Abstract
Positive transcription elongation factor b (P-TEFb) is the major metazoan RNA polymerase II (Pol II) carboxyl-terminal domain (CTD) Ser2 kinase, and its activity is believed to promote productive elongation and coupled RNA processing. Here, we demonstrate that P-TEFb is critical for the transition of Pol II into a mature transcription elongation complex in vivo. Within 3 min following P-TEFb inhibition, most polymerases were restricted to within 150 bp of the transcription initiation site of the active Drosophila melanogaster Hsp70 gene, and live-cell imaging demonstrated that these polymerases were stably associated. Polymerases already productively elongating at the time of P-TEFb inhibition, however, proceeded with elongation in the absence of active P-TEFb and cleared from the Hsp70 gene. Strikingly, all transcription factors tested (P-TEFb, Spt5, Spt6, and TFIIS) and RNA-processing factor CstF50 exited the body of the gene with kinetics indistinguishable from that of Pol II. An analysis of the phosphorylation state of Pol II upon the inhibition of P-TEFb also revealed no detectable CTD Ser2 phosphatase activity upstream of the Hsp70 polyadenylation site. In the continued presence of P-TEFb inhibitor, Pol II levels across the gene eventually recovered.
RNA polymerase II (Pol II) undergoes a series of critical modifications as it progresses through a cycle of transcription. The carboxyl-terminal domain (CTD) of the largest subunit of Pol II, which consists of tandem heptapeptide repeats with the consensus sequence Y1S2P3T4S5P6S7, is a major target for these modifications, which include phosphorylation (primarily of serines 2 and 5 of the consensus repeat), proline cis/trans isomerization, and glycosylation (36). Distinct phosphorylation marks are associated with different transcription stages: in metazoans, Pol II enters the preinitiation complex in a hypophosphorylated state; it progresses from the initiation site to promoter-proximal regions, where it is phosphorylated primarily at Ser5; it matures into a doubly (Ser2 and Ser5) phosphorylated, productively elongating Pol II; and upon passing the polyadenylation site, it displays reduced phosphorylation (7, 20, 30). These phosphorylation states allow Pol II to interact with distinct repertoires of proteins at different stages of the transcription cycle and are critical in the coordination of transcription and coupled pre-mRNA processing (44, 46).
The Cdk7 and Cdk9 kinases are responsible for most CTD serine phosphorylation in metazoans (39). Cdk7, a subunit of the general transcription factor TFIIH, phosphorylates the CTD preferentially at Ser5 (47, 53, 60). By contrast, Cdk9, which together with cyclin T composes the P-TEFb kinase complex (42), phosphorylates the CTD predominantly at Ser2 (39, 47, 57, 71). P-TEFb and Cdk7 both have positive roles in transcription and function at postinitiation steps (3, 22, 33, 56).
P-TEFb recruitment to an activated gene is concomitant with the efficient release of Pol II from a promoter-proximally paused to a productively elongating form (7). The promoter-proximally paused polymerase has initiated transcription and is associated with a short nascent RNA but remains just downstream of the promoter until appropriate cues fuel its release into productive elongation (49, 52). Promoter-proximal pausing of Pol II at several higher-eukaryote genes, including Drosophila melanogaster Hsp70 and mammalian c-myc and c-fos, has been well-characterized previously (10, 12, 27, 45, 48, 52, 59) and may provide a general checkpoint prior to productive elongation (reviewed in reference 54). P-TEFb is known to phosphorylate two complexes, in addition to Pol II, that are proposed to be required for promoter-proximal pausing: DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) sensitivity-inducing factor (DSIF) and negative elongation factor (17, 25, 65; reviewed in reference 54). The kinase activity of P-TEFb on these factors, as well as on the CTD of Pol II at Ser2, is thought to be required for the escape of Pol II from the pause region (17, 25, 32, 57, 62, 66). Indeed, the attenuation of P-TEFb activity can reduce the level of Pol II in downstream regions of fission yeast or human immunodeficiency virus type 1 genes, relative to Pol II levels at the 5′ end (15, 21). The recruitment of P-TEFb does appear to be a rate-limiting step for transcription: the tethering of Cdk9 or cyclin T to promoters activates gene expression, dependent on a functional P-TEFb kinase complex (5, 29, 31), and the inhibition of P-TEFb reduces total cellular gene expression in human cells (11). P-TEFb is also an essential cofactor for human immunodeficiency virus type 1 transcription (72). For some genes, however, the greatest effect of P-TEFb inhibition appears to be faulty pre-mRNA processing, and transcription defects are not always apparent under certain conditions (35, 39). Pre-mRNA processing is intimately linked with transcription and is dependent upon the Pol II CTD (44). Many members of the pre-mRNA-processing machinery are known to interact physically with the CTD of Pol II (44), and studies of Saccharomyces cerevisiae and higher eukaryotes indicate that the phosphorylated CTD stimulates RNA-processing events (23, 24, 51, 63).
Flavopiridol (FP) is a potent and highly specific inhibitor of P-TEFb kinase activity (11, 39). We have previously examined the effect of fairly long FP treatments (∼20 min) on the transcription of the D. melanogaster heat shock gene Hsp70 (39). Given that P-TEFb is rapidly recruited to and tracks with Pol II along activated Hsp70 (7), we were surprised to find that these long FP treatments had only minor effects on Hsp70 transcription, although there were clear defects in pre-mRNA 3′-end processing (39). In this study, we examined the immediate effects of FP treatment on activated Hsp70 transcription and, in doing so, provided direct evidence for the necessity of P-TEFb activity in vivo in the transition of Pol II into a fully competent elongation complex. We found that, in the first few minutes following FP addition, elongating Pol II and its associated elongation factors cleared from the induced Hsp70 gene but that, strikingly, the level of Pol II at the 5′ end of the gene, corresponding to initiating and promoter-proximally paused polymerases, remained largely unaffected. We exploited this observation to map the positions of the promoter-proximally stalled polymerases under activating conditions and showed that these early elongation complexes stably associated with the gene. The rapid kinetics of FP-mediated P-TEFb inhibition also allowed an assessment of Ser2-specific CTD phosphatase activity during transcription elongation in vivo and the identification of the gene region where CTD phosphatases act. Interestingly, with time after FP addition, cells can at least partially overcome the FP-induced effects on transcription, in that Pol II association with the main body of the Hsp70 gene recovers to near normal levels. The latter observation explains why only a modest Pol II elongation defect was observed following the longer FP treatments in our prior study (39).
