Abstract
Eukaryotic genome integrity is safeguarded by two highly conserved protein kinases that are called ATR and ATM for humans and Mec1 and Tel1 for Saccharomyces cerevisiae. Although they share sequence similarities and substrates, these protein kinases perform different specialized functions. In particular, Mec1 plays a key role in the DNA damage checkpoint response, whereas Tel1 primarily is involved in telomere homeostasis, and its checkpoint function is masked by the prevailing activity of Mec1. In order to understand how this specificity is achieved, we searched for TEL1 mutations able to compensate for the lack of Mec1 functions. Here, we describe seven independent dominant TEL1-hy alleles that are able to suppress, to different extents, both the hypersensitivity to genotoxic agents and the checkpoint defects of Mec1-deficient cells. Most of these alleles also cause telomere overelongation. In vitro kinase activity was increased compared to that of wild-type Tel1 in the Tel1-hy385, Tel1-hy394, Tel1-hy680, and Tel1-hy909 variants, but its activity was not affected by the TEL1-hy184 and TEL1-hy628 mutations and was slightly reduced by the TEL1-hy544 mutation. Thus, the phenotypes caused by at least some Tel1-hy variants are not simply the consequence of improved catalytic activity. Further characterization shows that Tel1-hy909 not only can sense and signal a single double-stranded DNA break, unlike wild-type Tel1, but also contributes more efficiently than Tel1 to single-stranded DNA accumulation at double-strand ends, thus enhancing Mec1 signaling activity. Moreover, it causes unscheduled checkpoint activation in unperturbed conditions and upregulates the checkpoint response to small amounts of DNA lesions. Finally, Tel1-hy544 can activate the checkpoint more efficiently than wild-type Tel1, while it causes telomere shortening, indicating that the checkpoint and telomeric functions of Tel1 can be separable.
Eukaryotic cells ensure genetic integrity after DNA damage or the inhibition of DNA replication through a complex network of surveillance mechanisms, known as DNA damage checkpoints (56). These protective mechanisms are signal transduction pathways specialized in detecting abnormal DNA structures and conveying the damage signal to transducers that, in turn, transmit it to numerous downstream effectors. The DNA damage checkpoints serve at least two primary purposes: (i) to arrest the cell cycle in response to DNA damage, thereby coordinating cell cycle progression with DNA repair capacity; and (ii) to regulate both the transcription of DNA damage response genes and the activation of various repair/recombination proteins that help cells to survive genotoxic stress (reviewed in references 22 and 45).
Key players of the DNA damage checkpoint signal transduction pathways belong to a protein kinase family that includes mammalian ATM (ataxia telangiectasia mutated) and ATR (ataxia telangiectasia and Rad3-related) as well as Saccharomyces cerevisiae Tel1 and Mec1 (reviewed in references 22 and 45). Mec1 functions in a complex with Ddc2 (35, 41, 55) that is functionally related to S. pombe Rad26 and human ATRIP, which interact with Rad3 and ATR, respectively (9, 10). A complete Mec1-dependent activation of downstream targets requires Rad24 and the Ddc1-Rad17-Mec3 complex (23, 24, 34, 56), which binds damaged DNA independently of Mec1/ATR in both yeast and human cells (18, 27, 59). Once DNA perturbations are sensed, the checkpoint signals are propagated through evolutionarily conserved protein kinases, which are called Rad53 and Chk1 for S. cerevisiae and Chk2 and Chk1 for humans. Rad53 and Chk1 activation is not governed by their simple interaction with Mec1 or Tel1 but rather requires a stepwise process. In particular, it has been proposed that the Mec1-dependent phospho-Rad9 protein (44) acts first as an adaptor to promote Mec1-Rad53 interaction and Mec1-mediated Rad53 phosphorylation/activation (49). Mec1-dependent phospho-Rad9 then completes the Rad53 activation process by facilitating in trans Rad53 autophosphorylation, perhaps by increasing the local Rad53 concentration on the Rad9 surface (12). The Rad9 counterpart in the response to replication stress is Mrc1, which also actively participates in DNA replication itself (2, 17, 33, 51).
Human ATR and its S. cerevisiae ortholog, Mec1, are thought to recognize single-stranded DNA (ssDNA) regions that arise during the replication of damaged template or after double-stranded DNA break (DSB) processing and mainly are thought to transduce the signals emanating from UV damage and stalled replication forks (16, 58). Conversely, both human ATM and its S. cerevisiae ortholog, Tel1, appear to bind DSBs via a highly conserved trimeric complex, known as MRN (Mre11-Rad50-Nbs1) for humans and MRX (Mre11-Rad50-Xrs2) for S. cerevisiae (11, 32). MRN also is required for full ATM activation and for efficient interaction between ATM and some of its substrates (19, 20). Human ATM has been shown to be activated primarily by DSBs caused by ionizing radiation and to be required for ATR recruitment and subsequent ATR-dependent checkpoint activation (1, 16, 30, 58). On the other hand, Tel1 checkpoint functions are not so obvious, as tel1Δ cells show neither hypersensitivity to DNA-damaging agents nor major defects in genome stability and checkpoint activation (29, 31). However, Tel1, but not Mec1, specifically is required to phosphorylate the histone isoform H2AX after DSB formation during the G1 cell cycle phase, suggesting that Tel1 has Mec1-independent functions in the response to DNA damage (47). Moreover, although the budding yeast checkpoint response to DSBs depends primarily on Mec1 (6, 15), Tel1 contributes to generate 3′-ended ssDNA, leading to Mec1-dependent checkpoint activation (26). Finally, Tel1 can activate the checkpoint response to DSBs independently of Mec1, although its signaling activity becomes apparent only in the presence of multiple DSBs, and it is disrupted by DSB termini resection (26). This suggests that Tel1's contribution to checkpoint activation is masked by the prevailing activity of Mec1. Consistently, TEL1 deletion exacerbates the checkpoint defects and increases the rate of genome rearrangements in mec1 mutants (31, 42). Furthermore, Tel1 can trigger Rad53 phosphorylation in response to DNA damage, although Tel1-dependent Rad53 phosphorylation is minor and can be detected only in the absence of Mec1 at the end of S phase (7).
Indeed, Tel1, together with the MRX complex, primarily is implicated in maintaining the proper length and structure of linear chromosome termini, which are called telomeres (reviewed in reference 54). For instance, Tel1 or ATM inactivation results in shortened telomeres and increased chromosomal end-to-end fusions by nonhomologous end joining (4, 14, 28, 36, 38, 39, 48). Moreover, both Tel1 and ATM exhibit a cell-cycle-dependent association with telomeres (50, 53). Recruitment of the telomerase catalytic subunit Est2 and of its accessory protein Est1 is severely reduced in cells lacking Tel1 (13). This suggests that Tel1 promotes the telomere association of telomerase, although its critical substrates have yet to be identified. On the other hand, yeast cells can rely on Mec1 to maintain short but stable telomeres in the absence of Tel1, indicating that the Mec1 function at telomeres is redundant with or masked by that of Tel1 (39).
Thus, although Mec1 and Tel1 share substrates, they appear to be specialized in responding to DNA lesions and in regulating telomere length, respectively. This specificity might be due to differences in their intrinsic kinase activity, in their ability to recognize DNA lesions, and/or their ability to interact with their final targets. On the other hand, the ability of Tel1 to sense and transduce the DNA damage signal can be enhanced by increasing the amount of Tel1 (8, 42), prompting us to ask whether mutations in the TEL1 gene might compensate for the lack of Mec1 checkpoint functions. We describe here the identification and characterization of TEL1-hy mutant alleles suppressing the hypersensitivity to genotoxic agents and the checkpoint defects of Mec1-deficient cells. Most TEL1-hy alleles also cause telomere overelongation, indicating that the corresponding Tel1-hy variants are more efficient than wild-type Tel1 not only in activating the checkpoint but also in promoting telomere elongation. We also provide evidence that the levels of in vitro kinase activity of some Tel1-hy variants are higher than that of the wild type, and this may account for their enhanced ability to activate the checkpoint and induce telomere elongation.
MATERIALS AND METHODS
Search for tel1 mutants.
Strain YLL1414, carrying the LEU2 gene located 65 bp downstream of the TEL1 stop codon, was generated by transforming W303 cells with a PCR product obtained using primers PRP571 (5′-GAT GCT TTA TAC TTC TTT TTT TTG TTT ATA GAA ATA CGA AAA GTA ATG TAC AAA CCA TTG GCC CTA CCC ACA TAT G-3′) and PRP572 (5′-CAG TAA TGG TAG CTT CAC TAT CAG CGA TTG TAT TTT TGG GCA CAT CAT CAC ATT GAT GCA GGC TAA CCG GAA CCT G-3′) (TEL1 sequences are underlined) and a plasmid carrying the Kluyveromyces lactis LEU2 gene as the template. Genomic DNA of strain YLL1414 then was used as the template, together with primers PRP583 (5′-GCA CGA ATG AAA GAG TAC CGA CAA GCA T-3′) and PRP584 (5′-TGG GTT CTA GAT GCA AGG AAC GAA ACT G-3′), to amplify by PCR (under mutagenic conditions) a TEL1 region spanning position +5140 to position +8560 from the TEL1 translation start codon. Therefore, the resulting PCR amplification products contained the LEU2 gene flanked by TEL1 sequences spanning positions +5140 to +8429 (including 65 bp of 3′ noncoding sequence) on one side and positions +8430 to +8560 (3′ noncoding sequence) on the other side. Thirty independent PCR mixtures (100 μl) were prepared, each containing 5 U of EuroTaq DNA polymerase (Euroclone), 10 ng of template genomic DNA, 500 ng each primer, 0.5 mM each deoxynucleoside triphosphate (dATP, dTTP, and dCTP), 0.1 mM dGTP, 0.5 mM MnCl2, 10 mM β-mercaptoethanol, 10 mM Tris-HCl (pH 9), 50 mM KCl, and 1.5 mM MgCl2. A mec1Δ sml1Δ strain was transformed with the PCR products in order to replace the corresponding TEL1 wild-type sequences with the mutagenized PCR products. One thousand transformants were selected on synthetic complete (SC) plates lacking leucine and then were assayed by drop tests for the ability to grow at 25°C on yeast extract-peptone-dextrose (YEPD) plates containing either the alkylating agent methyl methanesulfonate (MMS; 0.005%) or the DNA synthesis inhibitor hydroxyurea (HU; 5 mM).
Yeast strains and media.
