Abstract
Cells of the budding yeast Saccharomyces cerevisiae sense extracellular amino acids and activate expression of amino acid permeases through the SPS-sensing pathway, which consists of Ssy1, an amino acid sensor on the plasma membrane, and two downstream factors, Ptr3 and Ssy5. Upon activation of SPS signaling, two transcription factors, Stp1 and Stp2, undergo Ssy5-dependent proteolytic processing that enables their nuclear translocation. Here we show that Ptr3 is a phosphoprotein whose hyperphosphorylation is increased by external amino acids and is dependent on Ssy1 but not on Ssy5. A deletion mutation in GRR1, encoding a component of the SCFGrr1 E3 ubiquitin ligase, blocks amino acid-induced hyperphosphorylation of Ptr3. We found that two casein kinase I (CKI) proteins, Yck1 and Yck2, previously identified as positive regulators of SPS signaling, are required for hyperphosphorylation of Ptr3. Loss- and gain-of-function mutations in PTR3 result in decreased and increased Ptr3 hyperphosporylation, respectively. We found that a defect in PP2A phosphatase activity leads to the hyperphosphorylation of Ptr3 and constitutive activation of SPS signaling. Two-hybrid analysis revealed interactions between the N-terminal signal transduction domain of Ssy1 with Ptr3 and Yck1. Our findings reveal that CKI and PP2A phosphatase play antagonistic roles in SPS sensing by regulating Ptr3 phosphorylation.
How cells sense nutrients and adjust the intracellular machinery to utilize them is a fundamental biological question. Nutrients, such as glucose, amino acids, and fatty acids, provide cells with energy and building materials for biosynthesis. Nutrients can also serve as signaling molecules to mediate multiple signal transduction pathways (13, 18, 30). In mammals, studies on nutrient sensing have been concentrated on cell types that have prominent nutrient-sensing functions, such as pancreatic β cells for glucose sensing (20). In yeast (Saccharomyces cerevisiae), G protein-coupled receptors, nonfunctional nutrient transporter homologs, and functional nutrient transporters have been shown to function as extracellular nutrient sensors to elicit intracellular signal transduction pathways (13). Yeast cells are able to sense extracellular nutrients and activate respective transporters to facilitate nutrient import (12). In the budding yeast, there are two such well-studied nutrient-sensing pathways: one is for glucose and deploys several hexose transporters, such as Hxt1 (24, 31); and the other is for amino acid sensing and utilizes several amino acid transporters, including Agp1 (8, 14, 17).
In the glucose-sensing pathway, there are two plasma membrane glucose sensors, Rgt2 and Snf3, that function to transduce extracellular glucose signals to inhibit a transcriptional repressor, Rgt1 (13). Activation of the glucose signal transduction pathway also requires two casein kinase I (CKI) proteins, Yck1 and Yck2, which are proposed to phosphorylate two negative regulatory factors, Std1 and Mth1, leading to their ubiquitination via the SCFGrr1 ubiquitin ligase and their subsequent degradation via proteasomes (16, 22, 32).
Amino acids are sensed through the amino acid sensor Ssy1, which is a homolog of functional amino acid permeases and is localized on the plasma membrane (8, 14, 17). When extracellular amino acids bind to Ssy1, two related transcription factors, Stp1 and Stp2, are activated through Ptr3, Ssy5, and Grr1, a component of the SCFGrr1 ubiquitin ligase (1, 4). Because signals generated by Ssy1 are transmitted through Ptr3 and Ssy5, this route for amino acid sensing is also called the SPS-sensing pathway. Both Ptr3 and Ssy5 have been reported to be peripheral plasma membrane proteins (11, 17). In two-hybrid assays, Ptr3 has been shown to interact with itself and with Ssy5 (5). These studies have led to the suggestion that Ssy1, Ptr3, and Ssy5 form a plasma membrane-localized signal transduction complex. Three recent reports indicate that Ssy5 is a novel chymotrypsin-like protease, which undergoes a proteolytic processing event to generate an N-terminal prodomain and a C-terminal activity domain (1, 3, 26). After processing, the prodomain and the activity domain remain associated. It has been proposed that such an association may inhibit the protease activity of the catalytic domain. Down-regulation of the Ssy5 prodomain in response to amino acids has been proposed as an activation mechanism for the SPS-sensing pathway (3). The activated catalytic domain of Ssy5 removes an N-terminal inhibitory sequence from Stp1 and Stp2, which allows them to enter the nucleus and activate target gene expression (1, 3, 26). The CKI proteins Yck1 and Yck2 have been reported to be positive regulators of the SPS-sensing pathway (1, 32), and Rts1, a regulatory subunit of the PP2A phosphatase complex, negatively regulates SPS sensing (9).