MATERIALS AND METHODS
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described by Boehm et al. (2003) (7), with modifications: D. melanogaster Kc cells were grown in SFX-Insect serum-free medium (HyClone). Cells were heat shocked by raising the culture temperature to 36.5°C in a 42°C water bath for 1 to 2 min before transferring the flask to a 36.5°C water bath for the remainder of the heat shock. Cells were cross-linked with 1% formaldehyde for 1 min (see Fig. 1 and 2). Alternatively, a volume of 48°C medium equal to the volume of the cell suspension was added to the cells for an instantaneous heat shock, followed by incubation at 36.5°C for the appropriate time. A volume equal to that of the original cell culture of 4°C medium was added immediately prior to cross-linking by the addition of 16% paraformaldehyde (Electron Microscopy Sciences) to a final concentration of 1% (see Fig. 3). Sonication was performed as described by Adelman et al. (2005) (1). Extract from 2.5 × 106 cells was diluted 20-fold in ChIP dilution buffer for each immunoprecipitation. Three low-salt, three high-salt, two LiCl, and two Tris-EDTA washes were performed. The amount of antibody per immunoprecipitation was as follows: for total Pol II, 4 μl of rabbit anti-Rpb3 (1); for Ser5-phosphorylated Pol II, 3 μl of H14 (Covance); for Ser2-phosphorylated Pol II, 6 μl of H5 (Covance); for P-TEFb, 2 μl of rabbit anti-cyclin T (a gift from David H. Price) (29); and for other factors, 2 μl of guinea pig anti-Spt5 (4), 3 μl of guinea pig anti-TFIIS (1), 4 μl of guinea pig anti-Spt6 (4), and 4 μl of guinea pig anti-CstF50 (in-house preparation).
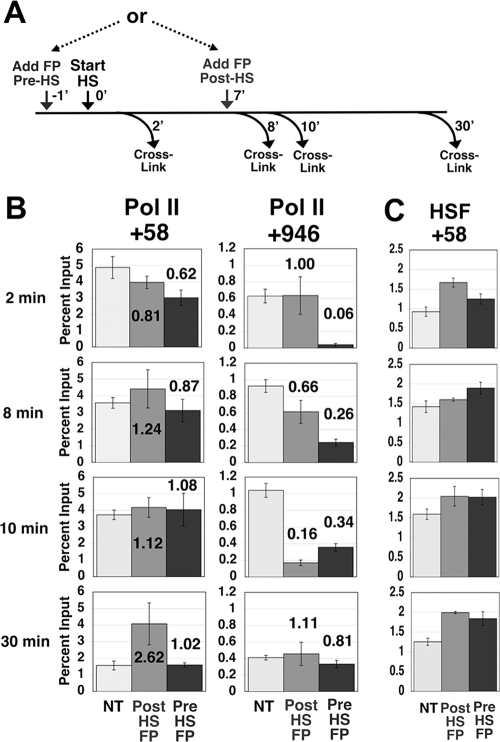
FIG. 1.
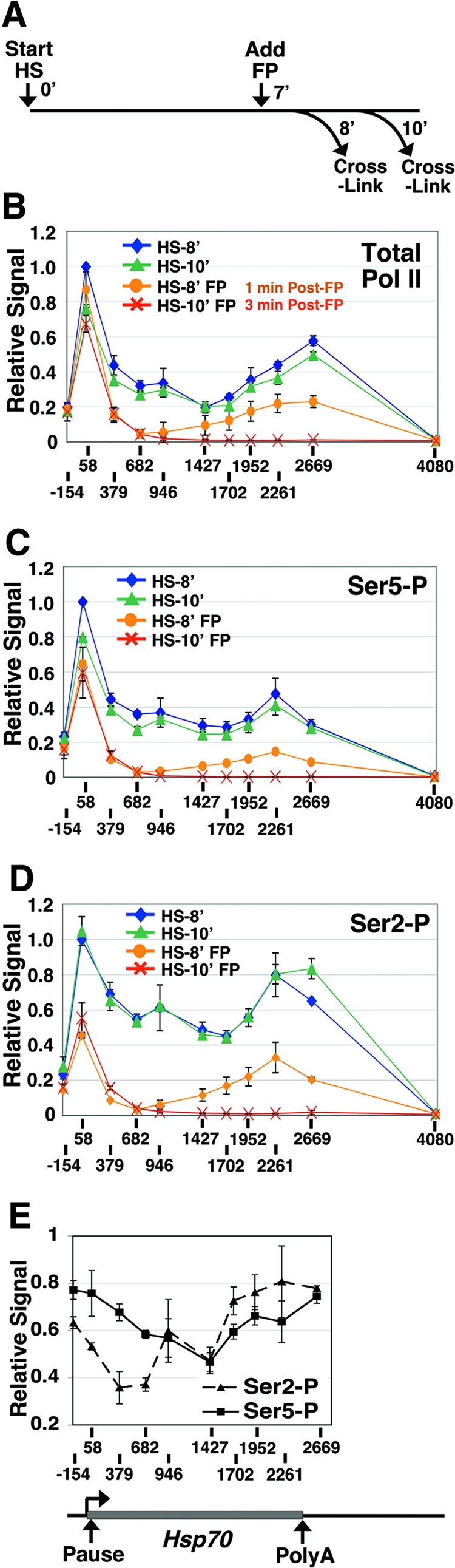
Depletion of elongating Pol II from Hsp70 immediately after P-TEFb inhibition. (A) Experimental scheme depicting the time after gene activation (start heat shock [HS]), FP addition (add FP), and formaldehyde cross-linking (cross-link). 0′, 7′, 8′, and 10′ represent time zero and 7-, 8-, and 10-min time points, respectively. (B to D) Levels of total Pol II (detected with an antibody to the Rpb3 subunit) (B) and Ser5 (C)- and Ser2 (D)-phosphorylated Pol II across the Hsp70 gene as detected by ChIP after 8 (HS-8′) or 10 (HS-10′) min of gene activation and in the presence or absence of FP administered at 7 min of gene activation. The x axis gives the position relative to the Hsp70 transcription start site (+1). The level of input material immunoprecipitated is plotted relative to the input value for HS-8′ at +58, which is set to 1. Vertical bars represent the ranges in results from two independent experiments. Ser5-P and Ser2-P represent Ser5- and Ser2-phosphorylated Pol II. (E) Relative levels of Ser2- and Ser5-phosphorylated Pol II at 1 min after FP addition. The input values for Ser2- and Ser5-phosphorylated Pol II were normalized to total Pol II, and values for the FP-treated samples (HS-8′ FP) were then divided by the respective values for the untreated samples (HS-8′). Error bars represent the square root of the sum of the squares of the errors associated with the numerator and denominator of the ratio. In all panels, the lines connect points within a data series and do not necessarily imply levels between data points. A schematic of Hsp70 is shown below the chart in panel E.
FIG. 2.
