of smell may also play into the DR response. Work by Pletcher and colleagues have shown that yeast odorants can partially reverse the effects of DR on lifespan (Libert et al., 2007; see also Pletcher et al., 2002). This result suggests that the perception of nutrient availability leads to changes that can affect aging. Consistent with this hypothesis, mutations in the odorant receptor Orb83b, which is broadly expressed in olfactory neurons, can alter metabolism and extend lifespan (Libert et al., 2007).
It is known that organs age at different rates, yet how these organ aging processes occur and are coordinated is not well established. In keeping with different organs having tissue-specific signatures of aging, a genomic analysis of aging in different tissues has revealed unique as well as common signatures (Zhan et al., 2007; see also Pletcher et al., 2002). For example, Zhou and colleagues recently showed that different tissues have some genes in common that are altered with age (Zhan et al., 2007). Many of the genes include factors involved in the regulation of energy production as well as protein and amino acid biosynthesis. As InR-TOR is a critical pathway for regulating both growth and metabolism, these results are in keeping with the role of InR-TOR signaling as a global coordinator of organ and organismal aging. Furthermore, there were also individual genetic signatures associated with the aging of individual tissues. This result suggests that there are non-autonomous regulators that may coordinate the aging of individual organs with organismal aging. One example of non-autonomous regulation comes from the study of HMGCR, an enzyme involved in the production of juvenile hormone (JH) after stimulation by insulin signaling (Tatar, 2004). Loss of InR affects growth non-autonomously by decreasing HMGCR and JH levels in the organism (Belgacem and Martin, 2007). This loss of HMGCR and InR in the corpus allatum leads to changes in size and sexually dimorphic changes in locomotor activity due to loss of JH. This result is also consistent with the non-autonomous control of aging by InR signaling because the increased lifespan due to loss of InR function requires JH production (Tatar, 2004).
Subcellular organelles are also important in the age-dependent functional decline of organ systems. The nuclear envelope contributes to organismal aging as laminopathies show degenerative and progeric phenotypes. A Drosophila model of laminopathy includes Lamin A mutants, which show accelerated senescent phenotypes such as muscle degeneration and a shortened lifespan (Muñoz-Alarcón et al., 2007). The function of the proteosome also declines with age, which suggests that the ability to degrade proteins is altered with age in both Drosophila and mammals. For example, the 26S proteosome switches to the 20S form with age, but the mechanism by which this impacts organismal and tissue aging is not clear (Vernace et al., 2007). It is possible that this change affects the ability of the proteosome to remove damaged proteins, which can contribute to aging and degenerative diseases. Mitochondria are also subcellular organelles, whichcontribute to aging through the generation of ATP and ROS. Work by Sheldahl and colleagues has shown the mitochondrial control of aging via different haplotypes depends on the nuclear DNA genetic background, suggesting the relationship between the mitochondria and nuclear background is more complex than previously envisioned (Rand et al., 2006).
In sum, we are at the beginning of an understanding of how individual organs age as well as how this process is coordinated by the InR-TOR nutrient endocrine system. Future work using the Drosophila model promises to provide a more comprehensive picture of both organismal and tissue aging at genetic, cellular, and physiological levels.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Paul Shaw, Department of Anatomy and Neurobiology, Washington University, School of Medicine, 660, S. Euclid Ave, Campus Box 8108, St. Louis, MO 63110, ph: (314) 362-2703, fax: (314) 362-3446, shawp@pcg.wustl.edu.
Karen Ocorr, Development and Aging Program, Neuroscience, Aging and Stem Cell Research Center, Burnham Institute for Medical Research, 10901 North Torrey Pines Road, La Jolla, CA 92037, kocorr@burnham.org.
Rolf Bodmer, Development and Aging Program, Neuroscience, Aging and Stem Cell Research Center, Burnham Institute for Medical Research, 10901 North Torrey Pines Road, La Jolla, CA 92037, ph: (858) 795-5295, rolf@burnham.org.
Sean Oldham, Signal Transduction Program, Cancer Center, Burnham Institute for Medical Research, 10901 North Torrey Pines Road, La Jolla, CA 92037 ph: (858) 795-5234, fax: (858) 795-5298, soldham@burnham.org.