It is not clear how Ssy1 generates downstream signals and how those signals are transmitted via Ptr3, Ssy5, and Grr1 to activate Stp1/2. It is also unclear how Yck1/2 or Rts1 functions in the SPS-sensing pathway. Here we show that Ptr3 is a phosphoprotein and that its phosphorylation plays a critical regulatory role in SPS sensing. In response to amino acids, Ssy1-dependent hyperphosphorylation of Ptr3, likely via Grr1 and Yck1/2, leads to pathway activation. The Rts1-containing PP2A phosphatase complex negatively regulates SPS sensing by increasing the dephosphorylation of Ptr3. Mutations in the conserved threonine residue 525, a consensus phosphorylation site for CKI proteins, abolish both Ptr3 function and Ptr3 hyperphosphorylation. In contrast, an activating allele of PTR3 results in Ptr3 hyperphosphorylation. We identified interactions between Ssy1's N-terminal extension with both Ptr3 and Yck1. Our results suggest that the hyperphosphorylation of Ptr3 is a key activation step of the SPS amino acid-sensing pathway.
MATERIALS AND METHODS
Strains and plasmids.
Yeast strains and plasmids used in this study are listed in Tables 1 and 2.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source/reference |
|---|---|---|
| ZLY1917 | MATaura3-52 ptr3-Δ15 AGP1::AGP1-lacZ::kanMX4 | This study |
| HKY31 | MATaura3-52 lys2-Δ201 ptr3-Δ15 | 17 |
| ZLY2142 | MATaura3-52 ptr3-Δ15::PTR3-myc6::URA3 AGP1::AGP1-lacZ::kanMX4 | This study |
| ZLY2916 | MATaura3-52 ptr3-Δ15::PTR3-myc6::URA3 AGP1::AGP1-lacZ::kanMX4 grr1Δ::kanMX4 | This study |
| RBY923 | MATaura3-52 lys2-Δ201 ptr3-Δ15 ssy1::hisG | This study |
| RBY875 | MATaura3-52 lys2-Δ201 ptr3-Δ15 ssy5Δ::kanMX4 | This study |
| RBY951 | MATaura3-52 lys2-Δ201 ptr3-Δ15 ssy1::hisG ssy5Δ::kanMX4 | This study |
| ZLY175 | MATaura3-52 lys2-Δ201 grr1Δ::kanMX4 | This study |
| RBY661 | MATaura3 AGP1::AGP1-lacZ::kanMX4 | This study |
| ZLY1915 | MATaura3-52 ssy1-Δ13 AGP1::AGP1-lacZ::kanMX4 | This study |
| LRB341 | MATahis3 leu2 ura3-52 | 25 |
| LRB264 | MATahis3 leu2 ura3-52 yck1-1::URA3 | 25 |
| ZLY2021 | MATahis3 leu2 ura3-52 yck1-1::ura3::kanMX4 | This study |
| LRB343 | MATahis3 leu2 ura3-52 yck2::HIS3 | 25 |
| LRB362 | MATahis3 leu2 ura3-52 yck1-1::ura3 yck2-2ts | 25 |
| ZLY044 | MATaura3-52 AGP1::AGP1-lacZ::kanMX4 | This study |
| ZLY2507 | MATaura3-52 rts1Δ::kanMX4 AGP1::AGP1-lacZ::kanMX4 | This study |
| ZLY2512 | MATaura3-52 rts1Δ::kanMX4 ptr3-Δ15::PTR3-myc6::URA3 AGP1::AGP1-lacZ::kanMX4 | This study |
| ZLY2848 | MATaura3-52 ssy1-Δ13 ptr3-Δ15::PTR3-myc6::URA3 lys2-Δ201 | This study |
| ZLY2820 | MATaura3-52 ssy1-Δ13 rts1Δ::kanMX4 ptr3-Δ15::PTR3-myc6::URA3 | This study |
| ZLY2535 | MATaura3-52 rts1Δ::kanMX4 | This study |
| ZLY2536 | MATaura3-52 rts1Δ::kanMX4 ssy1-Δ 13 | This study |
| ZLY652 | MATα ura3 lys2 leu2 HXT1-lacZ::LEU2 | This study |
| ZLY2538 | MATα ura3 lys2 leu2 HXT1-lacZ::LEU2 rts1Δ::kanMX4 | This study |
| ZLY2518 | MATahis3 leu2 ura3-52 yck1-1::ura3 yck2-2ts | This study |
| ZLY2627 | MATahis3 leu2 ura3-52 yck1-1::ura3 yck2-2ts rts1Δ::kanMX4 | This study |
| ZLY1939 | MATaura3-52 ssy5Δ::kanMX4 AGP1::AGP1-lacZ::kanMX4 | This study |
| RBY909 | MATaura3-52 lys2-Δ 201 ssy5Δ::kanMX4 | This study |
| RBY873 | MATaura3-52 lys2-Δ201 ssy1-Δ13 ssy5Δ::kanMX4 | This study |
| RBY721 | MATaura3-52 | This study |
| HKY20 | MATaura3-52 lys2-Δ201 ssy1-Δ13 | 17 |