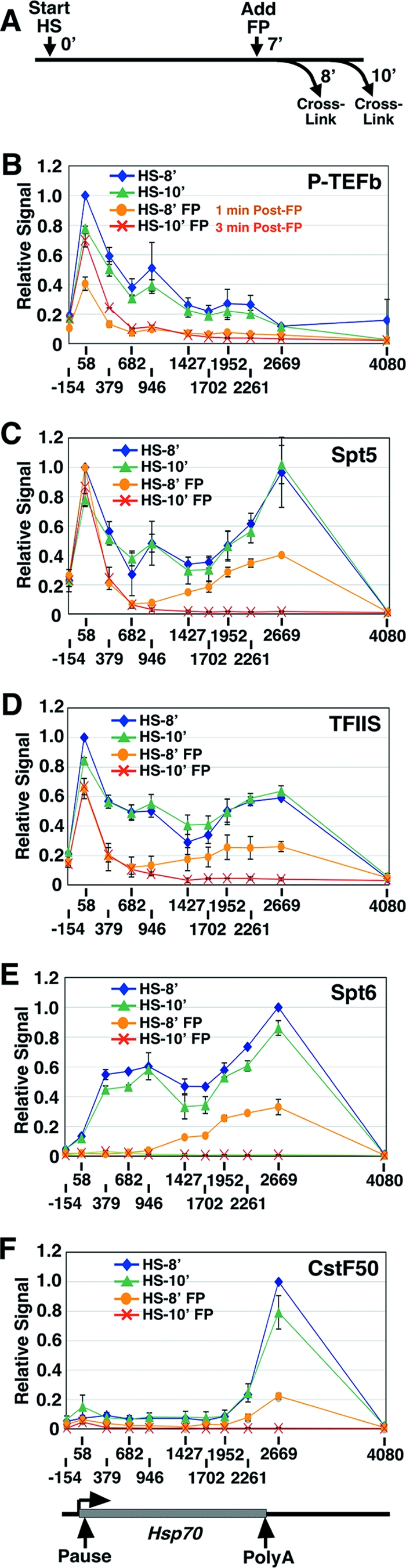
Elongation and RNA-processing factors clear from Hsp70 with kinetics matching that of Pol II. (A) Experimental scheme depicting the time after gene activation (start heat shock [HS]), FP addition (add FP), and formaldehyde cross-linking (cross-link). 0′, 7′, 8′, and 10′ represent time zero and 7-, 8-, and 10-min time points. (B to F) Levels of P-TEFb (B), Spt5 (C), TFIIS (D), Spt6 (E), and CstF50 (F) across the Hsp70 gene as detected by ChIP after 8 (HS-8′) or 10 (HS-10′) min of gene activation and in the presence or absence of FP administered at 7 min of gene activation. A schematic of Hsp70 is shown below the chart in panel F. The input value for HS-8′ at the peak of factor association (+58 for panels B, C, and D and +2669 for panels E and F) is set to 1. Vertical bars represent the ranges of results from two independent experiments. The lines connect points within a data series and do not necessarily imply levels between data points.
FIG. 3.
Pol II levels recover with time after FP addition. (A) Experimental scheme depicting the time of FP addition (add FP) and formaldehyde cross-linking (cross-link) in relation to the start of gene activation (start heat shock [HS]). −1′, 0′, 7′, 8′, 10′, and 30′ represent 1 min pre-heat shock, time zero, and 7-, 8-, 10-, and 30-min time points. (B and C) Levels of input DNA immunoprecipitated by an antibody against the Rpb3 subunit of Pol II (total Pol II) (B) or HSF (C) in the +58 (B, left graphs, and C) or +946 (B, right graphs) region of Hsp70 after 2, 8, 10, and 30 min of gene activation and in the absence (no treatment [NT]; light gray bars) or the presence of FP administered at 7 min after gene activation (post-HS-FP; dark gray bars) or 1 min before gene activation (pre-HS-FP; black bars). The numbers on the graphs in panel B represent the amount of change (n-fold) in the Pol II signal after FP treatment. Standard error bars for results from three independent experiments are shown.
Quantitative real-time PCR analysis.
Real-time PCR was performed using SYBR green I (Cambrex) PCR mix (0.8% glycerol, 0.006 M Tris [pH 8], 0.025 M KCl, 0.0025 M MgCl2, 0.075 M trehalose, 0.1% Tween, 0.1 mg of nonacetylated ultrapure bovine serum albumin/ml, 0.067× SYBR green I, 0.1 mM deoxynucleoside triphosphates, 50 U of Taq DNA polymerase/ml) on an Opticon thermocycler and a continuous fluorescence detector (MJ Research) or an ABI Prism 7900 sequence detection system (Applied Biosystems). A standard curve was derived from the analysis of serially diluted input material; all values were obtained in the linear range of amplification. The oligonucleotides used were as follows: Hsp70 −154 F, 5′-GCCAGAAAGAAAACTCGAGAAA, and Hsp70 −154 R, 5′-GACAGAGTGAGAGAGCAATAGTACAGAGA; Hsp70 +58 F, 5′-CAATTCAAACAAGCAAAGTGAACAC, and Hsp70 +58 R, 5′-TGATTCACTTTAACTTGCACTTTA; Hsp70 +379 F, 5′-CACCACGCCGTCCTACGT, and Hsp70 +379 R, 5′-GGTTCATGGCCACCTGGTT; Hsp70 +682 F, 5′-ATATCTGGGCGAGAGCATCACA, and Hsp70 +682 R, 5′-GTAGCCTGGCGCTGGGAGTC; Hsp70 +946 F, 5′-CATCGACGAGGGATCTCTGTTC, and Hsp70 +946 R, 5′-GGCGCGAGGGTTGGA; Hsp70 +1427 F, 5′-CTGTGCAGGCCGCTATCC, and Hsp70 +1427 R, 5′-GCGCTCGATCAGCTTGGT; Hsp70 +1702 F, 5′-GGGTGTGCCCCAGATAGAAG, and Hsp70 +1702 R, 5′-TGTCGTTCTTGATCGTGATGTTC; Hsp70Ab +1952 F, 5′-TGGACGAGGCTGACAAGAACT, and Hsp70Ab +1952 R, 5′-ACCGGATAGTGTCGTTGCACTT; Hsp70Ab +2261 F, 5′-TGTTCATCAATGGGTTATAACATATGGGTT, and Hsp70Ab +2261 R, 5′-AAGACTTGGTAATTAGGTAATACTATTGTT; Hsp70Ab +2669 F, 5′-TCGCAGACACCGCATTTGT, and Hsp70Ab +2669 R, 5′-ACCAATTGCAACAGAGACTGGAA; and Hsp70Ab +4080 F, 5′-TGGAAACTGCCTCCAACAACTG, and Hsp70Ab +4080 R, 5′-AGACGCACGAGACCAATCTGTA. The oligonucleotide names refer to the central bases of the resulting amplicons.
Terminated nuclear run-on assays.
After treatment (with or without FP and with or without instantaneous heat shock), Kc cells were rapidly cooled by the addition of treated cells to 150 ml of ice-cold medium. Cells were washed and resuspended in buffer A (10 mM Tris-Cl [pH 8.0], 300 mM sucrose, 3 mM CaCl2, 2 mM MgAc2, 0.1% Triton X-100, 0.5 mM dithiothreitol) at 108 cells/ml. Nuclei were extracted, washed with buffer A, and resuspended in buffer D (50 mM Tris-Cl [pH 8.0], 25% glycerol, 5 mM MgAc2, 0.1 mM EDTA, 5 mM dithiothreitol) (108 cells/100 μl). Nuclei were mixed with 115 μl of run-on buffer (7.2 mM Tris-Cl [pH 8.0], 4.4 mM MgAc2, 1.4 mM MnCl2, 150 mM KCl, 3 mM ATP, 3 mM CTP, 0.4 mM 3′-deoxy-GTP [TriLink BioTechnologies], 100 μCi of [32P]UTP [3,000 Ci/mmol], 40 U of Superase-In [Ambion], 0.6% N-lauroyl-sarcosine). Run-on assays proceeded for 10 min at room temperature before a 30-min RQ1 DNase digestion at 37°C, followed by the addition of 235 μl of stop solution (20 mM Tris-Cl [pH 7.4], 2% sodium dodecyl sulfate, 10 mM EDTA) and 40 μl of RNA-grade proteinase K and incubation at 55°C for 30 min. Purified RNAs were hybridized to biotinylated probe as described by Rasmussen and Lis (1993) (49) and were captured with streptavidin-coated magnetic beads (300 μg; Dynal Biotech). Beads were washed four times with wash buffer (10 mM Tris-Cl [pH 7.5], 50 mM NaCl, 5 mM EDTA, 0.5 mg of yeast tRNA/ml) and resuspended in 6 μl of gel-loading buffer II (Ambion). RNAs were separated by electrophoresis through 8% acrylamide-7 M urea sequencing gels. To calculate the 1.5- to 2.5-fold increase in the number of Pol II molecules on the promoter-proximal region from the noninducing conditions to the conditions of heat shock plus FP treatment, we estimated (from our previously published analysis [49]) that this pair of labeled and chain-terminating nucleotides would detect 35% of paused Pol II in non-heat-shocked cells and 55% of the Pol II (if randomly distributed) through the first 120 bases of Hsp70.