All strains used in this study are listed in Table 1 and are derivatives of W303 (MATa or MATα; ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 ssd1), with the exceptions of strains JKM139, YLL1939, DMP4740/10B, and DMP4894/10A, the genotypes and sources of which are described below. All strains carrying the deletion of MEC1 also contained the deletion of SML1, which suppresses cell lethality caused by the absence of Mec1 (57).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| YLL1414 | MATaTEL1::LEU2 | This study |
| YLL488 | MATasml1Δ::KANMX4 | 7 |
| YLL490 | MATamec1Δ::HIS3 sml1Δ::KANMX4 | 7 |
| YLL936 | MATamre11Δ::HIS3 | This study |
| TEL1-hy184 | MATaTEL1-hy184::LEU2 | This study |
| TEL1-hy385 | MATaTEL1-hy385::LEU2 | This study |
| TEL1-hy394 | MATaTEL1-hy394::LEU2 | This study |
| TEL1-hy544 | MATaTEL1-hy544::LEU2 | This study |
| TEL1-hy628 | MATaTEL1-hy628::LEU2 | This study |
| TEL1-hy680 | MATaTEL1-hy680::LEU2 | This study |
| TEL1-hy909 | MATaTEL1-hy909::LEU2 | This study |
| TEL1-hy184 sml1Δ | MATaTEL1-hy184::LEU2 sml1Δ::KANMX4 | This study |
| TEL1-hy385 sml1Δ | MATaTEL1-hy385::LEU2 sml1Δ::KANMX4 | This study |
| TEL1-hy394 sml1Δ | MATaTEL1-hy394::LEU2 sml1Δ::KANMX4 | This study |
| TEL1-hy544 sml1Δ | MATaTEL1-hy544::LEU2 sml1Δ::KANMX4 | This study |
| TEL1-hy628 sml1Δ | MATaTEL1-hy628::LEU2 sml1Δ::KANMX4 | This study |
| TEL1-hy680 sml1Δ | MATaTEL1-hy680::LEU2 sml1Δ::KANMX4 | This study |
| TEL1-hy909 sml1Δ | MATaTEL1-hy909::LEU2 sml1Δ::KANMX4 | This study |
| TEL1-hy184 mec1Δ sml1Δ | MATaTEL1-hy184::LEU2 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| TEL1-hy385 mec1Δ sml1Δ | MATaTEL1-hy385::LEU2 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| TEL1-hy394 mec1Δ sml1Δ | MATaTEL1-hy394::LEU2 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| TEL1-hy544 mec1Δ sml1Δ | MATaTEL1-hy544::LEU2 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| TEL1-hy628 mec1Δ sml1Δ | MATaTEL1-hy628::LEU2 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| TEL1-hy680 mec1Δ sml1Δ | MATaTEL1-hy680::LEU2 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| TEL1-hy909 mec1Δ sml1Δ | MATaTEL1-hy909::LEU2 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| DMP2995/1B | MATaddc2Δ::KANMX4 sml1Δ:: KANMX4 | This study |
| DMP4967/2D | MATaddc2Δ::KANMX4 sml1Δ:: KANMX4 TEL1-hy909::LEU2 | This study |
| DMP4971/9D | MATaxrs2-11::KanMX4 sml1Δ::LEU2 | This study |
| DMP4973/10C | MATaxrs2-11::KANMX4 mec1Δ::HIS3 sml1Δ::LEU2 | This study |
| KRY22 | MATaTEL1-HA | 25 |
| DMP4690/9A | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-HA | This study |
| YLL1974 | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy385-HA::LEU2 | This study |
| YLL1975 | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy394-HA::LEU2 | This study |
| YLL1976 | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy544-HA::LEU2 | This study |
| YLL1977 | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy628-HA::LEU2 | This study |
| YLL1978 | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy680-HA::LEU2 | This study |
| YLL1979 | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy909-HA::LEU2 | This study |
| YLL2104 | MATamec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy184-HA::LEU2 | This study |
| JKM139 | MATahmlΔ hmrΔ ho ade3::GAL1-HO | 21 |
| DMP4740/10B | MATahmlΔ hmrΔ ho ade3::GAL1-HO TEL1-hy909::LEU2 | This study |
| DMP4894/10A | MATahmlΔ hmrΔ ho ade3::GAL1-HO mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| YLL1939 | MATahmlΔ hmrΔ ho ade3::GAL1-HO mec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy909::LEU2 | This study |
| DMP2056/7B | MATaddc1Δ::KANMX4 | 24 |
| DMP4712/1D | MATaTEL1-hy909::LEU2 ddc1Δ::KANMX4 | This study |
| YLL509 | MATarad53Δ::HIS3 sml1Δ::KANMX4 | 7 |
| DMP4713/7B | MATarad53Δ::HIS3 sml1Δ::KANMX4 TEL1-hy909::LEU2 | This study |
| DMP1911/1C | MATarad9Δ::URA3 | This study |
| DMP4711/3C | MATarad9Δ::URA3 TEL1-hy909::LEU2 | This study |
| DMP4726/6C | MATarad9Δ::URA3 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| DMP4726/12A | MATarad9Δ::URA3 mec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy909::LEU2 | This study |
| YLL1310 | MATamrc1Δ::HIS3 | This study |
| DMP4714/2D | MATamrc1Δ::HIS3 TEL1-hy909::LEU2 | This study |
| DMP4742/4C | MATamrc1Δ::HIS3 rad9Δ::URA3 sml1Δ::KANMX4 | This study |
| DMP4732/1A | MATamrc1Δ::HIS3 rad9Δ::URA3 sml1Δ::KANMX4 TEL1-hy909::LEU2 | This study |
| DMP4774/16C | MATamrc1Δ::HIS3 mec1Δ::HIS3 sml1Δ::KANMX4 | This study |
| DMP4774/41D | MATamrc1Δ::HIS3 mec1Δ::HIS3 sml1Δ::KANMX4 TEL1-hy909::LEU2 | This study |
| DMP4831/34D | MATamrc1Δ::HIS3 mec1Δ::HIS3 sml1Δ::KANMX4 rad9Δ::URA3 TEL1-hy909::LEU2 | This study |
TEL1-hy sml1Δ strains were meiotic segregants from crosses between strains TEL1-hy mec1Δ sml1Δ MATa and W303 MATα. Homozygous mec1Δ/mec1Δ sml1Δ/sml1Δ diploid strains, each of which was heterozygous for one TEL1-hy allele, were obtained by crossing a MATα mec1Δ sml1Δ strain with TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, or TEL1-hy909 strains, all carrying the MATa mec1Δ and sml1Δ alleles.
Strains carrying the deletion(s) of RAD9, MEC1, SML1, MRC1, DDC1, DDC2, RAD53, and/or MRE11 were obtained by the one-step PCR disruption method. Sequences for all primers used for this purpose are available upon request. Yeast strain JKM139 (MATa hmlΔ hmrΔ ade1 lys5 leu2-3,112 trp1::hisG ura3-52 ho ade3::GAL-HO), kindly provided by J. Haber (Brandeis University, Waltham, MA) (21), was used to disrupt the MEC1 and SML1 genes and to replace the TEL1 chromosomal copy with the TEL1-hy909 allele, giving rise to strains YLL1939, DMP4740/10B, and DMP4894/10A. Strains DMP4971/9D, DMP4973/10C, and TEL1-hy mec1Δ sml1Δ xrs2-11 were obtained by crossing strain KSC1563 (MATa-inc ADH4cs::HIS2 ade1 his2 leu2 trp1 ura3 xrs2-11::KANMX4 sml1Δ::LEU2), kindly provided by K. Sugimoto (University of New Jersey, Newark) (32), with MATα TEL1-hy mec1Δ sml1Δ strains. Strain KRY22, carrying a fully functional TEL1-HA allele at the TEL1 chromosomal locus (the hemagglutinin [HA] tag coding sequence was located 2,394 bp downstream of the TEL1 start codon) was kindly provided by T. Petes (University of North Carolina, Chapel Hill) (25). Strain DMP4690/9A was a meiotic segregant from a cross between strain KRY22 and a MATα mec1Δ sml1Δ strain. Strains YLL2104, YLL1974, YLL1975, YLL1976, YLL1977, YLL1978, and YLL1979, all carrying both the mec1Δ and the sml1Δ alleles and one HA-tagged TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, or TEL1-hy909 allele at the TEL1 chromosomal locus, respectively, were obtained by transforming strain DMP4690/9A with PCR products obtained using genomic DNA of TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, and TEL1-hy909 strains as the template together with primers PRP583 (5′-GCA CGA ATG AAA GAG TAC CGA CAA GCA T-3′) and PRP585 (5′-GAA AAC AAA CAG CTC AGT CCA TCC TTC C-3′). Wild-type, mec1Δ, and TEL1-hy mec1Δ strains carrying a CEN4 MEC1 plasmid were constructed by transforming wild-type and TEL1-hy strains with plasmid pML166 (CEN4 MEC1 URA3). The MEC1 gene then was deleted by a one-step PCR disruption method. The accuracy of all gene replacements and integrations was verified by Southern blot or PCR analysis. Standard yeast genetic techniques and media are described in reference 40. Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, 50 mg/liter adenine) supplemented with 2% glucose (YEPD) or 3% glycerol, 2% lactic acid, and 0.005% glucose (designated YEP+lac) or with 3% glycerol, 2% lactic acid, and 2% galactose (designated YEP+lac+gal).
Western blot analysis.
Protein extracts for Western blot analysis were prepared by trichloroacetic acid precipitation (24). Protein extracts were resolved on 10% or 7.5% polyacrylamide gels to detect Rad53 and Tel1-HA proteins, respectively. Proteins were transferred to Protran membranes (Schleicher & Schuell), and blots were probed with anti-HA monoclonal antibody 12CA5 or with polyclonal antibodies against Rad53, kindly provided by J. Diffley (Clare Hall, London, United Kingdom) and C. Santocanale (Nerviano Medical Sciences-Oncology, Milan, Italy). Secondary antibodies were purchased from Amersham, and proteins were visualized by an enhanced chemiluminescence system according to the manufacturer's instructions.
Immunoprecipitation and kinase assay.
Protein extracts for the immunoprecipitations were prepared in a lysis buffer containing 50 mM HEPES (pH 7.4), 100 mM KCl, 0.1 mM EDTA (pH 7.5), 0.2% Tween-20, 1 mM dithiothreitol (DTT), 25 mM NaFl, 100 μM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride, 25 mM β-glycerophosphate, and a protease inhibitor cocktail (Roche Diagnostics). After the addition of a 1:1 volume of acid-washed glass beads and breakage, equal amounts of protein of the different clarified extracts were incubated for 2 h at 4°C with 75 μl of a 50% (vol/vol) protein A-Sepharose resin covalently linked to 12CA5 monoclonal antibody. Resins then were washed three times in the lysis buffer and were resuspended in 450 μl of a kinase buffer containing 10 mM HEPES (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT. Resuspended resins (150 μl) were dried, followed by the addition of 11.5 μl of kinase buffer, 1.5 μl of 20 μM unlabeled ATP, 10 μCi of 32P-labeled ATP, and 1 μl of phosphorylated heat- and acid-stable protein I (PHAS-I; 1 μg/μl; Stratagene). Kinase reactions were incubated at 30°C for 30 min. Sodium dodecyl sulfate (SDS) gel-loading buffer (15 μl) was added to the resins, and bound proteins were resolved by SDS-18% polyacrylamide gel electrophoresis and visualized after exposure of the gels to autoradiography films. The residual 300 μl of each resuspended resin was dried, resuspended in 10 μl of loading buffer, and subjected to Western blot analysis with anti-HA antibody.