References
- Akerstedt T, Nilsson PM. Sleep as restitution: an introduction. J. Intern. Med. 2003;254:6–12. doi: 10.1046/j.1365-2796.2003.01195.x. [DOI] [PubMed] [Google Scholar]
- Allikian MJ, Bhabha G, Dospoy P, Heydemann A, Ryder P, Earley JU, Wolf MJ, Rockman HA, McNally EM. Reduced lifespan with heart and muscle dysfunction in Drosophila sarcoglycan mutants. Hum. Mol. Genet. 2007 Sept 12; doi: 10.1093/hmg/ddm254. ePub ahead of print. [DOI] [PubMed] [Google Scholar]
- Belgacem YH, Martin J-R. Hmgcr in the corpus allatum controls sexual dimorphism of locomotor activity and body size via the insulin pathway in Drosophila. PLoS One. 2007;2(1):e187. doi: 10.1371/journal.pone.0000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JM, Hwangbo DS, Corby-Harris V, Promislow DE. The functional costs and benefits of dietary restriction in Drosophila. Aging Cell. 2007;6:63–71. doi: 10.1111/j.1474-9726.2006.00261.x. [DOI] [PubMed] [Google Scholar]
- Bushey D, Tononi HKG, Cirelli C. Changes in lifespan in hyperkinetic short sleeping mutant flies. Sleep. 2007;30:a370. [Google Scholar]
- Buysse DJ, Ganguli M. Can sleep be bad for you? Can insomnia be good? Arch. Gen. Psychiatry. 2002;59:137–138. doi: 10.1001/archpsyc.59.2.137. [DOI] [PubMed] [Google Scholar]
- Cajochen C, Munch M, Knoblauch V, Blatter K, Wirz-Justice A. Age-related changes in the circadian and homeostatic regulation of human sleep. Chronobiol. Int. 2006;23:461–474. doi: 10.1080/07420520500545813. [DOI] [PubMed] [Google Scholar]
- Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci. Biobehav. Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Cooke JR, Ancoli-Israel S. Sleep and its disorders in older adults. Psychiatr. Clin. North Am. 2006;29:1077–1093. doi: 10.1016/j.psc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Dijk DJ, Lockley SW. Integration of human sleep-wake regulation and circadian rhythmicity. J. Appl. Physiol. 2002;92:852–862. doi: 10.1152/japplphysiol.00924.2001. [DOI] [PubMed] [Google Scholar]
- Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jacobson J, Vinti G, Leevers SJ, Partridge L. Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell. 2007;6:429–438. doi: 10.1111/j.1474-9726.2007.00290.x. [DOI] [PubMed] [Google Scholar]
- Greenspan RJ, Tononi G, Cirelli C, Shaw PJ. Sleep and the fruit fly. Trends Neurosci. 2001;24(3):142–145. doi: 10.1016/s0166-2236(00)01719-7. [DOI] [PubMed] [Google Scholar]
- Hartse KM. Sleep in insects and nonmammalian vertebrates. In: Kryger MH, Roth T, Dement WC, editors. Principals and Practice of Sleep Medicine. 2nd Ed. W.B. Saunders; Philadelphia: 1994. pp. 95–104. [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25(1):129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC-P, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat. Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Lu S, Kume K, Yin JC-P, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J. Biol. Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signaling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Jentsch TJ. Neuronal KCNQ potassium channels: physiology and role in disease. Nat. Rev. Neurosci. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. PNAS. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kripke DF, Garfinkel L, Wingard DL, Klauber MR, Marler MR. Mortality associated with sleep duration and insomnia. Arch. Gen. Psych. 2002;59:131–136. doi: 10.1001/archpsyc.59.2.131. [DOI] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part II: the aging heart in health: links to heart disease. Circulation. 2003;107:346–354. doi: 10.1161/01.cir.0000048893.62841.f7. [DOI] [PubMed] [Google Scholar]
- Libert S, Zwiener J, Chu X, Vanvoorhies W, Roman G, Pletcher SD. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- Lim HY, Bodmer R, Perrin L. Drosophila Aging 2005/2006. Exp. Geront. 2006;41:1213–1216. doi: 10.1016/j.exger.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech RL, Bodmer R, Oldham SM. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Manabe K, Matsui T, Yamaya M, Sato-Nakagawa T, Okamura N, Arai H, Sasaki H. Sleep patterns and mortality among elderly patients in a geriatric hospital. Gerontology. 2000;46:318–322. doi: 10.1159/000022184. [DOI] [PubMed] [Google Scholar]