Imaging of live salivary-gland nuclei.
The fly line expressing an enhanced green fluorescent protein-tagged Rpb3 under the control of the Gal4 upstream activation sequence with Gal4 expressed speifically in the salivary glands (C147-Gal4), the preparation of salivary glands, and methods of multiphoton microscopy imaging and fluorescence recovery after photobleaching (FRAP) have been described previously (69). The tissue temperature was measured by a 100-μm thermoprobe (Physitemp Th5). FP (500 nM) was perfused into the FCS2 chamber (Bioptechs) at the indicated time point after heat shock (see Fig. 5A). The Rpb3-enhanced green fluorescent protein intensities within the 87A and 87C puffs during FRAP were quantified as described by Phair et al. (2004) (43). Unintentional photobleaching during the FRAP experiment as a consequence of imaging has been observed to be within 10% of total intensities and was corrected in the FRAP intensity plots during normalization (but may appear on the raw images shown) (see Fig. 5B).
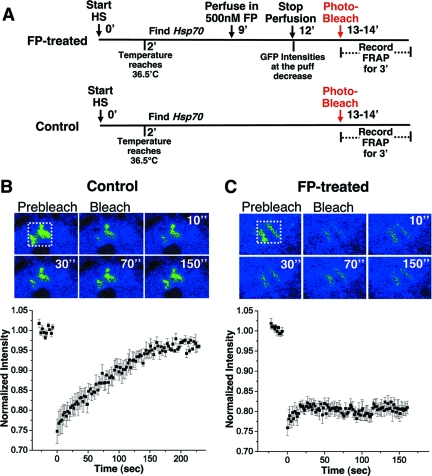
FIG. 5.
The 5′-end-restricted polymerases stably associate with activated Hsp70 following FP treatment. (A) Illustration of the experimental scheme. Numbers indicate time points. HS, heat shock. (B and C) FRAP experiment with live polytene nuclei expressing GFP-tagged Pol II in the absence (B) or presence (C) of FP. Representative images of the Hsp70 loci before photobleaching (prebleach), immediately after photobleaching (bleach), and then at 10, 30, 70, and 150 s after photobleaching (top panels) are shown. Photobleached regions are bordered with dotted white lines. Distinct appearances of Hsp70 loci from the optical sections in images in panels B and C are due to different orientations of the two adjacent heat shock puffs at 87A and 87C. The intensity plots (bottom panels) quantify the FRAP (panel B, n = 5; panel C, n = 4). Error bars are shown.
RESULTS
P-TEFb inhibition rapidly depletes elongating Pol II from activated Hsp70.
During a heat shock response, the D. melanogaster heat shock gene Hsp70 is fully occupied by transcriptionally engaged Pol II molecules. These molecules include paused polymerases in the promoter-proximal region (18), as well as productive elongation complexes throughout the transcription unit (40). The P-TEFb kinase is critical for Ser2 phosphorylation of the CTD (39), a step that occurs when promoter-proximally paused Pol II enters into productive elongation (7). To examine the immediate effects of P-TEFb inhibition on Hsp70 transcription, we treated D. melanogaster cells with FP, a highly specific P-TEFb kinase inhibitor (11, 39), during active transcription and examined the effects on Pol II levels—by using formaldehyde cross-linking and ChIP—just 1 and 3 min later. In treated cells, FP was added to a final concentration of 500 nM at 7 min after gene activation; the level of Pol II on Hsp70 was then assessed at 8 and 10 min after gene activation (Fig. 1A). The concentration of FP used was equivalent to the approximate concentration of P-TEFb in the nucleus and was sufficient to strongly inhibit Ser2 phosphorylation in vivo (39).
In the absence of FP, Pol II was detected at high levels across the Hsp70 transcription unit (Fig. 1B, HS-8′ and HS-10′). Notably, there were two peaks of Pol II association with the gene. The first, 5′-end peak corresponded to initiating and promoter-proximally paused polymerases (with some contribution of early productively elongating Pol II), as has been observed previously (7), and the second, 3′-end peak was located just after the polyadenylation site, which is around +2400 relative to the transcription start site at +1. The 5′ peak was consistent with the idea that release from the paused polymerase continues to be rate limiting for transcription even after gene induction (18, 41). The 3′ peak was consistent with the slowing down of polymerases in this region during 3′-end processing of the RNA and transcription termination (46).
One minute after the addition of FP, the level of Pol II detected in the body of Hsp70 was reduced most dramatically in the regions detected by the +379, +682, and +946 probes (Fig. 1B). In contrast, Pol II levels at +58 appeared to be relatively unaffected. The peak of polymerase remaining on the 3′ half of the transcription unit likely represents the last wave of productively elongating Pol II as it cleared from the gene. The capture of polymerases still in the transcription unit at this time point (1 min after FP addition) was expected given that the in vivo transcription rate of Pol II is ∼1.2 kb/min (40). By just 3 min after FP treatment, the polymerases were absent from the middle and downstream regions of the gene, although the 5′ peak was largely unaffected (Fig. 1B). These data demonstrate that P-TEFb activity is required in vivo for the transition of Pol II from the paused to the productively elongating state but that it is not required for polymerases that are already productively elongating to complete the transcription of the gene, and these findings substantiate previous analyses (14, 33, 34).
Levels of phosphorylated Ser2 in the 5′ region are reduced.
We also examined the phosphorylation status of the CTD of Pol II before and after FP treatment. As seen previously, in the absence of FP and under inducing conditions, the level of Ser2 phosphorylation remained high through the gene, whereas the detection of Pol II phosphorylated at Ser5 decreased slightly toward the 3′ end in proportion to the decrease in total Pol II levels (Fig. 1B to D) (7). In agreement with results from the human p21 gene (20), the levels of Ser2- and Ser5-phosphorylated polymerases relative to total Pol II levels were reduced beyond the polyadenylation site, indicating that CTD phosphatase activity was present, at least in this region of Hsp70.
Following FP treatment, the relative level of Ser5 phosphorylation appeared to be largely unaffected. The level of Ser5-phosphorylated Pol II closely resembled that of total Pol II (Fig. 1B and C). This result is consistent with the Ser2 residue being the preferential CTD target of P-TEFb (39, 47, 57, 71).