Other techniques.
Synchronization experiments were performed as described in reference 24. DSB end resection at the MAT locus in JKM139 derivative strains was analyzed on alkaline agarose gels as described in reference 47, using a single-stranded probe complementary to the unresected DSB strand. This probe was obtained by in vitro transcription using Promega Riboprobe system T7 and plasmid pML514 as a template (26). Plasmid pML514 was constructed by inserting into the pGEM-7Zf EcoRI site a 900-bp fragment containing part of the MATα locus (coordinates 200870 to 201587 on chromosome III) that was obtained by PCR using yeast genomic DNA as a template and PRP643 (5′-CGG AAT TCC CTG GTT TTG GTT TTG TAG AGT GG-3′) and PRP644 (5′-CGG AAT TCG AAA CAC CAA GGG AGA GAA GAC-3′) as primers. To measure telomere length, yeast DNA was prepared according to standard methods and digested with XhoI. The resulting DNA fragments were separated by gel electrophoresis in a 0.8% agarose gel and transferred to a GeneScreen nylon membrane (New England Nuclear, Boston, MA), followed by hybridization with a 32P-labeled poly(GT) probe. Standard hybridization conditions were used.
RESULTS
TEL1-hy mutations partially suppress the hypersensitivity to genotoxic agents caused by the lack of Mec1.
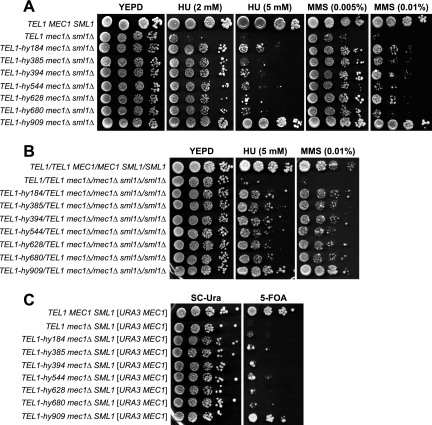
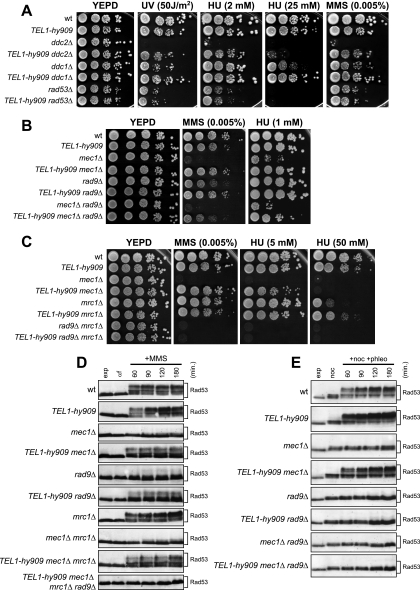
In order to gain insights into Mec1 and Tel1 regulation and interconnections, we searched for Tel1 variants able to substitute for Mec1 checkpoint functions. Random PCR mutagenesis of the C-terminal Tel1-coding sequence was followed by the replacement of the corresponding wild-type sequence with the mutagenized fragments in mec1Δ cells kept viable by the deletion of the SML1 gene. Transformant clones showing decreased sensitivity to HU and MMS compared to that of the untransformed strain then were chosen for further analysis (see Materials and Methods). This procedure allowed us to isolate seven independent mutants that we called TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, and TEL1-hy909. As shown in Fig. 1A, the TEL1-hy mec1Δ sml1Δ triple mutants formed colonies on plates containing 2 mM HU and 0.005% MMS more efficiently, although to different extents, than the corresponding mec1Δ sml1Δ strain expressing wild-type Tel1. This indicates that the corresponding Tel1-hy variants can partially substitute for Mec1 in the response to DNA damage. Only the TEL1-hy909 allele was able to efficiently suppress the hypersensitivity of mec1Δ sml1Δ cells in the presence of higher HU and MMS doses (Fig. 1A).
FIG. 1.
Hypersensitivity to genotoxic agents and cell viability of mec1Δ cells carrying the TEL1-hy alleles. (A) Serial dilutions of wild-type (W303; TEL1 MEC1 SML1), TEL1 mec1Δ sml1Δ (YLL490), TEL1-hy184 mec1Δ sml1Δ, TEL1-hy385 mec1Δ sml1Δ, TEL1-hy394 mec1Δ sml1Δ, TEL1-hy544 mec1Δ sml1Δ, TEL1-hy628 mec1Δ sml1Δ, TEL1-hy680 mec1Δ sml1Δ, and TEL1-hy909 mec1Δ sml1Δ strains, exponentially growing in YEPD, were spotted on plates with or without MMS or HU at the indicated concentrations. (B) Serial dilutions (1:10) of wild-type (W303; TEL1/TEL1 MEC1/MEC1 SML1/SML1), TEL1/TEL1 mec1Δ/mec1Δ sml1Δ/sml1Δ, and TEL1-hy/TEL1 mec1Δ/mec1Δ sml1Δ/sml1Δ diploid strains, exponentially growing in YEPD, were spotted on plates with or without MMS or HU at the indicated concentrations. (C) Serial dilutions of wild-type (W303; TEL1 MEC1 SML1), TEL1 mec1Δ, TEL1-hy184 mec1Δ, TEL1-hy385 mec1Δ, TEL1-hy394 mec1Δ, TEL1-hy544 mec1Δ, TEL1-hy628 mec1Δ, TEL1-hy680 mec1Δ, and TEL1-hy909 mec1Δ strains, all carrying a wild-type copy of the SML1 gene and the MEC1 gene on a centromeric URA3 plasmid, were spotted on SC-Ura (SC without uracil) plates and on SC plates with 5-FOA.
The suppression of mec1Δ cells’ hypersensitivity to genotoxic treatments was dominant. In fact, mec1Δ/mec1Δ sml1Δ/sml1Δ TEL1-hy/TEL1 diploid cells carrying the different TEL1-hy alleles formed colonies on HU- and MMS-containing plates more efficiently than otherwise isogenic mec1Δ/mec1Δ sml1Δ/sml1Δ TEL1/TEL1 diploid cells (Fig. 1B), suggesting that all of the TEL1-hy alleles encoded gain-of-function Tel1 variants.
Because the lack of Mec1 causes cell death in the presence of functional Sml1 (57), we asked whether the TEL1-hy alleles were able to compensate for the Mec1 essential function. For this purpose, we replaced the TEL1 chromosomal copy with the TEL1-hy alleles in a haploid ura3 mec1Δ SML1 strain. This strain was kept viable by a centromeric plasmid carrying wild-type MEC1 and the URA3 marker, with the presence of the latter inhibiting cell growth on 5-fluoroorotic acid (5-FOA). As shown in Fig. 1C, haploid mec1Δ cells did not form colonies on 5-FOA-containing plates even when they carried the TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, and TEL1-hy680 alleles. On the contrary, the ability of mec1Δ cells to survive in the absence of the MEC1 URA3 plasmid was restored by the presence of the TEL1-hy909 allele. In fact, the efficiency of TEL1-hy909 mec1Δ cells to form colonies on 5-FOA-containing plates was similar to that of cells carrying chromosomal wild-type MEC1 (Fig. 1C). Therefore, only the TEL1-hy909 allele can suppress the lethal effect of the absence of Mec1.
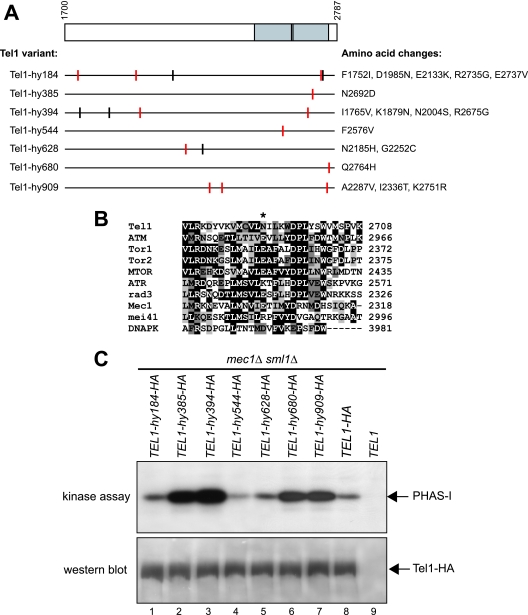
Determination and comparison of the entire wild-type and mutant TEL1 coding sequences revealed that most TEL1-hy alleles carried multiple-base-pair substitutions causing changes of different amino acid residues in the mutagenized C-terminal region (Fig. 2A). The contribution of the single-amino-acid changes to the mutant phenotypes remains to be established, while the alignment of the amino acid sequences of Tel1 and human ATM indicated that most TEL1-hy mutations caused changes of highly conserved residues (Fig. 2A). Only the TEL1-hy385, TEL1-hy544, and TEL1-hy680 alleles carried single-base-pair substitutions, resulting in the amino acid changes N2692D, F2576V, and Q2764H, respectively. Although these amino acid substitutions do not alter the G2611, D2612, N2616, and D2631 residues that have been shown to be essential for Tel1 kinase activity (25), they are located inside the C-terminal domain spanning positions 2460 to 2755 (Fig. 2A) that is shared by mammalian ATM, ATR, DNA-PK (DNA protein kinase), and mTOR (mammalian target of rapamycin) and the yeast protein kinases Rad3 (S. pombe), Tor1, Tor2, and Mec1 (S. cerevisiae) (43). Interestingly, an acidic residue (E or D) is found in Mec1, ATM, Tor1, Tor2, mTOR, Rad3, and DNA-PK amino acid sequences at the position corresponding to the Tel1-hy385 N2692D substitution (Fig. 2B).
FIG. 2.