| S288c | MATaura3 | Butow lab stock |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source/reference |
|---|---|---|
| pZL473 | pRS416 (CEN URA3) containing PTR3 | This study |
| pZL1179 | pRS416 containing PTR3-myc; expresses Ptr3 with a 6× myc tag inserted between Ptr3 residues 157 and 158 | This study |
| pEG-KG-YCK1 | 2 μm URA3 Leu2-d plasmid containing GAL1-GST-YCK1 | 36 |
| pEG-KG-YCK2 | 2 μm URA3 Leu2-d plasmid containing GAL1-GST-YCK2 | 36 |
| pZL1834 | pRS416 containing STP1-HA; expresses Stp1 with 3× HA tagged at the C-terminal end | This study |
| pZL1949 | pRS416 containing HIS6-PTR3-myc; expresses Ptr3 with a six-His tag at the N-terminal end and a 9× myc tag inserted between residues 157 and 158 of Ptr3 | This study |
| pZL2107 | pRS416 containing HIS6-PTR3(S321A)-myc | This study |
| pZL2112 | pRS416 containing HIS6-PTR3(T635A)-myc | This study |
| pZL2122 | pRS416 containing HIS6-PTR3(T525A)-myc | This study |
| pZL2125 | pRS416 containing HIS6-PTR3(T525D)-myc | This study |
| pZL2132 | pRS416 containing HIS6-PTR3(T525E)-myc | This study |
| pZL2323 | Yip352 containing PTR3(Q439R)-myc | |
| pZL736 | pRS416 containing SSY5 | This study |
| pZL1668 | pRS416 containing SSY5-HA; expresses Ssy5 with a 3× HA tag at the C-terminal end | This study |
| pZL1239 | pRS417 (CEN LYS2) containing PTR3-myc; expresses Ptr3 with a 9× myc tag inserted between residues 157 and 158 of Ptr3 | This study |
| pZL835 | pRS416 containing PTR3-HA; expresses Ptr3 with a 3× HA tag inserted between Ptr3 residues 157 and 158 | This study |
| pZL840 | pRS416 containing HA-SSY5; expresses Ssy5 with a 6× HA tag at the N-terminal end of Ssy5a | This study |
| pZL465 | pWCJ (CEN URA3) containing AGP1-lacZ | This study |
| pZL2230 | pRS417 containing STP1-HA; expresses Stp1p with 3× HA tagged at the C-terminal end | This study |
| pGBKT7 | A yeast two-hybrid plasmid carrying Gal4 DNA binding domain encoding sequence | Clontech Laboratories, Inc. |
| pGBKT7-Lst8 | pGBKT7 containing the LST8 open reading frame (ORF) | This study |
| pGBKT7-SSY1N | pGBKT7 carrying DNA sequence encoding amino acid residues 1-284 of Ssy1 | This study |
| pACTII | A yeast two-hybrid plasmid carrying Gal4 DNA binding domain encoding sequence | Clontech Laboratories, Inc. |
| pACTII-LST8 | pACTII carrying the LST8 ORF | This study |
| pACTII-PTR3 | pACTII carrying the PTR3 ORF | This study |
| pACTII-SSY5 | pACTII carrying the SSY5 ORF | This study |
| pACTII-YCK1 | pACTII carrying the YCK1 ORF | This study |
The primer pair gtcaACTAGTGTCACGGCGAATCGATCTAT and gtcaACGCGTCATCGGTATATCGAGTTTAC (lowercase letters indicate nucleotides introduced to facilitate digestion of PCR products by restriction endonucleases) was used to amplify a 1,003-bp DNA sequence containing the SSY5 promoter region plus the start codon by PCR. The primer pair gtcaACGCGTGTCAGATTTTTTGGTTTAAACAAGAAAAAGAACGA and gtcaGTCGACCTGTGAACCAAGGTACCTTC was used to amplify the SSY5 ORF plus a 376-bp 3′ untranslated region sequence. These two DNA fragments were digested with SpeI and MluI and with MluI and SalI, respectively, and cloned into SpeI- and SalI-digested pRS416 to form pZL737, thus introducing an MluI site right after the start codon of SSY5. Two tandem repeats of HA3 flanked by MluI sites were cloned into the MluI site of pZL737 to form pZL840.