After a 1-min FP treatment, the polymerases that were still elongating through the gene were also still phosphorylated at Ser2 (Fig. 1D). This result was expected if P-TEFb activity was inhibited subsequent to its action on these polymerases, a likely situation given the short time frame after FP addition and the observation that a substantial fraction of Ser2 residues are phosphorylated in the 5′ region (7) (Fig. 1D). This finding also suggests that there was minimal Ser2 phosphatase activity on these polymerases, at least before the polyadenylation site, and is consistent with polymerases’ not requiring continual phosphorylation by P-TEFb to proceed with transcription elongation. Finally, following a 1- or 3-min FP treatment, the level of Ser2 phosphorylation was dramatically reduced in the region corresponding to paused polymerases (+58) whereas Ser5 phosphorylation levels were only modestly affected (Fig. 1C and D). A comparison of relative Ser2 and Ser5 phosphorylation levels clearly showed that there was a reduction in Ser2 phosphorylation at the 5′ end of the gene 1 min after FP treatment but that Ser2 levels in the middle and 3′ regions of the gene remained high (Fig. 1E). Importantly, the reduction in Ser2 phosphorylation in the 5′ region correlated with the reduced escape of these polymerases from the promoter-proximal region (Fig. 1B and D).
P-TEFb levels across Hsp70 are reduced immediately following FP treatment.
We also examined the distribution of P-TEFb across the Hsp70 gene before and after FP treatment (Fig. 2A and B). As observed previously, in the absence of FP, P-TEFb was present at the 5′ end and tracked with Pol II through to the 3′ region of the activated gene (Fig. 2B) (7). Interestingly, expanding the analysis in the present study to include P-TEFb levels up to and beyond the end of the mRNA-encoding unit revealed that P-TEFb dissociated from the gene close to the polyadenylation site. This dissociation of P-TEFb preceded the reduction in phosphorylated-Ser2 levels past the polyadenylation site (Fig. 1D) and may be important for transcription termination and the dissociation of the polymerase from the template DNA.
Following 1 min of FP treatment, a marked reduction in P-TEFb levels across the gene was observed (Fig. 2B). This result correlates with the reduced levels of Ser2-phosphorylated Pol II in the 5′ region of Hsp70 at this time point (Fig. 2B). Surprisingly, by 3 min after FP treatment, the level of P-TEFb associated with the gene had partially recovered: P-TEFb was detected in the 5′ region at near pre-FP levels but was still absent from the rest of the gene, where Pol II was also absent (Fig. 2B). This pattern may be the beginning of an adaptive response (see below and Discussion).
Association of elongation and RNA-processing factors with Hsp70 is dependent on Pol II.
The depletion of elongating Pol II from the middle and the 3′ end of Hsp70 following a 3-min FP treatment provided a unique opportunity to assess the dependence of several transcription factors on the presence of Pol II for their association with the gene. We examined the distribution of members from several classes of elongation factors along Hsp70 in the absence or presence of FP. Spt5 is a subunit of the bipartite DSIF complex, which is thought to be required for promoter-proximal pausing (54, 61), whereas TFIIS prevents transcription arrest by stimulating nascent-transcript cleavage (13) and is required for the efficient release of Pol II from the promoter-proximal pause site (1). In the absence of FP, Spt5 and TFIIS are present in the promoter-proximal region and track with Pol II through the gene (Fig. 2C and D) (1, 4). By contrast, Spt6, a factor involved in nucleosome disassembly and reassembly (54), is absent from the promoter-proximal region, which is free of nucleosomes, and instead localizes to the middle and downstream regions of Hsp70 during activated transcription (55) (Fig. 2E). CstF50 is a subunit of the 3′ RNA-processing complex cleavage stimulation factor (CstF) (46), and the peak of CstF50 association was at the 3′ end of the active Hsp70 gene (Fig. 2F).
All factors cleared from the body of the Hsp70 gene following FP treatment with kinetics that paralleled that of Pol II (Fig. 2). This result appears to be specific to elongation and downstream-acting factors: the level of the activator heat shock factor (HSF) was relatively unchanged by FP addition (data not shown). Interestingly, levels of factors known to associate with the promoter-proximal paused polymerase (Spt5 and TFIIS) (1, 4) remained high in the promoter-proximal region, even when depleted from the middle and 3′ regions of the gene.
Pol II levels across Hsp70 recover with time.
The results presented in Fig. 1 were somewhat surprising given that Pol II does not display dramatic elongation defects at Hsp70 following a 20-min FP treatment that is administered before heat shock (39). The more dramatic decrease in Pol II levels on the Hsp70 gene observed in the present study may be a consequence of administering the FP in the middle of a heat shock treatment, instead of before heat shock, or it may be the result of examining the Pol II levels after a short (3-min) FP treatment rather than a long one (20 min), during which cells may have time to invoke compensatory mechanisms that reverse some of the effects of the drug. To address this question, we again utilized ChIP to monitor Pol II levels and we expanded the number of time points examined. We treated the cells with FP at 7 min after, or 1 min prior to, gene activation with an instantaneous heat shock (Fig. 3A). We chose to focus on two regions of the Hsp70 gene: the region corresponding to the paused polymerase at +58, where little effect following FP treatment is observed, and a region in the middle of the Hsp70 transcription unit, at +946, where large differences in the levels of Pol II in untreated and FP-treated samples were observed (Fig. 1). For each individual experiment, samples at all time points were taken from the same starting population of heat-shocked cells.
In the absence of FP, the levels of polymerase observed at the +58 and +946 regions at each of the heat shock time points agreed with previous data (Fig. 3B) (7). The reduced Pol II level at 30 min after gene induction was consistent with the eventual attenuation of Hsp70 gene expression by ∼50% under continued thermal stress (37). For the post-heat shock-FP treatment data set (Fig. 3B), the level of Pol II at the 2-min heat shock time point, which occurred before drug addition, was unaffected, as expected. However, 1 min after FP addition, at the 8-min heat shock time point, there was a decrease in the level of Pol II at the +946 region while the level in the +58 region was relatively unchanged. By 3 min after FP addition (10 min after heat shock), Pol II levels at the +946 region were dramatically reduced while those at the +58 region remained unaffected. Twenty minutes later, 30 min after heat shock, the level of Pol II at the +946 region was similar to that observed in the untreated sample.
When FP was added 1 min before heat shock, the level of polymerase at the +58 region was relatively unaffected at all time points (Fig. 3B, left panels). Conversely, the level of polymerase at the +946 region was dramatically reduced 3 min after the addition of FP (after 2 min of heat shock) (Fig. 3B, right panels). Strikingly, the level of Pol II at the +946 region increased with time after FP treatment until the 30-min time point, when nearly equivalent levels of polymerase in the untreated and FP-treated samples were observed (Fig. 3B). Thus, these data show that Pol II levels on the body of the Hsp70 gene were reduced immediately after FP treatment but recovered within the time course of 10 to 30 min posttreatment.
The levels of the Hsp70 activator HSF at the +58 region were also determined as a control. As can be seen in Fig. 3C, HSF levels were relatively unaffected by FP addition, as expected.
Overall, this analysis is consistent with our previous data (39) in that long (≥20-min) 500 nM FP treatments do not reveal major Pol II elongation defects at the D. melanogaster Hsp70 gene. This result also indicates that cells compensate for the FP-mediated inhibition of Pol II maturation into the productively elongating form, or that the FP is inactivated, in the course of minutes.
Polymerases remaining on Hsp70 after FP treatment are restricted to the promoter-proximal region.