Tel1-hy amino acid changes and in vitro kinase activity. (A) The C-terminal part of wild-type Tel1, spanning amino acid residues 1700 to 2787, is schematically depicted in the top part, with a gray box highlighting the C-terminal domain (between amino acids 2460 and 2755) that is shared by a number of PI3-like kinases, including mammalian ATM, ATR, DNA-PK, and MTOR; S. pombe Tel1 and Rad3; and S. cerevisiae Tel1, Mec1, and Tor1. Black lines inside the gray block indicate the G2611, D2612, N2616, and D2631 amino acid residues that have been shown to be essential for Tel1 kinase activity (25). Black horizontal lines represent the same C-terminal region in the Tel1-hy variants indicated on the left, with the corresponding amino acid changes listed on the right. Vertical lines indicate the position of the amino acid changes, with red lines highlighting changes affecting conserved amino acid residues based on a ClustalW alignment of the whole Tel1 and human ATM amino acid sequences. (B) ClustalW alignment of Tel1 amino acid sequence between residues 2679 and 2708, spanning the conserved PI3 kinase domain, with the corresponding sequences of PI3-like kinases indicated on the left. Identical amino acids are indicated by black boxes, and similar ones are indicated by different shades of gray. An asterisk indicates the position corresponding to the N2692D alteration in Tel1-hy385. (C) Immunoprecipitation and in vitro kinase assays of HA-tagged Tel1 and Tel1-hy proteins. The strains used were TEL1-hy184-HA (YLL2104), TEL1-hy385-HA (YLL1974), TEL1-hy394-HA (YLL1975), TEL1-hy544-HA (YLL1976), TEL1-hy628-HA (YLL1977), TEL1-hy680-HA (YLL1978), TEL1-hy909-HA (YLL1979), TEL1-HA (DMP4690/9A), and TEL1 (YLL490), all carrying the deletions of both MEC1 and SML1. Kinase assays and Western blot analysis (see Materials and Methods) were performed on equal amounts of anti-HA immunoprecipitates of protein extracts from exponentially growing untreated cells. Products of a kinase reaction using γ-32P-labeled ATP were analyzed by SDS-polyacrylamide gel electrophoresis (kinase assay). All of the immunoprecipitates also were subjected to Western blot analysis using anti-HA antibodies (western blot).
In vitro kinase activity of Tel1-hy variants.
The functional differences between wild-type Tel1 and the Tel1-hy species might be due to differences in their amounts and/or in their intrinsic kinase activities. We therefore constructed mec1Δ sml1Δ strains in which the chromosomal wild-type TEL1 or TEL1-hy allele was tagged with HA epitopes. The TEL1-hy-HA alleles still suppressed the hypersensitivity to genotoxic agents of mec1Δ sml1Δ cells (data not shown), indicating that the HA tag did not alter the Tel1-hy suppressor effects. We then used anti-HA antibodies to immunoprecipitate either Tel1 or Tel1-hy variants in order to measure their kinase activities in vitro. As shown in Fig. 2C, immunoprecipitated wild-type Tel1 (lane 8) phosphorylated PHAS-I, a known artificial substrate of the ATM kinase family (25). PHAS-I in vitro phosphorylation specifically was dependent on Tel1, since it was not detectable when the assayed immunoprecipitate was prepared from a strain expressing untagged TEL1 (Fig. 2C, lane 9). The amount of phosphorylated PHAS-I was similar to that observed with wild-type Tel1 in the assays containing Tel1-hy184-HA and Tel1-hy628-HA, whereas it was considerably higher in those performed with Tel1-hy385-HA, Tel1-hy394-HA, Tel1-hy680-HA, and Tel1-hy909-HA immunoprecipitates (Fig. 2C, top). On the contrary, PHAS-I phosphorylation appeared to take place less efficiently in the presence of Tel1-hy544-HA (Fig. 2C, top). The in vitro kinase activity differences among the immunoprecipitates were not due to different amounts of Tel1 or Tel1-hy proteins. In fact, equivalent amounts of wild-type Tel1-HA and of its mutant variants were present in the immunoprecipitates under analysis (Fig. 2C, bottom). Since all of these immunoprecipitates were obtained starting from the same protein amounts (see Materials and Methods), these results also rule out the possibility that different levels of the corresponding Tel1 species are present in the different strains. Thus, increased kinase activity cannot explain the effects of Tel1-hy184-HA, Tel1-hy628-HA, or Tel1-hy544-HA, while it may explain those of Tel1-hy385-HA, Tel1-hy394-HA, Tel1-hy680-HA, and Tel1-hy909-HA, although we cannot completely exclude the possibility that these variants might stimulate PHAS-I phosphorylation by other kinases possibly present in the immunoprecipitates.
The TEL1-hy alleles partially suppress the checkpoint defects caused by the lack of Mec1.
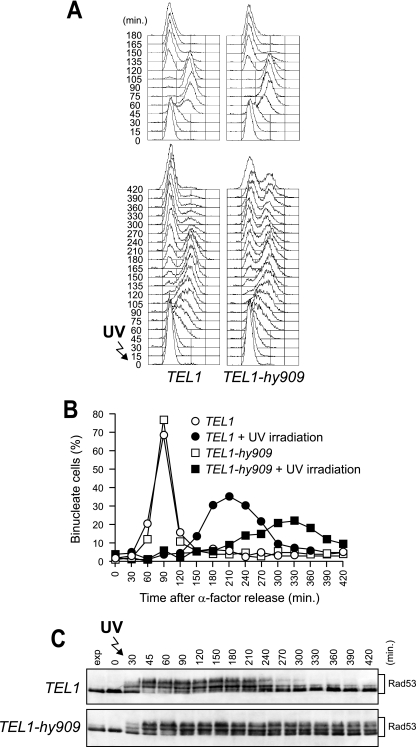
We asked whether the TEL1-hy alleles also could suppress the checkpoint defects of mec1Δ sml1Δ cells (referred to as mec1Δ cells hereafter). In order to address this point, we analyzed the response of TEL1-hy mec1Δ cells to UV irradiation in G1, which causes a Mec1-dependent checkpoint activation, leading to the phosphorylation of the checkpoint kinase Rad53 (42) and a slowing down of S-phase entry and progression.
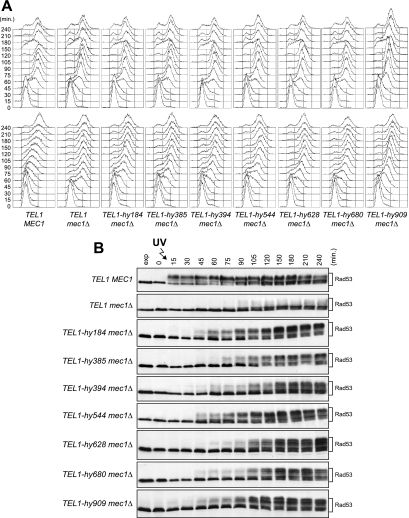
When α-factor-arrested wild-type, mec1Δ, and TEL1-hy mec1Δ cells were released from G1 arrest under unperturbed conditions, they all completed DNA replication within 60 min after release, indicating that the TEL1-hy alleles do not affect unperturbed cell cycle kinetics (Fig. 3A, top). On the contrary, when samples of the same G1-arrested cell cultures were UV irradiated immediately before release, the completion of DNA replication was slower in mec1Δ cells carrying the TEL1-hy alleles (90 to 120 min after release) than in similarly treated mec1Δ cells with wild-type TEL1 (60 min after release) (Fig. 3A, bottom). The slowing down of S-phase entry and progression in UV-irradiated TEL1-hy mec1Δ cells correlated with more efficient DNA damage checkpoint activation compared to that of similarly treated mec1Δ TEL1 cell cultures, in which the amount of phosphorylated Rad53 was much smaller than that in TEL1-hy mec1Δ cell cultures (Fig. 3B). Although the TEL1-hy alleles suppressed the checkpoint defects of mec1Δ cells, the extent of both cell cycle progression delay and Rad53 phosphorylation were not as efficient as that in wild-type cells in any of the mutants (Fig. 3). Thus, the Tel1-hy mutant variants can only partially compensate for the lack of Mec1 functions in UV-induced checkpoint activation.
FIG. 3.
G1/S DNA damage checkpoint in TEL1-hy mec1Δ cells. Cell cultures of wild-type (W303; TEL1 MEC1), TEL1 mec1Δ sml1Δ (YLL490), TEL1-hy184 mec1Δ sml1Δ, TEL1-hy385 mec1Δ sml1Δ, TEL1-hy394 mec1Δ sml1Δ, TEL1-hy544 mec1Δ sml1Δ, TEL1-hy628 mec1Δ sml1Δ, TEL1-hy680 mec1Δ sml1Δ, and TEL1-hy909 mec1Δ sml1Δ strains, logarithmically growing in YEPD at 25°C, were synchronized in G1 with α-factor, UV irradiated (30 J/m2), and released from α-factor at time zero in YEPD at 25°C. Samples were withdrawn at the indicated times after α-factor release to analyze the DNA content by a fluorescence-activated cell sorter in unirradiated (A, top) and UV-irradiated (A, bottom) cell cultures, as well as Rad53 phosphorylation in UV-irradiated cell cultures by Western blot analysis with anti-Rad53 antibodies (B). Time zero corresponds to cell samples withdrawn immediately before UV irradiation and release from α-factor. exp, exponentially growing cells.
It has been shown that UV irradiation in G1 in the absence of Mec1 activates a Tel1-dependent checkpoint only when cells complete S phase, suggesting that the replication of UV-damaged DNA generates signals that can be detected by Tel1 (7). Interestingly, we found that significant amounts of phosphorylated Rad53 were detectable in UV-irradiated TEL1-hy mec1Δ cells still undergoing S phase, whereas, as expected, Rad53 phosphorylation became slightly detectable only concomitantly with S-phase completion in similarly treated mec1Δ cells (Fig. 3B). This suggests that the Tel1-hy variants can be activated either in cell cycle phases not allowing wild-type Tel1 activation or in response to DNA lesions that are not sensed by the wild-type kinase.
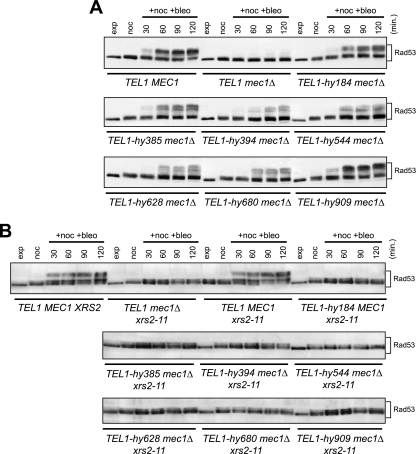
The effects of the TEL1-hy alleles were not limited to the checkpoint triggered by UV irradiation. In fact, when we analyzed Rad53 phosphorylation in cells that were kept arrested in G2 by nocodazole treatment in the presence of the radiomimetic drug bleomycin, we found that significant amounts of phosphorylated Rad53 were present in all of the TEL1-hy mec1Δ cell cultures, whereas they were below the detection level in mec1Δ cells (Fig. 4A). Moreover, the extent of Rad53 phosphorylation in different bleomycin-treated TEL1-hy mec1Δ strains seemed to correlate with the extent of suppression of mec1Δ cells’ MMS and HU hypersensitivity by the different TEL1-hy alleles. In particular, levels of bleomycin-induced Rad53 phosphorylation were similar in wild-type and TEL1-hy909 mec1Δ cells (Fig. 4A), which also showed a quite similar ability to form colonies in the presence of HU and MMS (Fig. 1A).