Growth media and growth conditions.
Yeast strains were grown at 30°C in YPD medium (1% yeast extract, 2% peptone, and 2% dextrose), minimal SD medium (0.67% yeast nitrogen base [YNB] and 2% dextrose), SPD medium (0.67% YNB without amino acids and ammonium sulfate, 0.1% proline, and 2% glucose), YNBcasRaffGal (0.67% YNB, 1% Casamino Acids, 2% raffinose, and 2% galactose), or YNBCasD (0.67% YNB, 1% Casamino Acids, and 2% dextrose). When required, lysine or histidine was added at a final concentration of 0.003% to the culture medium.
β-Galactosidase activity assays for lacZ reporter gene expression.
Liquid cultures were inoculated with a pool of several independent transformants in media indicated in the figure legends, and cells were grown for at least six generations to an optical density at 600 nm of 0.5 to 0.8. The preparation of cell extracts and β-galactosidase assays were carried out as described by Adams et al. (2).
Cell extracts, phosphatase treatment, immunoblotting, and immunoprecipitation.
Total cellular proteins were prepared by lysing yeast cells in 1.85 N NaOH-7.5% β-mercaptoethanol on ice for 10 min, followed by precipitation with trichloroacetic acid (TCA) at a final concentration of 8%. The TCA pellets were neutralized with 1 M unbuffered Tris and resuspended in 1× sodium dodecyl sulfate (SDS) loading buffer. For phosphatase treatment, neutralized TCA-precipitated total cellular proteins were incubated with 400 units of lambda protein phosphatase (New England Biolabs, Inc.) in a final volume of 20 μl at 30°C for 90 min. When indicated, phosphatase inhibitors (1 μM sodium orthovanadate, 10 mM β-glycerol phosphate, 10 mM sodium pyrophosphate, and 10 mM NaF) were added to inhibit lambda protein phosphatase activity. Immunoblotting and coimmunoprecipitation experiments were performed as described previously (19). Briefly, for immunoprecipitation experiments, total cell extracts were obtained in immunoprecipitation buffer (20 mM HEPES-KOH, pH 7.4, 120 mM NaCl, 0.5% Triton X-100, 1 μM sodium orthovanadate, 10 mM NaF, 10 mM pyrophosphate, 10 mM β-glycerol phosphate, and protease inhibitors). Eight hundred-microliter cell extracts (3 μg/μl protein) were incubated at 4°C for 1 h with monoclonal anti-myc antibody (9E10), after which 50 μl of a 50% slurry of protein G-Sepharose (Roche) was added for each reaction then further incubated at 4°C for 2 h. The immune complexes bound to the Sepharose beads were released by boiling in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer after being washed three times with immunoprecipitation buffer. The released immune complexes were analyzed by Western blotting. myc- and hemagglutinin (HA)-tagged proteins were probed with anti-myc antibody 9E10 and with anti-HA antibody 3F10 or 12CA5, respectively.
RESULTS
PTR3 is a phosphoprotein.
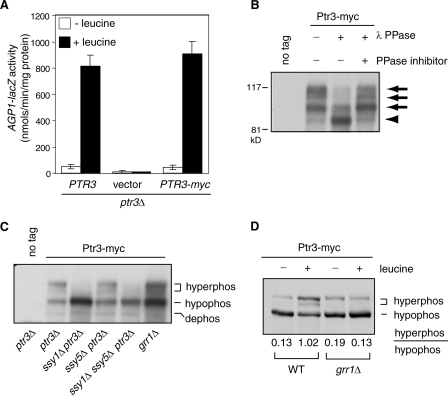
Ptr3 exists as several electrophoretic mobility variants detected by Western blotting (11), but the nature of those variants is unclear. To determine whether different forms of Ptr3 are due to phosphorylation, we tagged Ptr3 with a 6× myc tag inserted between residues 157 and 158 of the protein. The resultant PTR3-myc construct is fully functional based on its ability to complement a ptr3Δ mutation, which affects the expression of an AGP1-lacZ reporter gene in cells grown in the presence or absence of leucine (Fig. 1A). Total cellular proteins were separated by SDS-PAGE and detected using monoclonal anti-myc antibody. Figure 1B shows that Ptr3 exists as four bands, although the fastest-mobility form of Ptr3 is sometimes difficult to discern. Treatment of total cellular proteins with lambda protein phosphatase leads to a reduction of the three slower-mobility forms of Ptr3-myc and a corresponding increase of the fastest-mobility form. Phosphatase inhibitors abolish the effect of lambda protein phosphatase, indicating that all three slower-mobility forms of Ptr3-myc are due to phosphorylation.