To ascertain precisely the location of Pol II molecules on Hsp70 following a short FP treatment, we sized the nascent transcripts by using terminated nuclear run-on assays (49). In the run-on assays, engaged polymerases from isolated nuclei were stimulated, in the presence of Sarkosyl, to proceed with transcription (run on) by the addition of nucleoside triphosphates. The Sarkosyl prevents new transcription initiation events and washes away many transcriptional impediments to Pol II. In the experiment presented in Fig. 4, the UTP was radiolabeled and the GTP was replaced with a chain-terminating GTP analog. Therefore, the polymerases could only run on to the next site of GTP incorporation, and after the isolation of the run-on transcripts from the Hsp70 gene, the position of the polymerases could be determined within several nucleotides.
FIG. 4.
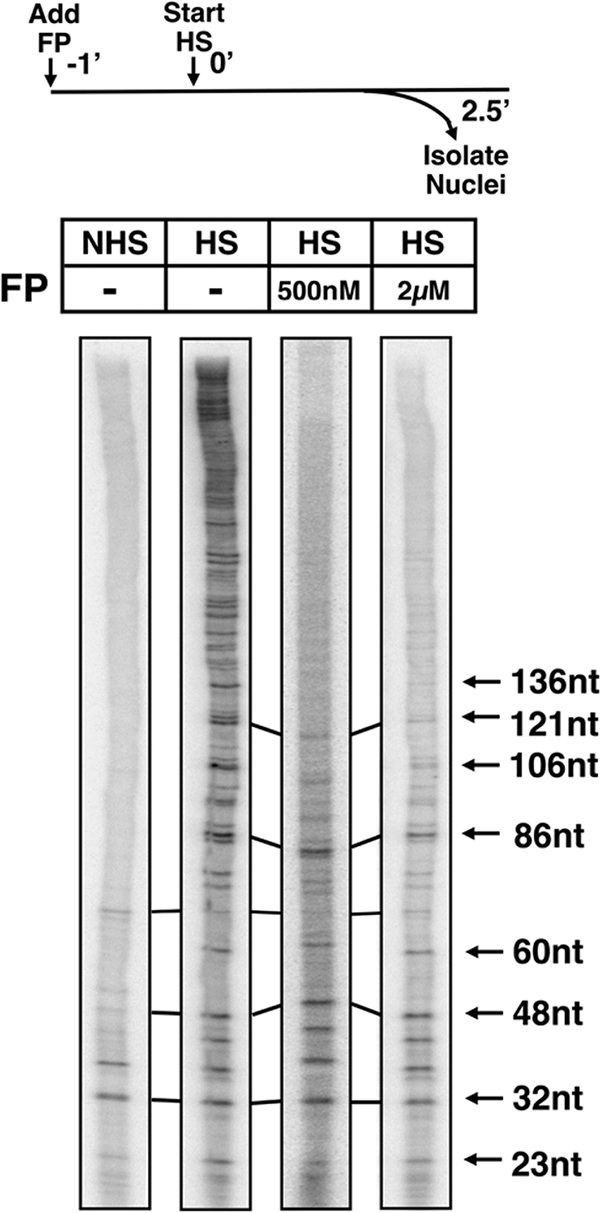
Polymerases remaining on Hsp70 3.5 min post-FP treatment are promoter proximally restricted. Terminating nuclear run-on assays were performed under noninducing conditions (no heat shock [NHS]) or after 2.5 min of gene activation (heat shock [HS]) and in the absence (−) or presence of 500 nM or 2 μM FP added 1 min before heat shock. The experimental scheme is illustrated (top panel). The sizes of the Hsp70 RNA transcripts are indicated. nt, nucleotides; −1′, 0′, and 2.5′ represent 1 min pre-heat shock, time zero, and 2.5-min time point.
We first performed a terminated nuclear run-on assay under non-heat shock conditions to reexamine the distribution of Pol II at the uninduced Hsp70 gene. Under noninducing conditions, a polymerase is located in the 5′ region of Hsp70 in an engaged but paused state (18, 52). As observed previously (49), the paused polymerases under noninducing conditions map from +21 to +35 nucleotides relative to the Hsp70 transcription start site (Fig. 4, NHS).
This same analysis was then performed during active Hsp70 transcription, following 2.5 min of heat shock induction. This early heat shock time point was chosen so that the Hsp70 RNA levels would not yet be high enough to saturate the transcript isolation step of the experiment. As can be seen in Fig. 4, transcripts in a range of sizes were isolated from the induced gene, indicative of the engagement of polymerases in transcription throughout the Hsp70 transcription unit. Next, we examined the location of polymerases at the 2.5-min heat shock time point following the addition of 500 nM or 2 μM FP 1 min before heat shock. In the presence of both concentrations of FP, short transcripts (up to ∼90 nucleotides) with intensities comparable to those under the untreated heat shock conditions were detected; however, the detection of nascent transcripts longer than 90 nucleotides rapidly and progressively decreased. This result indicates that polymerases do not extend efficiently into the gene following P-TEFb inhibition and that RNAs rarely reach a length of more than 150 nucleotides (Fig. 4). The quantification of the run-on signal from four independent experiments revealed a 1.5- to 2.5-fold increase in the number of polymerases on the promoter-proximal region (+1 to +120) under inducing conditions and in the presence of FP relative to the number present under noninducing conditions. One polymerase occupies the gene prior to induction (19); therefore, approximately two polymerases occupy the promoter-proximal region under inducing conditions. This value corresponds well with the reproducible increase in the ChIP signal in the pause region observed upon gene activation (1, 7).
Overall, we conclude that a large fraction of Pol II remaining on the Hsp70 gene after a short FP treatment was engaged and promoter proximal. The polymerases on the activated gene occupy more sites in the promoter-proximal region in the presence of FP than do polymerases on the uninduced gene, where only one paused Pol II molecule is present (19). More polymerases are rapidly recruited to Hsp70 upon gene activation (7), and additional locations may serve as promoter-proximal pause sites under inducing conditions. Nonetheless, these promoter-proximal polymerases on the induced Hsp70 gene appear to be unable to escape efficiently into productive elongation without P-TEFb activity.
The promoter-proximal polymerases are stably associated with the gene.
We next decided to characterize the dynamic properties of the polymerases remaining in the Hsp70 promoter-proximal region immediately following FP treatment. These polymerases might stably associate with the gene, or they might participate in a continuous cycle of initiation, stalling, and premature termination. To address the stability of these polymerases, we employed the technique of FRAP analysis of D. melanogaster salivary-gland tissue expressing a green fluorescent protein (GFP)-tagged Rpb3 subunit of Pol II (69). FRAP enables an analysis of the rate of exchange of Pol II on the Hsp70 gene. We activated Hsp70 transcription prior to FP addition, so that the Hsp70 loci could be rapidly identified on the polytene chromosomes by their unique and characteristic chromosomal decondensation (“puffing”) pattern and strong Pol II recruitment, which occurs upon Hsp70 induction (69). We exploited the observation that a short FP treatment clears productively elongating polymerases from the Hsp70 gene, leaving only those polymerases in the promoter-proximal region.