FIG. 4.
Bleomycin-induced DNA damage checkpoint in TEL1-hy mec1Δ cells. (A) Exponentially growing (exp) cell cultures of wild-type (W303; TEL1 MEC1), TEL1 mec1Δ sml1Δ (YLL490), TEL1-hy184 mec1Δ sml1Δ, TEL1-hy385 mec1Δ sml1Δ, TEL1-hy394 mec1Δ sml1Δ, TEL1-hy544 mec1Δ sml1Δ, TEL1-hy628 mec1Δ sml1Δ, TEL1-hy680 mec1Δ sml1Δ, and TEL1-hy909 mec1Δ sml1Δ strains were synchronized in G2 with nocodazole (15 μg/ml) (noc) and resuspended in YEPD containing nocodazole in the presence of 20 mU/ml bleomycin (+noc +bleo). Protein extracts prepared from cell samples collected at the indicated times after the addition of bleomycin were subjected to Western blot analysis with anti-Rad53 antibodies. (B) Exponentially growing cell cultures of wild-type (W303; TEL1 MEC1 XRS2), TEL1 xrs2-11 mec1Δ sml1Δ (DMP4973/10C), TEL1 MEC1 xrs2-11 sml1Δ (DMP4971/9D), TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, and TEL1-hy909 strains, all carrying deletions of MEC1, SML1, and the xrs2-11 allele, were synchronized with nocodazole (noc) and resuspended in YEPD containing nocodazole in the presence of 20 mU/ml bleomycin (+noc +bleo). Protein extracts prepared from cell samples collected at the indicated times were subjected to Western blot analysis with anti-Rad53 antibodies.
Because ATM/Tel1 functions are known to depend on the activity of the MRN/MRX complex (32, 46), we evaluated whether the Tel1-hy variants could escape the MRX requirement to exert their suppression effects. MRX is thought to recruit Tel1 to the damaged sites through the interaction between the C-terminal motif of Xrs2 and Tel1 (32, 46). We therefore studied checkpoint activation in the TEL1-hy mec1Δ strains carrying the xrs2-11 mutation, which causes a lack of the Xrs2 C-terminal part and specifically affects Tel1-Xrs2 interaction (32). As shown in Fig. 4B, bleomycin-induced Rad53 phosphorylation was not detectable in any TEL1-hy mec1Δ xrs2-11 strain, indicating that the Tel1-hy variants still require MRX to be recruited to the DNA damage sites.
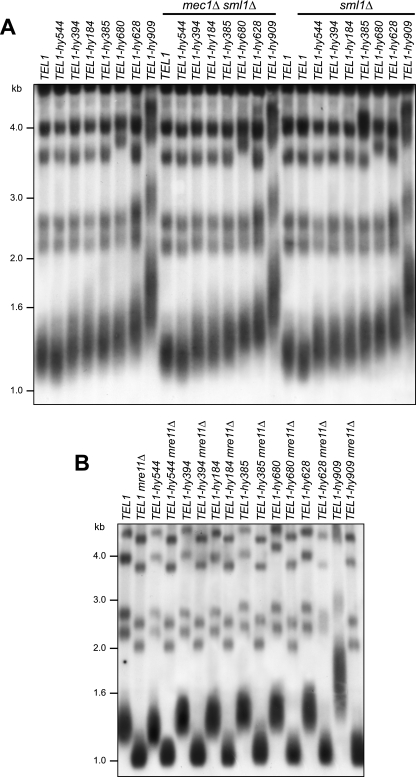
Tel1-hy proteins affect telomere homeostasis.
Tel1/ATM appears to be particularly important for telomere length maintenance in yeast, Drosophila melanogaster, and mammalian cells, in which its inactivation results in shortened telomeres and increased chromosomal end-to-end fusions by nonhomologous end joining (4, 28, 39, 48). This prompted us to ask whether the Tel1-hy proteins could influence telomere elongation. We therefore measured telomere length in the TEL1-hy mutants, either in the absence or in the presence of Mec1 or Sml1, by Southern blot analysis of XhoI-digested DNA using a poly(GT/CA) probe. Although to different extents, telomeres turned out to be longer than those of the wild type in all of the TEL1-hy mutants but TEL1-hy544, in which they appeared slightly shorter than those of the wild type (Fig. 5A). Since Tel1-hy544 is the only Tel1-hy protein showing a reduced ability to phosphorylate PHAS-I in vitro, its decreased kinase activity might account for the telomere length defects in TEL1-hy544 cells.
FIG. 5.
Telomere length in TEL1-hy mutants. (A) Genomic DNA was prepared from wild-type (W303; TEL1), TEL1 mec1Δ sml1Δ (YLL490), and TEL1 sml1Δ (YLL488) cell cultures and from cultures of cells carrying the TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, and TEL1-hy909 alleles, either alone or together with the deletion of MEC1 and/or SML1, exponentially growing in YEPD for more than 80 generations. DNA was digested with XhoI and hybridized with a poly(GT) telomere-specific probe. (B) Genomic DNA was prepared from wild-type (W303; TEL1) and TEL1 mre11Δ (YLL936) cell cultures and from cultures of cells carrying the TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, and TEL1-hy909 alleles, as well as either the mre11Δ or MRE11 allele, exponentially growing in YEPD for more than 80 generations. DNA was digested with XhoI and hybridized with a poly(GT) telomere-specific probe.
Similar to what was observed for the Tel1-hy-mediated checkpoint response, telomere overelongation in the TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy628, TEL1-hy680, and TEL1-hy909 mutants still was dependent on the MRX complex (Fig. 5B). Moreover, it was not affected by the absence of Mec1 or Sml1 (Fig. 5A), and it was telomerase dependent (data not shown).
TEL1-hy909 suppressor effects require Rad53 and its mediators, Rad9 and Mrc1.
The strong suppressor effects of the TEL1-hy909 allele on mec1Δ cells’ defects prompted us to characterize it in more detail. To this end, we first asked which other checkpoint proteins were required for the Tel1-hy909-mediated DNA damage response (Fig. 6). As shown in Fig. 6A, this variant suppressed the hypersensitivity to DNA-damaging agents of cells lacking either Ddc2, which physically interacts with Mec1 and is required for all Mec1 functions (35), or Ddc1, which stimulates Mec1 activity together with Rad17 and Mec3 (24, 34). These results are consistent with the ability of Tel1-hy909 to bypass Mec1 functions, while the inability of the TEL1-hy909 allele to suppress the HU, MMS, or UV hypersensitivity of rad53Δ sml1Δ cells (Fig. 6A) indicates that Rad53 still is required for the Tel1-hy909-mediated response to DNA damage.
FIG. 6.
Tel1-hy909 requires Rad53 and its mediators, Rad9 and Mrc1, for DNA damage response. (A) Serial dilutions of wild-type (wt; W303), TEL1-hy909, ddc2Δ sml1Δ (DMP2995/1B), TEL1-hy909 ddc2Δ sml1Δ (DMP4967/2D), ddc1Δ (DMP2056/7B), TEL1-hy909 ddc1Δ (DMP4712/1D), rad53Δ sml1Δ (YLL509), and TEL1-hy909 rad53Δ sml1Δ (DMP4713/7B) cell cultures, exponentially growing in YEPD, were spotted on plates with or without MMS or HU at the indicated concentrations. One YEPD plate was exposed at the indicated UV dose. (B) Serial dilution of wild-type, TEL1-hy909, mec1Δ sml1Δ (YLL490), TEL1-hy909 mec1Δ sml1Δ, rad9Δ (DMP1911/1C), TEL1-hy909 rad9Δ (DMP4711/3C), rad9Δ mec1Δ sml1Δ (DMP4726/6C), and TEL1-hy909 rad9Δ mec1Δ sml1Δ (DMP4726/12A) cell cultures, exponentially growing in YEPD, were spotted on plates with or without MMS or HU at the indicated concentrations. (C) Serial dilution of wild-type, TEL1-hy909, mec1Δ sml1Δ (YLL490), TEL1-hy909 mec1Δ sml1Δ, mrc1Δ (YLL1310), TEL1-hy909 mrc1Δ (DMP4714/2D), rad9Δ mrc1Δ sml1Δ (DMP4742/4C), and TEL1-hy909 rad9Δ mrc1Δ sml1Δ (DMP4732/1A) cell cultures, exponentially growing in YEPD, were spotted on plates with or without MMS or HU at the indicated concentrations. (D) Exponentially growing (exp) cell cultures of wild-type, TEL1-hy909, mec1Δ sml1Δ (YLL490), TEL1-hy909 mec1Δ sml1Δ, rad9Δ (DMP1911/1C), TEL1-hy909 rad9Δ (DMP4711/3C), mrc1Δ (YLL1310), mec1Δ mrc1Δ sml1Δ (DMP4774/16C), TEL1-hy909 mec1Δ mrc1Δ sml1Δ (DMP4774/41D), and TEL1-hy909 mec1Δ mrc1Δ rad9Δ sml1Δ (DMP4831/34D) strains were arrested in G1 with α-factor (αf) and released into the cell cycle in YEPD containing 0.01% MMS (+MMS). Protein extracts prepared from cell samples collected at the indicated times after MMS addition were subjected to Western blot analysis with anti-Rad53 antibodies. (E) Exponentially growing cell cultures of wild-type, TEL1-hy909, mec1Δ sml1Δ (YLL490), TEL1-hy909 mec1Δ sml1Δ, rad9Δ (DMP1911/1C), TEL1-hy909 rad9Δ (DMP4711/3C), rad9Δ mec1Δ sml1Δ (DMP4726/6C), and TEL1-hy909 rad9Δ mec1Δ sml1Δ (DMP4726/12A) strains were arrested in G2 with nocodazole (noc) and resuspended in YEPD containing nocodazole in the presence of 3 μg/ml phleomycin (+phleo +noc). Protein extracts prepared from cell samples collected at the indicated times after phleomycin addition were subjected to Western blot analysis with anti-Rad53 antibodies.