FIG. 1.
Ptr3 is a phosphoprotein. (A) A 6× myc-tagged Ptr3 is functional. A ptr3Δ deletion strain with an integrated AGP1-lacZ reporter gene (ZLY1917) was transformed with PTR3 (pZL473), vector (pRS416), or PTR3-myc (pZL1179) plasmid as indicated. Transformants were grown in SD medium with or without 0.02% leucine, and β-galactosidase activities were conducted as described in Materials and Methods. (B) Ptr3 is phosphorylated. The ptr3Δ strain transformed with PTR3 (pZL473) or PTR3-myc (pZL1179) plasmids used as described for panel A was grown in SD medium with 0.02% leucine to mid-log phase. TCA-precipitated total cellular proteins were treated with lambda protein phosphatase (λ PPase) as indicated. When required, phosphatase inhibitors (see Materials and Methods) were added to inhibit λ PPase. After treatment, proteins were separated by SDS-PAGE and Ptr3-myc was probed with anti-myc antibody. Arrows and the arrowhead indicate phosphorylated and dephosphorylated forms of Ptr3-myc, respectively. (C) Ptr3 hyperphosphorylation is blocked in ssy1Δ cells but not in ssy5Δ or grr1Δ cells. Yeast strains (ptr3Δ, HKY31; ssy1Δ ptr3Δ, RBY923; ssy5Δ ptr3Δ, RBY875, ssy1Δ ssy5Δ ptr3Δ, RBY951; grr1Δ, ZLY175) transformed with a centromeric plasmid carrying either PTR3 (no tag; pZL473) or PTR3-myc (pZL1179) as indicated were grown in SD medium supplemented with 0.02% leucine. Ptr3-myc was probed with anti-myc antibody. “hyperphos,” “hypophos,” and “dephos” indicate hyperphosphorylated, hypophosphorylated, and dephosphorylated forms of Ptr3-myc, respectively. (D) Leucine-induced hyperphosphorylation of Ptr3 requires Grr1. A wild-type (WT) strain and a grr1Δ mutant strain carrying an integrated copy of PTR3-myc (WT, ZLY2142; grr1Δ, ZLY2916) were grown in SD medium with or without 0.02% leucine. Total cellular proteins from equal amounts of cells were separated by SDS-PAGE and Ptr3-myc was probed with anti-myc antibody. The numbers (average from three independent experiments) indicate the ratios of the levels of hyperphosphorylated and hypophosphorylated forms of Ptr3-myc.
It has been reported that mobility variants of Ptr3 change in response to a mutation in SSY1 but not in SSY5 (11). With our identification of Ptr3 as a phosphoprotein, we examined Ptr3 phosphorylation states in various mutants defective in amino acid sensing, including an ssy1Δ, an ssy5Δ, an ssy1Δ ssy5Δ double, and a grr1Δ mutation. Figure 1C shows that in cells grown in the presence of 0.02% leucine, the ssy1Δ single and the ssy1Δ ssy5Δ double mutation abolish the appearance of the hyperphosphorylated forms of Ptr3-myc but not the hypophosphorylated form, whereas neither an ssy5Δ mutation nor a grr1Δ mutation blocks the appearance of hyperphosphorylated forms of Ptr3p. However, there appears to be a reduced ratio of hyperphosphorylated forms of Ptr3p to their hypophosphorylated counterpart in grr1Δ cells. In the absence of leucine, the basal level of Ptr3 hyperphosphorylation is also abolished by an ssy1Δ mutation (data not shown).
Protein phosphorylation plays critical regulatory roles in many signal transduction pathways (15, 21). To determine whether Ptr3 phosphorylation is subject to regulation in the SPS-sensing pathway, we examined Ptr3 phosphorylation in wild-type cells carrying an integrated copy of PTR3-myc in which the cells were grown in minimal medium with or without 0.02% leucine. Total cellular proteins were obtained, separated by SDS-PAGE, and probed with anti-myc antibody. Figure 1D shows that leucine treatment leads to increased amounts of two hyperphosphorylated forms of Ptr3-myc, with a corresponding decrease of the hypophosphorylated form, suggesting that activation of amino acid sensing results in the hyperphosphorylation of Ptr3. Quantitative analysis of band intensity in Fig. 1D revealed that the ratio of hyperphosphorylated forms of Ptr3 to their hypophosphorylated counterpart is 7.8-fold greater for cells grown in the presence of leucine than for cells grown in the absence of leucine. Because there appears to be a reduced ratio of hyperphosphorylated forms of Ptr3 to its hypophosphorylated counterpart when expressed from a centromeric plasmid in grr1Δ cells (Fig. 1C), we determined the phosphorylation states of Ptr3-myc expressed from the genomic PTR3 locus in grr1Δ cells. Among four independent grr1Δ mutants analyzed, one of which is shown in Fig. 1D, the leucine-induced hyperphosphorylation of Ptr3 was completely blocked by a grr1Δ mutation. The expression of GRR1 from a centromeric plasmid in grr1Δ cells restores leucine-induced hyperphosphorylation of Ptr3 (data not shown). Together, these data suggest that extracellular amino acids cause a Grr1-dependent increase of Ptr3 hyperphosphorylation.