- Muñoz-Alarcón A, Pavlovic M, Wismar J, Schmitt B, Eriksson M, Kylsten P, Dushay MS. Characterization of lamin mutation phenotypes in Drosophila and comparison to human laminopathies. PLoS One. 2007;2(6):e532. doi: 10.1371/journal.pone.0000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Reeves N, Wessells R, Fink M, Chen H-SV, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony J, Bodmer R. KNCQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. PNAS. 2007a;104:3943–3498. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Perrin L, Lim HY, Qian L, Wu X, Bodmer R. Genetic control of heart function and aging in Drosophila. Trends Cardiovasc. Med. 2007b;17:177–182. doi: 10.1016/j.tcm.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Crawley T, Gibson G, Bodmer R. Genetic variations for cardiac dysfunction in Drosophila. PLoS ONE. 2007c;2:e601. doi: 10.1371/journal.pone.0000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Pandi-Perumal SR, Seils LK, Kayumov L, Ralph MR, Lowe A, Moller H, Swaab DF. Senescence, sleep, and circadian rhythms. Ageing Res. Rev. 2002;1:559–604. doi: 10.1016/s1568-1637(02)00014-4. [DOI] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Rand DM, Fry A, Sheldahl L. Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics. 2006;172:329–341. doi: 10.1534/genetics.105.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Israel S, Toledo R, Shaw P. Evaluating a Drosophila model of insomnia. Sleep. 2004;27:A384. [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, Baker D, Ruohola-Baker H. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. Embo. J. 2007;26:481–93. doi: 10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiromani PJ, Lu J, Wagner D, Thakkar J, Greco MA, Basheer R, Thakkar M. Compensatory sleep response to 12 h wakefulness in young and old rats. Am. J. Physiol. Regulatory Integrative Comp. Physiol. 2000;278:R125–133. doi: 10.1152/ajpregu.2000.278.1.R125. [DOI] [PubMed] [Google Scholar]
- Spiegel K, Leproult R, Van Cauter E. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354:1435–1439. doi: 10.1016/S0140-6736(99)01376-8. [DOI] [PubMed] [Google Scholar]
- Tamakoshi K, Ohno Y. Self-reported sleep duration as a predictor of all-cause mortality: results from JACC study, Japan. Sleep. 2004;27:51–54. [PubMed] [Google Scholar]
- Tatar M. The neuroendocrine regulation of Drosophila aging. Exp. Gerontol. 2004;39:1745–1750. doi: 10.1016/j.exger.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Tobler I. Effect of forced locomotion on the rest-activity cycle of the cockroach. Behav. Brain Res. 1983;8:351–360. doi: 10.1016/0166-4328(83)90180-8. [DOI] [PubMed] [Google Scholar]
- Towbin JA. Molecular genetic basis of sudden cardiac death. Pediatr. Clin. North Am. 2004;51:1229–55. doi: 10.1016/j.pcl.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Van Cauter E, Leproult R, Plat L. Age-related changes in slow wave sleep and REM sleep and relationship with growth hormone and cortisol levels in healthy men. JAMA. 2000;284:861–868. doi: 10.1001/jama.284.7.861. [DOI] [PubMed] [Google Scholar]
- Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 2007;21:2672–2682. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat. Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- Wessells RJ, Bodmer R. Age-related cardiac deterioration: insights from Drosophila front. Biosci. 2007;12:39–48. doi: 10.2741/2047. [DOI] [PubMed] [Google Scholar]
- Wilson RH, Morgan TJ, Mackay TF. High-resolution mapping of quantitative trait loci affecting increased life span in Drosophila melanogaster. Genetics. 2006;173:1455–1463. doi: 10.1534/genetics.105.055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. PNAS. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall M. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zhan M, Yamaza H, Sun Y, Sinclair J, Li H, Zhou S. Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome Res. 2007;17:1236–1243. doi: 10.1101/gr.6216607. [DOI] [PMC free article] [PubMed] [Google Scholar]