Both Mec1- and Tel1-dependent Rad53 activations require damage-specific or S-phase-specific mediators. In particular, Rad9 mediates the response to DNA damage in G1 or in G2 by acting first as an adaptor allowing Mec1-mediated Rad53 phosphorylation and activation (49) and subsequently acting as a scaffold facilitating in trans Rad53 autophosphorylation, thus completing the Rad53 activation process (12). Conversely, Mrc1 is known to mediate Rad53 activation in response to replication stress (2, 17, 33, 51). As shown in Fig. 6B, both TEL1-hy909 rad9Δ and TEL1-hy909 rad9Δ mec1Δ cells formed colonies on HU- or MMS-containing plates more efficiently than rad9Δ and rad9Δ mec1Δ cells, respectively. This indicates that the Tel1-hy909-dependent suppression of mec1Δ cells’ hypersensitivity to MMS and HU can take place to some extent even in the absence of Rad9. However, the efficiency to form colonies in the presence of genotoxic agents of TEL1-hy909 rad9Δ mec1Δ cells was lower than that of TEL1-hy909 mec1Δ cells (Fig. 6B), indicating that Rad9 still plays a role in the Tel1-hy909-mediated DNA damage response. Since both MMS and HU interfere with replication fork progression and Mrc1 is involved in the checkpoint response to replicative stress (2, 17, 33, 51), both Mrc1 and Rad9 may contribute to the action of Tel1-hy909 in the response to DNA perturbations. Consistent with this hypothesis, the TEL1-hy909 allele, which suppressed both the MMS hypersensitivity of rad9Δ cells (Fig. 6B) and the HU hypersensitivity of mrc1Δ cells (Fig. 6C), did not restore the ability to form colonies in the presence of any tested MMS or HU concentration of mrc1Δ rad9Δ double mutant cells, which were much more sensitive to both MMS and HU than either single mutant (Fig. 6C). Moreover, the amount of MMS-induced Rad53 phosphorylation, which was increased in both TEL1-hy909 rad9Δ and TEL1-hy909 mrc1Δ mec1Δ cells compared to that of rad9Δ and mrc1Δ mec1Δ cells, respectively, was undetectable in TEL1-hy909 mec1Δ rad9Δ mrc1Δ cells (Fig. 6D).
On the contrary, Rad9 seems to play a major role, compared to that of Mrc1, in mediating the Tel1-hy909 response to DSB-inducing agents. In fact, Rad53 phosphorylation clearly was detectable in TEL1-hy909 mec1Δ cells treated with the radiomimetic drug phleomycin but was not detectable in similarly treated TEL1-hy909 rad9Δ and TEL1-hy909 mec1Δ rad9Δ cells (Fig. 6E). Taken together, these data indicate that the DNA damage response through Tel1-hy909 still requires the Rad9 or Mrc1 mediator, depending on the types of DNA lesions.
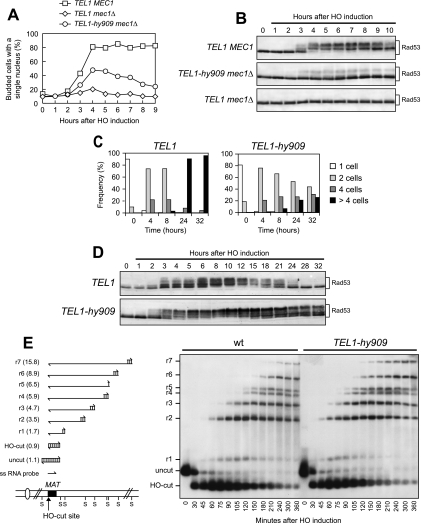
The TEL1-hy909 allele partially suppresses checkpoint defects of mec1Δ cells and prevents adaptation in response to a single irreparable DSB.
Cells carrying a single irreparable DSB are known to undergo a checkpoint-mediated cell cycle block that primarily is dependent on Mec1 (6, 15, 26), which recognizes replication protein A-coated ssDNA regions arising from DSB processing (58). The knowledge that wild-type Tel1 is not able to signal a single DSB even in the absence of Mec1 (26) prompted us to examine checkpoint activation after the induction of a single irreparable DSB in mec1Δ and TEL1-hy909 mec1Δ cells.
A single DSB can be generated at the MAT locus of JKM139 derivative strains by expressing the site-specific HO endonuclease gene from a galactose-inducible promoter (21). The HO cut cannot be repaired by homologous recombination, because the homologous donor sequences HML and HMR are deleted in JKM139 (21). As shown in Fig. 7A and B, the generation of a single DSB caused most wild-type cells to arrest, as expected, as mononucleate large-budded cells with heavily phosphorylated Rad53. On the contrary, mec1Δ cells under the same conditions neither arrested the cell cycle nor underwent significant Rad53 phosphorylation after HO induction, indicating that, in the absence of Mec1, wild-type Tel1 could not trigger checkpoint activation in response to a single HO-induced DSB (Fig. 7A and B). Strikingly, a single HO-induced DSB instead was sufficient to trigger Rad53 phosphorylation in TEL1-hy909 mec1Δ cells that also transiently accumulated as mononucleate large-budded cells (Fig. 7A and B). Thus, the Tel1-hy909 variant can partially substitute for Mec1 in activating the checkpoint in response to a single DSB, suggesting that this mutant protein might recognize DSBs more efficiently than wild-type Tel1. Alternatively, because the ability of wild-type Tel1 to sense and transduce the DSB signal becomes apparent by increasing the amount of either Tel1 or DSB (26), the enhanced Tel1-hy909 kinase activity may account for its acquired ability to activate the checkpoint in response to a single DSB.
FIG. 7.
TEL1-hy909 cell response to a single irreparable DSB. (A and B) Cell cultures of wild-type (JKM139; TEL1 MEC1), TEL1 mec1Δ sml1Δ (DMP4894/10A), and TEL1-hy909 mec1Δ sml1Δ (YLL1939) strains, exponentially growing in YEP+lac (time zero), were transferred to YEP+lac+gal to induce HO expression. Samples were collected at the indicated times after galactose addition to determine the percentage of mononucleate large-budded cells (A) and for Western blot analysis of protein extracts with anti-Rad53 antibodies (B). (C) YEP+lac G1-arrested cell cultures of wild-type (JKM139; TEL1) and isogenic TEL1-hy909 (DMP4740/10B) strains, both expressing wild-type MEC1, were spotted on galactose-containing plates, which were incubated at 30°C (time zero). At the indicated time points, 200 cells for each strain were analyzed to determine the frequency of single cells and of cells forming microcolonies of two, four, or more than four cells. (D) Cell cultures of strains shown in panel C, exponentially growing in YEP+lac (time zero), were transferred to YEP+lac+gal to induce HO expression. Protein extracts from aliquots withdrawn at the indicated times after galactose addition were subjected to Western blot analysis with anti-Rad53 antibodies. (E) YEP+lac nocodazole-arrested cell cultures of wild-type JKM139 and isogenic TEL1-hy909 (DMP4740/10B) strains (time zero) were transferred to YEP+lac+gal to induce HO expression in the presence of nocodazole. (E, left) Schematic representation of the region immediately centromere distal to the MAT HO site and of the DSB and 5′- to 3′-resection products detectable with the indicated ssRNA probe after alkaline gel electrophoresis of SspI (S)-digested DNA. The probe is specific for the MAT locus and reveals a 1.1-kb fragment from the uncut MAT locus. When HO cuts the MAT locus, a smaller 0.9-kb HO-cut fragment is produced. 5′ to 3′ resection progressively eliminates SspI sites, generating larger ssDNA SspI fragments (r1 through r7) detected by the probe. (E, right) Genomic DNA prepared from samples taken at the indicated times during the experiment was digested with SspI and run on an alkaline agarose gel, followed by gel blotting and hybridization with the ssRNA probe shown on the left. wt, wild type.
Although wild-type Tel1 is not able to activate the checkpoint after a single DSB, it participates in Mec1-dependent DSB-induced checkpoint activation by increasing the efficiency of ssDNA accumulation at DSB ends (26). We therefore asked whether the Tel1-hy909 variant could influence the checkpoint response to a single unrepaired DSB in cells expressing functional Mec1 (Fig. 7C to E). It has been shown that cells carrying a single irreparable DSB undergo a Mec1-dependent cell cycle block, but then they adapt to the damage, decrease Rad53 checkpoint kinase activity, and reenter the cell cycle (37, 52). As shown in Fig. 7C, when G1-arrested cells were spotted on galactose-containing plates to induce HO expression, most JKM139 wild-type cells overrode the checkpoint-mediated cell cycle arrest within 24 h, producing microcolonies of four or more cells, while most isogenic TEL1-hy909 cells still were arrested at the two-cell dumbbell stage after 32 h. Moreover, when the HO cut was induced into exponentially growing cell cultures of the same strains, Rad53 phosphorylation (the amount of which was higher in TEL1-hy909 cells than in wild-type cells) persisted for at least 30 h further in TEL1-hy909 cells that did not reenter the cell cycle, while it decreased in wild-type cells within 12 to 15 h, at which time most cells resumed cell cycle progression (Fig. 7C and D). Taken together, these data indicate that the Tel1-hy909 variant impairs adaptation to a single DSB.
Adaptation can be prevented by a twofold increase in the rate of 5′ to 3′ resection of broken chromosome ends (21), suggesting that escape from checkpoint activation after DSB formation depends on the extent of ssDNA created at broken ends. This observation, together with the knowledge that Tel1 contributes to ssDNA generation at DSBs (26), prompted us to ask whether the TEL1-hy909 allele could increase ssDNA accumulation to levels greater than that of Tel1. We therefore monitored the kinetics of both HO-cut and 3′-ended ssDNA formations (Fig. 7E) at the MAT locus after HO-cut induction in cell cultures that were blocked in mitosis with nocodazole to avoid specific cell cycle effects on DSB processing. Indeed, TEL1-hy909 cells accumulated 3′-ended resection products more efficiently than isogenic wild-type cells (Fig. 7E). Thus, the Tel1-hy909 variant may enhance Mec1 signaling activity by stimulating 3′-ended ssDNA generation that, in turn, leads to Mec1 recruitment to DSB ends and Mec1-dependent checkpoint activation.
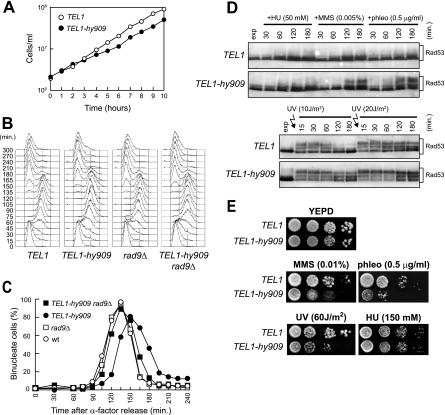
Tel1-hy909 effects on checkpoint activation under unperturbed conditions and after genotoxic treatments.