These results are consistent with the current view that Ptr3 functions downstream of Ssy1 but upstream of Ssy5 and further indicate that Grr1 functions upstream of Ptr3. Taken together, we have found that Ptr3 is a phosphoprotein and that the hyperphosphorylation of Ptr3 correlates with the activation of the SPS-sensing pathway.
Mutations in two CKI proteins, Yck1 and Yck2, block Ptr3 hyperphosphorylation.
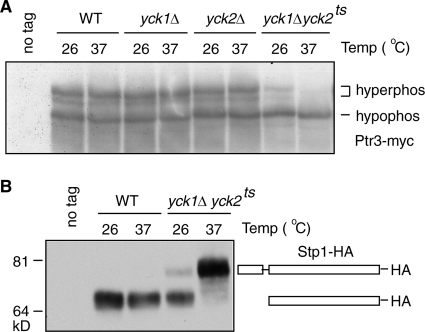
Two reports have indicated that Yck1 and Yck2, two yeast CKI proteins, are required for target gene expression in the SPS-sensing pathway (1, 32). Both kinases were also isolated in a genetic screening to identify kinases whose overexpression leads to a constitutive activation of AGP1-lacZ expression by use of a library of yeast kinases under the control of a galactose-inducible promoter (data not shown). It was proposed that Yck1 and Yck2 function as positive regulators by phosphorylating Stp1, resulting in the removal of an inhibitory N-terminal sequence that enables Stp1 to translocate into the nucleus (1). Because Ptr3 is also required for the processing and activation of Stp1 (4), we first asked whether Yck1 and Yck2 can regulate an upstream event, the phosphorylation of Ptr3, as a means of regulating the processing of Stp1. To this end, Ptr3-myc was examined in wild-type, yck1Δ, yck2Δ, and yck1Δ yck2ts mutant cells grown in the presence of leucine. At both permissive (26°C) and restrictive (37°C) temperatures, the yck1Δ or the yck2Δ single deletion mutation does not result in obvious changes in Ptr3 phosphorylation compared to what is observed for wild-type cells; however, at the restrictive temperature, the yck1Δ yck2ts double mutation results in the disappearance of both hyperphosphorylated forms of Ptr3-myc, similar to what is observed for ssy1Δ mutant cells (Fig. 2A and 1C). These data indicate that Yck1 and Yck2 play a redundant role in regulating the hyperphosphorylation of Ptr3. Consistent with previous findings, the yck1Δ yck2ts double mutation also blocks the processing of Stp1 in cells grown in the presence of leucine at 37°C (1) (Fig. 2B). Even at 26°C, the processing of Stp1 is already compromised in yck1Δ yck2ts cells (Fig. 2B), which correlates with a partial defect in the hyperphosphorylation of Ptr3 in yck1Δ yck2ts double mutant cells at this temperature (Fig. 2A). The correlation between the failure to hyperphosphorylate Ptr3 and the inactivation of the SPS-sensing pathway in the ssy1Δ single and the yck1Δ yck2ts double mutant suggests that hyperphosphorylation of Ptr3 is required for activation of the SPS-sensing pathway.
FIG. 2.
Yck1 and Yck2, two CKI proteins, are required for the hyperphosphorylation of Ptr3. (A) The hyperphosphorylation of Ptr3 is blocked in yck1Δ yck2ts double mutant cells. Wild-type (WT; LRB341), yck1Δ (ZLY2021), yck2Δ (LRB343), and yck1Δ yck2ts (LRB362) mutant cells expressing either nontagged Ptr3 (pZL473) or Ptr3-myc from a centromeric plasmid (pZL1179) were grown at the indicated temperatures (Temp) in SD medium supplemented with 0.02% leucine. Total cellular proteins were separated by SDS-PAGE and Ptr3-myc was probed with anti-myc antibody. “hyperphos” and “hypophos” indicate the hyperphosphorylated and hypophosphorylated forms of Ptr3-myc, respectively. (B) The yck1Δ yck2ts double mutation blocks proteolytic processing of Stp1. Wild-type (LRB341) and yck1Δ yck2ts mutant (LRB362) cells transformed with a centromeric plasmid carrying STP1-HA (pZL1834) were grown at the indicated temperatures in SD medium supplemented with 0.02% leucine. Total cellular proteins were separated by SDS-PAGE and Stp1-HA was probed with anti-HA antibody. The full-length Stp1-HA and the processed Stp1-HA are indicated diagrammatically in the figure.