The experimental scheme for the FRAP experiment with or without FP is illustrated in Fig. 5A. We first examined the recovery kinetics of Pol II in heat-shocked salivary-gland tissue in the absence of FP: the recovery of GFP fluorescence was detected within 3 min (Fig. 5B), in agreement with known elongation rates (40). Next, the FRAP kinetics of GFP-Pol II after the perfusion of FP into the microscope chamber was monitored. Following perfusion, a reduced level of fluorescence was detected at the Hsp70 gene loci (compare images in Fig. 5B and C, prebleach), consistent with the reduced levels of polymerase on the gene following a short FP treatment (Fig. 1). In FP-treated nuclei, we observed no recovery of fluorescence during the 3-min time course (Fig. 5C). We conclude that the polymerases remaining on the 5′ end of Hsp70 after a short FP treatment were not exchanging with unbleached polymerases in the nucleoplasm and were therefore stably associated with the gene.
DISCUSSION
P-TEFb activity is critical for the maturation of Pol II into a productive elongation complex.
We have found that P-TEFb inhibition leads to a rapid depletion of elongating Pol II from the Hsp70 gene (Fig. 1). Furthermore, the majority of polymerases remaining on the gene are restricted to the 5′ end and are transcriptionally engaged (Fig. 4). Together, these data indicate that P-TEFb is critical for the escape of Pol II into productive elongation in vivo. We also found that upon P-TEFb inhibition, levels of all elongation and RNA-processing factors so far tested (P-TEFb, Spt5, TFIIS, Spt6, and CstF50) were dramatically reduced, with kinetics indistinguishable from the depletion of elongating Pol II (Fig. 2). Therefore, at least a subset of transcription factors appear to depend on the continual presence of elongating Pol II for their association with chromatin.
The dependence of P-TEFb, Spt5, TFIIS, and CstF50 on Pol II for association with the Hsp70 gene was not surprising. P-TEFb, Spt5, and TFIIS all interact with Pol II (26, 32, 55, 58) and track with Pol II during activated transcription (1, 4, 7). CstF50 physically interacts with the CTD of Pol II (16), and CTD Ser2 phosphorylation is required for the association of cleavage and polyadenylation factors in S. cerevisiae (2). However, Spt6 was not necessarily expected to require Pol II elongation for its association with the gene. Spt6 directly interacts with histones (8, 64) and separates somewhat from Pol II on Hsp70: Spt6 does not colocalize with promoter-proximal Pol II but does colocalize with Pol II on the body of the gene (55). It is possible that Spt6 interacts cooperatively with Pol II and nucleosomes or that Spt6 interacts only with the productively elongating, Ser2-phosphorylated form of Pol II. Consistent with the latter possibility, Spt6 was recently shown to interact directly with Ser2-phosphorylated Pol II (70). Interestingly, however, the interaction of Spt6 with Pol II is not required for the positive elongation activity of Spt6 in vivo (70). Future studies using live-cell-imaging techniques (69) should shed light on the dynamics and mechanism of Spt6 recruitment to chromatin during transcription.
Promoter-proximally restricted Pol II has limited turnover.
We have also obtained evidence for the stable association of promoter-proximally stalled polymerases with the Hsp70 gene. Following a short FP treatment, the only transcriptionally engaged polymerases remaining on the gene were promoter proximal (Fig. 4) and the usual rapid recovery of fluorescence after photobleaching of GFP-tagged polymerase was abolished (Fig. 5). This observation may also apply to the promoter-proximally paused polymerase on Hsp70 in uninduced cells (28) and in the absence of FP and argues against rapid cycles of initiation, pausing, and premature termination. In further support of a stable association, the two main protein complexes believed to promote promoter-proximal pausing, DSIF and negative elongation factor, repress Pol II elongation in vitro but do not induce premature termination (50, 67). Furthermore, the Spt5 subunit of DSIF was shown to cooperate with the Tat activator in preventing premature RNA dissociation from Pol II in an in vitro transcription assay (9). Both Spt5 and low levels of the activator HSF are present at the Hsp70 gene under noninducing conditions (4, 7). We consider it unlikely, but cannot rule out, that the FP prevents polymerases from prematurely terminating. It is extremely unlikely that FP prevents new polymerases from initiating, since FP is a kinase inhibitor and initiating polymerases are most favorably unphosphorylated (30). The definitive test of the status of paused Pol II in untreated cells awaits additional technological advances that allow rapid mapping of the paused Pol II associated with uninduced, unpuffed loci.
P-TEFb is not required for productively transcribing Pol II.
Consistent with previous analyses (14, 34), we have found that active P-TEFb kinase activity is not required for Pol II that is already productively elongating to continue to do so: polymerases already elongating at the time of P-TEFb inhibition cleared from the middle and downstream regions of the gene (Fig. 1). Furthermore, these elongating polymerases still had substantial levels of Ser2 phosphorylation after FP addition, indicating that there was minimal Ser2 phosphatase activity upstream of the polyadenylation site.
Cells overcome the FP-induced inhibition of transcription elongation with time.
In the course of this study, we also found that Pol II levels across Hsp70 recovered with time after FP treatment (Fig. 3). This discovery explains why only a small reduction in Pol II density on heat shock genes was detected after 20 min of FP treatment in our previous work (39). In that work, we concluded that the major defect in Hsp70 expression following P-TEFb inhibition was at the level of 3′-end processing of RNA (39). We now show that there is a Pol II elongation defect immediately following P-TEFb inhibition. While the persistent mRNA-processing defects likely account for a significant reduction in Hsp70 mRNA levels following a long FP treatment (39), the initial elongation defect demonstrated here may also contribute to the low mRNA levels. Our previous work, in addition to demonstrating that severe mRNA-processing defects exist following P-TEFb inhibition, also showed that in the recovered phase, Ser2 phosphorylation levels are still low (39). Therefore, elongation and RNA processing may require different extents or patterns of phosphorylation of the CTD or other targets of P-TEFb.
There are several possible mechanisms for the recovery of Pol II levels after FP treatment. The FP may become inactivated with time, metabolized, or expelled from the cell. Alternatively, an FP-resistant kinase may compensate for reduced P-TEFb kinase activity. It is also possible that a very small residual activity of P-TEFb persists in the presence of FP and eventually enables recovery. Another intriguing possibility is that cellular levels of active P-TEFb are increased in response to FP treatment. In mammalian cells, two P-TEFb complexes exist: a large, inactive complex and a small, active complex (38, 68). The treatment of mammalian cells with P-TEFb inhibitors promotes the release of P-TEFb from the large complex to increase the pool of active P-TEFb (6, 38, 68). If this same mechanism exists in Drosophila, following FP treatment, P-TEFb may be released from the complex that sequesters it in an inactive form to create a larger population of active P-TEFb molecules. Consistent with this possibility, P-TEFb levels on the 5′ end of Hsp70 recovered after 3 min of FP treatment, as shown at the 10-min time point post-FP treatment in Fig. 2B. We did not yet see the recovery in Pol II elongation at this 10-min time point, and so the recovery of P-TEFb levels at this time may indicate the beginning of the rescue of Pol II elongation ability, which then occurs gradually over the course of the next 20 min. While support for this explanation exists, we cannot rule out a combination of the above-described possibilities contributing to the recovery of Pol II elongation ability with time after FP treatment.
The data presented here also show that, at least for some drugs, it is important to examine cells during very early periods of treatment to observe the immediate effect of the drug. The short FP treatments used here now provide evidence that P-TEFb is indeed required for the escape of Pol II into productive elongation at the Hsp70 gene. Other cases in which an elongation defect is not apparent in the presence of P-TEFb inhibitors may be so explained, or promoter-proximal pausing may not be a regulatory feature of the genes in question. In budding yeast, in which regulated promoter-proximal pausing is absent (28), the deletion of the Pol II CTD Ser2 kinase Ctk1 does not affect transcription elongation (2).