Inappropriate DNA damage checkpoint activation may be prevented by tightly regulating Mec1 and Tel1 activities. If this was the case, the Tel1-hy variants may cause unscheduled checkpoint activation by escaping the regulation of Tel1 activity. We noticed that the growth rate of TEL1-hy909 cells expressing functional Mec1 was slightly slower than that of the wild type (Fig. 8A), suggesting that the Tel1-hy909 variant elicits checkpoint activation even in the absence of exogenous DNA damage. To verify this possibility, G1-arrested wild-type (TEL1) and TEL1-hy909 cells were released into the cell cycle under unperturbed conditions. When 95% of cells had budded after release, α-factor was added back in order to prevent cells from entering a second cell cycle. As shown in Fig. 8B and C, TEL1-hy909 cells, which completed DNA replication with wild-type kinetics, underwent nuclear division about 15 min later than wild-type cells. This metaphase-to-anaphase transition delay likely was due to unscheduled checkpoint activation, since it was almost undetectable in TEL1-hy909 rad9Δ double mutant cells (Fig. 8B and C).
FIG. 8.
Checkpoint activation in TEL1-hy909 MEC1 cells during an unperturbed cell cycle or in response to mild genotoxic treatments. (A) Growth rate of asynchronous wild-type (W303; TEL1) and TEL1-hy909 cell cultures growing exponentially at 25°C. (B and C) Exponentially growing cell cultures of wild-type (W303; TEL1), TEL1-hy909, rad9Δ (DMP1911/1C), and TEL1-hy909 rad9Δ (DMP4711/3C) strains, all expressing wild-type MEC1, were arrested in G1 with α-factor (0) and released from the pheromone block in YEPD at 21°C. When 95% of cells had budded after release, 3 μg/ml α-factor was added back to all cultures. Samples were collected at the indicated times after α-factor release to analyze the DNA content by a fluorescence-activated cell sorter (B) and to determine the kinetics of nuclear division after propidium iodide staining (C). wt, wild type. (D) Cell cultures of exponentially growing (exp) wild-type (W303; TEL1) and TEL1-hy909 strains were UV irradiated (10 or 20 J/m2) or were resuspended in YEPD containing 50 mM HU (+HU), 0.005% MMS (+MMS), or 0.5 μg/ml phleomycin (+phleo). Protein extracts prepared from cell samples collected at the indicated times after genotoxic treatment were subjected to Western blot analysis with anti-Rad53 antibodies. (E) Serial dilution of wild type (W303; TEL1) and TEL1-hy909 cell cultures, exponentially growing in YEPD, were spotted on plates with or without phleomycin, MMS, and HU at the indicated concentrations. One YEPD plate was exposed to the indicated UV dose.
We then monitored checkpoint activation in both wild-type and TEL1-hy909 cells in response to mild genotoxic treatments. When exponentially growing wild-type and TEL1-hy909 cell cultures were exposed to low doses of MMS, phleomycin, or UV, the amount of Rad53 phosphorylation was higher in TEL1-hy909 cells than in similarly treated wild-type cells (Fig. 8D). This enhanced checkpoint activation correlated with the increased sensitivity of TEL1-hy909 cells to phleomycin, MMS, or UV irradiation (Fig. 8E). On the contrary, similar amounts of Rad53 phosphorylation were detected in the presence of 50 mM HU in both wild-type and TEL1-hy909 cells (Fig. 8D), which also displayed similar sensitivity to HU treatment (Fig. 8E). Thus, the Tel1-hy909 variant specifically upregulates checkpoint activation under mild DNA damage conditions, but it does not seem to affect the response to moderate DNA replication stress.
As shown in Fig. 9, checkpoint hyperactivation by Tel1-hy909 in the presence of functional Mec1 impairs the recovery from checkpoint-mediated cell cycle arrest. Exponentially growing cells were arrested in G1, UV irradiated, and released into the cell cycle. When 95% of cells had budded after release, α-factor was added back in order to prevent cells from entering a second cell cycle. As expected, G1/S checkpoint activation slowed down S-phase progression in UV-treated wild-type cells, which completed DNA replication within 150 min after release (Fig. 9A, bottom) and underwent Rad53 phosphorylation (Fig. 9C). The checkpoint then was turned off, since these cells completed the cell cycle, started to undergo nuclear division about 150 to 180 min after UV irradiation (Fig. 9B), and arrested in G1 after cell division due to α-factor readdition (Fig. 9A, bottom). On the contrary, most UV-treated TEL1-hy909 cells, which progressed through S phase and completed DNA replication with wild-type kinetics (Fig. 9A, bottom), had delayed metaphase-to-anaphase transitions (Fig. 9A and B). Moreover, the amount of phosphorylated Rad53, which decreased in wild-type cells concomitantly with reentry into the cell cycle, remained high in UV-treated TEL1-hy909 cells until the end of the experiment (Fig. 9C).
FIG. 9.
Prolonged UV-induced checkpoint activation in TEL1-hy909 cells expressing functional Mec1. Exponentially growing (exp) cell cultures of wild-type (W303; TEL1) and TEL1-hy909 strains, both expressing wild-type MEC1, were arrested in G1 with α-factor (0) and released from the pheromone block in YEPD or were UV irradiated (30 J/m2) prior to the release in YEPD. When 95% of cells had budded after release, 3 μg/ml α-factor was added back to all cultures. Samples of untreated and UV-treated cell cultures were collected at the indicated times after α-factor release to analyze the DNA content by a fluorescence-activated cell sorter (A) and to determine the kinetics of nuclear division after propidium iodide staining (B) and Rad53 phosphorylation by Western blot analysis with anti-Rad53 antibodies (C).
DISCUSSION
Mammalian ATM plays a primary role in sensing and transducing the signals emanating from DSBs and also is required for the generation of ionizing-radiation-induced replication protein A foci that lead to ATR recruitment and subsequent ATR-dependent checkpoint activation (1, 16, 30, 58). On the contrary, the checkpoint functions of the budding yeast ATM ortholog, Tel1, appear more furtive than those of ATM, as Tel1-deficient cells do not suffer from obvious hypersensitivity to DNA-damaging agents and do not show major checkpoint defects (7, 14, 26, 29). On the other hand, the finding that the lack of Tel1 exacerbates both the sensitivity to DNA-damaging agents and the checkpoint defects of cells lacking the ATR ortholog Mec1 (42) indicates that the Tel1 contribution to the checkpoint response is masked by the prevailing activity of Mec1. This suggests that Tel1 may differ from Mec1 and ATM in its intrinsic kinase activity, in its ability to interact with specific targets, and/or its ability to interact with damaged DNA. Indeed, high Tel1 levels suppress the hypersensitivity to DNA-damaging agents of mec1 mutants (8, 42), indicating that Tel1's ability to sense and transduce the DNA damage signals can be enhanced by increasing amounts of Tel1. This prompted us to ask whether it was possible to generate TEL1 mutations able to compensate for the lack of Mec1 checkpoint functions.
Here, we describe the new mutant alleles TEL1-hy184, TEL1-hy385, TEL1-hy394, TEL1-hy544, TEL1-hy628, TEL1-hy680, and TEL1-hy909, which we identified as dominant suppressors of the hypersensitivity to DNA-damaging agents of mec1Δ sml1Δ cells. Although to different extents, all of the TEL1-hy alleles also are able to partially suppress the inability of Mec1-deficient cells to trigger checkpoint activation in response to UV or bleomycin treatment. Moreover, with the exception of the TEL1-hy544 allele, all of the other TEL1-hy alleles cause telomere overelongation compared to that of wild-type TEL1, with TEL1-hy909 showing the longest telomeres. Thus, they can both activate the checkpoint in response to different kinds of DNA lesions and promote telomere elongation more efficiently than wild-type Tel1. Because Tel1 is thought to mediate the recruitment at telomeres of the Est2 catalytic subunit of telomerase and of the Est1 telomerase accessory protein (13), it is tempting to propose that Tel1-hy proteins promote the association of telomerase to telomeres more efficiently than wild-type Tel1, but further studies will be required to address this point. The MRX complex, which is known to recruit ATM/Tel1 to damaged sites (32), still is necessary to allow the Tel1-hy-mediated suppression of mec1Δ defects and telomere overelongation, indicating that both the checkpoint and telomeric functions of these variants still depend on MRX.
The differences between wild-type and Tel1-hy species may be ascribed to alterations in their amounts, in their intrinsic kinase activities, and in their abilities to interact with specific protein targets and/or with damaged DNA. While our data do not indicate significant changes in the amount of any Tel1-hy variant compared to that of the wild type, Tel1-hy385-HA, Tel1-hy394-HA, Tel1-hy680-HA, and Tel1-hy909-HA can phosphorylate in vitro the protein substrate PHAS-I more efficiently than wild-type Tel1, although they do so to different extents. Because Tel1 exerts all of its functions through phosphorylation events (25), Tel1-hy variants with an increased intrinsic kinase activity may display an enhanced ability to phosphorylate their targets, thus activating the checkpoint and promoting telomere elongation more efficiently than wild-type Tel1. Whereas the Tel1-hy394 and Tel1-hy909 variants carried multiple amino acid substitutions, the specific roles of which remain to be determined, the TEL1-hy385 and TEL1-hy680 mutations result in the single-amino-acid changes N2692D and Q2764H, respectively, inside the approximately 330-amino-acid C-terminal domain that is shared by a number of phosphatidylinositol 3 (PI3) protein kinases (43). It is tempting to speculate that these amino acid substitutions inside the conserved kinase domain facilitate the accessibility of target proteins, thus increasing the efficiency of their phosphorylation. On the other hand, human ATM is present as inert dimers or multimers that, after DNA damage, undergo a rapid intermolecular autophosphorylation, causing their dissociation in active monomers (3). If this regulatory mechanism also applied to its ortholog Tel1, the enhanced intrinsic kinase activity of Tel1-hy variants also could increase the ability to promote their autophosphorylation and activation.
An increased kinase activity cannot account for the enhanced DNA damage response mediated by Tel1-hy184, Tel1-hy544, and Tel1-hy628. In fact, neither Tel1-hy184 nor Tel1-hy628 show increased in vitro kinase activity compared to that of wild-type Tel1, whereas Tel1-hy544 kinase activity is even slightly lower than that of the wild type. Thus, the ability of Tel1-hy184, Tel1-hy544, and Tel1-hy628 to suppress mec1Δ cells’ hypersensitivity to genotoxic agents and checkpoint defects might be due to changes in their ability to interact with specific proteins and/or with damaged DNA. Because TEL1-hy184 and TEL1-hy628 cells undergo both checkpoint activation and telomere elongation more efficiently than the wild type, altered interactions with proteins in common between checkpoint and telomere length controls may account for the effects of Tel1-hy184 and Tel1-hy628. On the other hand, TEL1-hy544 cells activate the DNA damage checkpoint more efficiently than the wild type, but they are defective in maintaining telomere length, indicating that checkpoint and telomeric functions of Tel1 can be separable, as previously suggested (5). In this view, the reduced Tel1-hy544 kinase activity might explain the telomere length defects of TEL1-hy544 cells, whereas enhanced interactions with proteins specifically required to activate the checkpoint may account for their enhanced checkpoint response. Curiously, the Tel1-hy909 variant suppresses mec1Δ defects and elongates telomeres more efficiently than any other Tel1-hy species, but it displays an in vitro kinase activity similar to that of Tel1-hy680 and Tel1-hy385 and lower than that of Tel1-hy394. This suggests that the increased kinase activity cannot, by itself, account for the Tel1-hy909 effects.