Loss-of-function mutations in PTR3 correlate with its failure to be hyperphosphorylated.
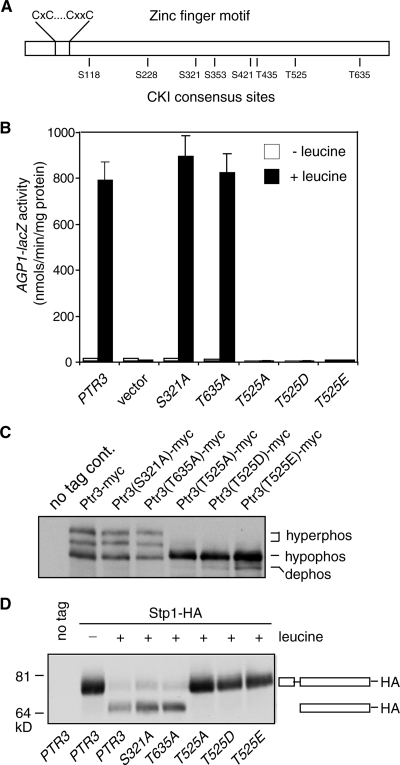
CKI proteins often target the consensus site D/ExxS/T (10). Ptr3 has 5 DxxS/T sites and 10 ExxS/T sites. Because the hyperphosphorylation of Ptr3 by Yck1 and Yck2 correlates with the activation of the SPS-sensing pathway, we hypothesize that such phosphorylation sites would be conserved among Ptr3 orthologs in diverse fungal species. A sequence alignment of Ptr3s from 12 fungal species revealed two highly conserved sites, the EIYS site for serine residue 321 and the ESAT site for threonine residue 635, and one completely conserved site, the EGIT site for threonine residue 525 (Fig. 3A). To determine whether these three sites are involved in the phosphorylation of Ptr3, point mutations were introduced to change these three residues to alanines to obtain the PTR3(S321A), PTR3(T525A), and PTR3(T635A) mutants. To determine the functionality of these PTR3 mutant constructs, ptr3Δ cells transformed with these mutant PTR3 constructs were analyzed for expression of AGP1-lacZ reporter gene; this analysis shows that the S321A and the T635A mutants are fully functional, while the T525A mutant is not, suggesting that threonine 525 is critical for Ptr3 function (Fig. 3B). Mutation of five other partially conserved CKI consensus sites revealed that none of them is critical for Ptr3 function (Fig. 3A; also see Fig. S1 in the supplemental material). To determine whether functional differences among CKI consensus site mutants are due to a difference in their capability to be hyperphosphorylated, we examined the phosphorylation state of Ptr3-myc carrying the loss-of-function T525A mutation and two inconsequential mutations, the S321A mutation and the T635A mutation. Figure 3C shows that the S321A and the T635A mutations have no effect on Ptr3 phosphorylation, while the T525A mutation abolished the hyperphosphorylation of Ptr3. In an attempt to create a dominant active form of Ptr3 by mimicking Ptr3 phosphorylation, threonine 525 was mutated to either aspartic acid or glutamic acid to form the PTR3(T525D) and PTR3(T525E) mutants, respectively. These two mutants, like the PTR3(T525A) mutant, are not functional (Fig. 3B) and are unable to be hyperphosphorylated (Fig. 3C). It is not uncommon that mutation of a serine or threonine residue to an acidic residue fails to mimic the effect of phosphorylation (7). It is also possible that threonine 525 of Ptr3 is not a phosphorylation site for Yck1/2. The addition of a 3× myc tag to the C-terminal end of Ptr3 also results in the loss of both Ptr3 function and Ptr3 hyperphosphorylation (data not shown). Together, these mutants show a clear correlation between Ptr3 hyperphosphorylation and Ptr3 functionality.
FIG. 3.