Acknowledgments
This work was supported by NIH grant GM25232 to J.T.L. and NSF grant CHE-0242328 to W.W.W. and J.T.L.
We thank D. H. Price for anti-cyclin T serum and Lis lab members for comments on the manuscript.
Z.N. and J.T.L. conceptualized the project; A.S. and J.T.L. wrote the manuscript; Z.N. obtained the data in Fig. 1 and 2 and purified recombinant CstF50 for antiserum production; A.S. obtained the data in Fig. 3; N.J.F. obtained the data in Fig. 4; J.Y. obtained the data in Fig. 5; J.-R.S. provided the FP; W.W.W. advised and provided support for the work presented in Fig. 5.
Footnotes
Published ahead of print on 10 December 2007.
REFERENCES
- 1.Adelman, K., M. T. Marr, J. Werner, A. Saunders, Z. Ni, E. D. Andrulis, and J. T. Lis. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17103-112. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 1367-76. [DOI] [PubMed] [Google Scholar]
- 3.Akoulitchev, S., T. P. Makela, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377557-560. [DOI] [PubMed] [Google Scholar]
- 4.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 142635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 967791-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biglione, S., S. A. Byers, J. P. Price, V. T. Nguyen, O. Bensaude, D. H. Price, and W. Maury. 2007. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 447-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boehm, A. K., A. Saunders, J. Werner, and J. T. Lis. 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 237628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 2721473-1476. [DOI] [PubMed] [Google Scholar]
- 9.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 221079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, S. A., A. N. Imbalzano, and R. E. Kingston. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 101479-1490. [DOI] [PubMed] [Google Scholar]
- 11.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 27631793-31799. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 231961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25375-380. [DOI] [PubMed] [Google Scholar]
- 14.Egyhazi, E., A. Ossoinak, A. Pigon, C. Holmgren, J. M. Lee, and A. L. Greenleaf. 1996. Phosphorylation dependence of the initiation of productive transcription of Balbiani ring 2 genes in living cells. Chromosoma 104422-433. [DOI] [PubMed] [Google Scholar]
- 15.Flores, O., G. Lee, J. Kessler, M. Miller, W. Schlief, J. Tomassini, and D. Hazuda. 1999. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc. Natl. Acad. Sci. USA 967208-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 151783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujinaga, K., D. Irwin, Y. Huang, R. Taube, T. Kurosu, and B. M. Peterlin. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giardina, C., M. Perez-Riba, and J. T. Lis. 1992. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 62190-2200. [DOI] [PubMed] [Google Scholar]
- 19.Gilmour, D. S., and J. T. Lis. 1986. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 63984-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guiguen, A., J. Soutourina, M. Dewez, L. Tafforeau, M. Dieu, M. Raes, J. Vandenhaute, M. Werner, and D. Hermand. 2007. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 261552-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 243-53. [DOI] [PubMed] [Google Scholar]
- 23.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 39593-96. [DOI] [PubMed] [Google Scholar]
- 24.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 131234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 202970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 226979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krumm, A., T. Meulia, M. Brunvand, and M. Groudine. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 62201-2213. [DOI] [PubMed] [Google Scholar]
- 28.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63347-356. [DOI] [PubMed] [Google Scholar]
- 29.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14792-803. [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, H., O. Flores, R. Weinmann, and D. Reinberg. 1991. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. USA 8810004-10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majello, B., G. Napolitano, A. Giordano, and L. Lania. 1999. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene 184598-4605. [DOI] [PubMed] [Google Scholar]
- 32.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 27127176-27183. [DOI] [PubMed] [Google Scholar]
- 33.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 27012335-12338. [DOI] [PubMed] [Google Scholar]
- 34.Marshall, N. F., and D. H. Price. 1992. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell. Biol. 122078-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medlin, J., A. Scurry, A. Taylor, F. Zhang, B. M. Peterlin, and S. Murphy. 2005. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 244154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 191401-1415. [DOI] [PubMed] [Google Scholar]
- 37.Mosser, D. D., N. G. Theodorakis, and R. I. Morimoto. 1988. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 84736-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414322-325. [DOI] [PubMed] [Google Scholar]
- 39.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 1355-65. [DOI] [PubMed] [Google Scholar]
- 40.O'Brien, T., and J. T. Lis. 1993. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 133456-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Brien, T., and J. T. Lis. 1991. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol. Cell. Biol. 115285-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 43.Phair, R. D., S. A. Gorski, and T. Misteli. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375393-414. [DOI] [PubMed] [Google Scholar]
- 44.Phatnani, H. P., and A. L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 202922-2936. [DOI] [PubMed] [Google Scholar]
- 45.Plet, A., D. Eick, and J. M. Blanchard. 1995. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene 10319-328. [PubMed] [Google Scholar]
- 46.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108501-512. [DOI] [PubMed] [Google Scholar]
- 47.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 27610913-10920. [DOI] [PubMed] [Google Scholar]
- 48.Raschke, E. E., T. Albert, and D. Eick. 1999. Transcriptional regulation of the Ig kappa gene by promoter-proximal pausing of RNA polymerase II. J. Immunol. 1634375-4382. [PubMed] [Google Scholar]
- 49.Rasmussen, E. B., and J. T. Lis. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 907923-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 27642601-42609. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rougvie, A. E., and J. T. Lis. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54795-804. [DOI] [PubMed] [Google Scholar]
- 53.Roy, R., J. P. Adamczewski, T. Seroz, W. Vermeulen, J. P. Tassan, L. Schaeffer, E. A. Nigg, J. H. Hoeijmakers, and J. M. Egly. 1994. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 791093-1101. [DOI] [PubMed] [Google Scholar]
- 54.Saunders, A., L. J. Core, and J. T. Lis. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7557-567. [DOI] [PubMed] [Google Scholar]
- 55.Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J. T. Lis. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 3011094-1096. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz, B. E., S. Larochelle, B. Suter, and J. T. Lis. 2003. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol. Cell. Biol. 236876-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 162135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sopta, M., R. W. Carthew, and J. Greenblatt. 1985. Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 26010353-10360. [PubMed] [Google Scholar]
- 59.Strobl, L. J., and D. Eick. 1992. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 113307-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trigon, S., H. Serizawa, J. W. Conaway, R. C. Conaway, S. P. Jackson, and M. Morange. 1998. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 2736769-6775. [DOI] [PubMed] [Google Scholar]
- 61.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 177395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wen, Y., and A. J. Shatkin. 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 131774-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winkler, M., T. aus Dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 748053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 171402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura, and H. Handa. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21227-237. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 9741-51. [DOI] [PubMed] [Google Scholar]
- 68.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414317-322. [DOI] [PubMed] [Google Scholar]
- 69.Yao, J., K. M. Munson, W. W. Webb, and J. T. Lis. 2006. Dynamics of heat shock factor association with native gene loci in living cells. Nature 4421050-1053. [DOI] [PubMed] [Google Scholar]
- 70.Yoh, S. M., H. Cho, L. Pickle, R. M. Evans, and K. A. Jones. 2007. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21160-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 205077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 112622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]