Only the TEL1-hy909 allele, which suppresses the hypersensitivity of mec1Δ sml1Δ cells more efficiently than any other analyzed Tel1-hy variant, also is able to partially rescue the cell lethality caused by the absence of Mec1. Consistent with its ability to bypass all known Mec1 functions, this allele also suppresses the hypersensitivity to genotoxic agents of cells lacking Ddc1 or Ddc2, which are known to act in concert with Mec1 in the DNA damage response (24, 34, 35). On the other hand, checkpoint activation induced by Tel1-hy909 has the same downstream requirements as wild-type Mec1 and Tel1. In fact, the Tel1-hy909-induced DNA damage response does not escape the requirement of Rad53 and of its mediators, Mrc1 and Rad9 (2, 12, 17, 33, 49, 51). Moreover, the Tel1-hy909 protein appears to sense and signal a single DSB, which cannot activate a Tel1-dependent checkpoint even in the absence of Mec1 (26). Because the ability of wild-type Tel1 to sense and transduce the DSB signal becomes apparent by increasing the amount of either Tel1 or DSB (8, 26, 42), the enhanced Tel1-hy909 kinase activity may account for its acquired ability to activate the checkpoint in response to a single DSB. Alternatively, because resection of DSB ends inhibits wild-type Tel1 signaling activity (26), it also is possible that the Tel1-hy909 protein recognizes DSBs more efficiently than wild-type Tel1.
We also found that Tel1-hy909 increases the efficiency of 3′-ended ssDNA accumulation, which in turn leads to Mec1 recruitment to DSB ends and Mec1-dependent checkpoint activation (21, 26, 58). Because wild-type Tel1 is known to contribute to DSB resection, and because this likely involves the activation of the MRX complex that is required to resect DSB ends (26), the Tel1-hy909 variant might display an enhanced ability to activate MRX, thus promoting DSB resection more efficiently than wild-type Tel1.
The potential to generate Tel1 mutant variants that are more active than wild-type Tel1 suggests that Tel1 activity is tightly regulated in order to avoid unscheduled checkpoint activation. Consistent with this hypothesis, we found that TEL1-hy909 cells expressing functional Mec1 exhibit a slow-growth phenotype and a checkpoint-mediated delay of the metaphase-to-anaphase transition under unperturbed conditions. Moreover, they show an increased sensitivity to DNA-damaging agents, possibly due to an upregulation of the checkpoint response in the presence of small amounts of DNA lesions. Finally, they impair checkpoint switch off. These negative consequences of having Tel1-hyperactive variants indicates that Tel1 has evolved in order to have a specific amount of activity to minimize the risk of inappropriately launching the checkpoint response.
Taken together, our data provide evidence that it is possible to generate Tel1 variants that trigger checkpoint activation and telomere elongation more efficiently than wild-type Tel1 and can at least partially replace Mec1 in the response to genotoxic treatments. Both the differences and the similarities in the phenotypes of Tel1/ATM- or Mec1/ATR-defective cells can be explained in the light of the specific roles of these proteins in the DNA damage response and telomere homeostasis. In this scenario, the characterization of these Tel1-hy variants provides new insights into the dynamic interactions between Tel1/ATM and Mec1/ATR and may help in the development of new strategies in the diagnosis and treatment of human diseases related to ATM and ATR dysfunctions.
Acknowledgments
We thank J. F. X. Diffley, J. Haber, K. Sugimoto, T. Petes, and C. Santocanale for providing reagents and all the members of our laboratory for useful discussions and criticisms.
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro, Cassa di Risparmio delle Provincie Lombarde (CARIPLO), to M.P.L. and from Cofinanziamenti 2005 MIUR/Università di Milano-Bicocca to M.P.L. and G.L.
Footnotes
Published ahead of print on 22 October 2007.
REFERENCES
- 1.Adams, K. E., A. L. Medhurst, D. A. Dart, and N. D. Lakin. 2006. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 253894-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3958-965. [DOI] [PubMed] [Google Scholar]
- 3.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421499-506. [DOI] [PubMed] [Google Scholar]
- 4.Bi, X., S. C. Wei, and Y. S. Rong. 2004. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr. Biol. 141348-1353. [DOI] [PubMed] [Google Scholar]
- 5.Chakhparonian, M., D. Faucher, and R. J. Wellinger. 2005. A mutation in yeast Tel1p that causes differential effects on the DNA damage checkpoint and telomere maintenance. Curr. Genet. 48310-322. [DOI] [PubMed] [Google Scholar]
- 6.Clerici, M., D. Mantiero, G. Lucchini, and M. P. Longhese. 2006. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 7212-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clerici, M., V. Baldo, D. Mantiero, F. Lottersberger, G. Lucchini, and M. P. Longhese. 2004. A Tel1/MRX-dependent checkpoint inhibits the metaphase-to-anaphase transition after UV irradiation in the absence of Mec1. Mol. Cell. Biol. 2410126-10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clerici, M., V. Paciotti, V. Baldo, M. Romano, G. Lucchini, and M. P. Longhese. 2001. Hyperactivation of the yeast DNA damage checkpoint by TEL1 and DDC2 overexpression. EMBO J. 206485-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signalling. Science 2941713-1716. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1393-398. [DOI] [PubMed] [Google Scholar]
- 11.Falck, J., J. Coates, and S. P. Jackson. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434605-611. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8129-136. [DOI] [PubMed] [Google Scholar]
- 13.Goudsouzian, L. K., C. T. Tuzon, and V. A. Zakian. 2006. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24603-610. [DOI] [PubMed] [Google Scholar]
- 14.Greenwell, P. W., S. L. Kronmal, S. E. Porter, J. Gassenhuber, B. Obermaier, and T. D. Petes. 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82823-829. [DOI] [PubMed] [Google Scholar]
- 15.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 4311011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith, J. Lukas, and S. P. Jackson. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 837-45. [DOI] [PubMed] [Google Scholar]
- 17.Katou, Y., M. Bando, Y. Kanoh, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. Tof1 and Mrc1 mediate S-phase checkpoint activation as part of a stable replication pausing complex. Nature 4241078-1083. [DOI] [PubMed] [Google Scholar]
- 18.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294867-870. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. H., and T. T. Paull. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 30493-96. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. H., and T. T. Paull. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308551-554. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94399-409. [DOI] [PubMed] [Google Scholar]
- 22.Longhese, M. P., D. Mantiero, and M. Clerici. 2006. The cellular response to chromosome breakage. Mol. Microbiol. 601099-1108. [DOI] [PubMed] [Google Scholar]
- 23.Longhese, M. P., R. Fraschini, P. Plevani, and G. Lucchini. 1996. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol. Cell. Biol. 163235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longhese, M. P., V. Paciotti, R. Fraschini, R. Zaccarini, P. Plevani, and G. Lucchini. 1997. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 165216-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallory, J. C., and T. D. Petes. 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 9713749-13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantiero, D., M. Clerici, G. Lucchini, and M. P. Longhese. 2007. Dual role for S. cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 152809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalfe, J. A., J. Parkhill, L. Campbell, M. Stacey, P. Biggs, P. J. Byrd, and A. M. Taylor. 1996. Accelerated telomere shortening in ataxia telangiectasia. Nat. Genet. 13350-353. [DOI] [PubMed] [Google Scholar]
- 29.Morrow, D. M., D. A. Tagle, Y. Shiloh, F. S. Collins, and P. Hieter. 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82831-840. [DOI] [PubMed] [Google Scholar]
- 30.Myers, J. S., and D. Cortez. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2819346-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104397-408. [DOI] [PubMed] [Google Scholar]
- 32.Nakada, D., K. Matsumoto, and K. Sugimoto. 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 161957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 171755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paciotti, V., G. Lucchini, P. Plevani, and M. P. Longhese. 1998. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 174199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 142046-2059. [PMC free article] [PubMed] [Google Scholar]
- 36.Pandita, T. K. 2002. ATM function and telomere stability. Oncogene 21611-618. [DOI] [PubMed] [Google Scholar]
- 37.Pellicioli, A., S. E. Lee, C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7293-300. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie, K. B., and T. D. Petes. 2000. The Mre11/Rad50/Xrs2 complex and the Tel1 function in a single pathway for telomere maintenance in yeast. Genetics 155475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 196065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Rouse, J., and S. P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signaling pathway in Saccharomyces cerevisiae. EMBO J. 195801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271357-360. [DOI] [PubMed] [Google Scholar]
- 43.Savitsky, K., A. Bar-Shira, S. Gilad, G. Rotman, Y. Ziv, L. Vanagaite, D. A. Tagle, S. Smith, T. Uziel, S. Sfez, et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 2861749-1753. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 91055-1065. [DOI] [PubMed] [Google Scholar]
- 45.Shiloh, Y. 2006. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 31402-410. [DOI] [PubMed] [Google Scholar]
- 46.Shima, H., M. Suzuki, and M. Shinohara. 2005. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics 17071-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. M. Bonner, J. H. Petrini, J. E. Haber, and M. Lichten. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 141703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva, E., S. Tiong, M. Pedersen, E. Homola, A. Royou, B. Fasulo, G. Siriaco, and S. D. Campbell. 2004. ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr. Biol. 141341-1347. [DOI] [PubMed] [Google Scholar]
- 49.Sweeney, F. D., F. Yang, A. Chi, J. Shabanowitz, D. F. Hunt, and D. Durocher. 2005. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 151364-1375. [DOI] [PubMed] [Google Scholar]
- 50.Takata, H., Y. Kanoh, N. Gunge, K. Shirahige, and A. Matsuura. 2004. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14515-522. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3966-972. [DOI] [PubMed] [Google Scholar]
- 52.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 901097-1106. [DOI] [PubMed] [Google Scholar]
- 53.Verdun, R. E., L. Crabbe, C. Haggblom, and J. Karlseder. 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell 20551-561. [DOI] [PubMed] [Google Scholar]
- 54.Viscardi, V., M. Clerici, H. Cartagena-Lirola, and M. P. Longhese. 2005. Telomeres and DNA damage checkpoints. Biochimie 87613-624. [DOI] [PubMed] [Google Scholar]
- 55.Wakayama, T., T. Kondo, S. Ando, K. Matsumoto, and K. Sugimoto. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8652-665. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, X., E. G. D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2329-340. [DOI] [PubMed] [Google Scholar]
- 58.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 3001542-1548. [DOI] [PubMed] [Google Scholar]
- 59.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17 dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]