Mutations in threonine residue 525 abolish the hyperphosphorylation of Ptr3 and inactivate Ptr3. (A) A schematic representation of CKI consensus sites and a conserved zinc finger motif in Ptr3. (B) β-Galactosidase activity assay of AGP1-lacZ reporter gene expression in ptr3Δ cells transformed with various PTR3 constructs. ptr3Δ cells (ZLY1917) were transformed with a plasmid carrying wild-type PTR3 or mutant ptr3 alleles as indicated [PTR3, pZL1949; PTR3(S321A), pZL2107; PTR3(T635A), pZL2112; PTR3(T525A), pZL2122; PTR3(T525D), pZL2125; PTR3(T525E), pZL2132]. The resultant transformants were grown in SD medium with or without 0.02% leucine, and β-galactosidase assays were conducted as described previously. (C) Mutations in threonine 525 block the hyperphosphorylation of Ptr3. Cells used for panel B were grown in SD medium supplemented with 0.02% leucine. Total cellular proteins were separated by SDS-PAGE. Wild-type Ptr3-myc and various mutant Ptr3-myc proteins were probed with anti-myc antibody. “hyperphos,” “hypophos,” and “dephos” indicate hyperphosphorylated, hypophosphorylated, and dephosphorylated forms of Ptr3-myc, respectively. (D) Blocks in Ptr3 hyperphosphorylation correlate with a failure to process Stp1. ptr3Δ cells (HKY31) carrying centromeric plasmids coexpressing Stp1-HA (pZL2230) and various PTR3 constructs as described for panel B were grown in SD medium with or without 0.02% leucine as indicated. Total cellular proteins were separated by SDS-PAGE and Stp1-HA was probed with anti-HA antibody. The full-length Stp1-HA and the processed Stp1-HA are indicated diagrammatically in the figure.
Our data clearly show that mutants that fail to hyperphosphorylate Ptr3 also fail to activate target gene expression. To determine whether this is due to a failure to process Stp1, we examined Stp1 processing in ptr3Δ cells expressing various PTR3 constructs. To this end, cells cotransformed with centromeric plasmids expressing Stp1-HA and various Ptr3 constructs were grown in SD medium in the presence or absence of leucine as indicated. Leucine treatment leads to the appearance of a processed form of Stp1-HA in cells expressing wild-type PTR3, the S321A mutant, or the T635 mutant, while Stp1p-HA fails to be processed in cells expressing the T525A, the T525D, or the T5252E mutant (Fig. 3D). Together, these data indicate that blocks in Ptr3 hyperphosphorylation result in a failure to process Stp1, which in turn leads to a failure to activate target gene expression.
Sequence alignment also revealed a highly conserved, putative zinc finger motif, CxCx13-21CxxC, close to the N-terminal end of Ptr3 (Fig. 3A). However, mutating those cysteine residues to serine in pairs did not affect AGP1-lacZ expression in cells grown in the presence or absence of leucine (data not shown), indicating that this zinc finger motif is unlikely to play an important role in regulating the amino acid-induced activation of Ptr3.
A constitutively active Ptr3 mutant is hyperphosphorylated.
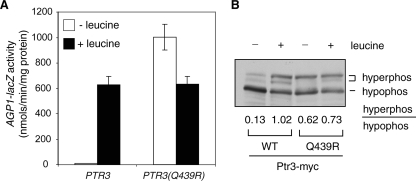
Poulsen et al. have reported the isolation of constitutive activation mutants of Ptr3 (27). If the hyperphosphorylation of Ptr3 is a key determination factor in the activation of SPS signaling, such constitutively active Ptr3 mutants should lead to Ptr3 hyperphosphorylation. To this end, we introduced two reported PTR3-5 and PTR3-17 mutations, which are due to Gln-to-Arg (Q439R) and Thr-to-Lys (T435K) substitutions, respectively, into PTR3. When expressed from a centromeric plasmid in ptr3Δ cells, the PTR3(Q439R) mutation constitutively activates expression of an integrated AGP1-lacZ reporter gene, while the PTR3(T435K) mutation only marginally activates AGP1-lacZ expression in the absence of amino acid induction (data not shown), possibly due to interference of a 12× His tag and a 3× myc tag introduced into Ptr3 or due to strain differences. We decided to focus on the Q439R mutation, which was then introduced into the PTR3-myc fusion gene and integrated into the genomic locus in ptr3Δ cells. The expression of an integrated AGP1-lacZ reporter gene is fully activated in the absence of leucine treatment in the resultant PTR3(Q439R) mutant cells (Fig. 4A), consistent with the findings of Poulsen et al. (27). Western blot analysis of Ptr3(Q439R)-myc revealed a four- to fivefold increase in the ratio of hyperphosphorylated Ptr3 to its hypophosphorylated counterpart in cells without leucine treatment (Fig. 4B). The correlation between Ptr3 hyperphosphorylation and the activation of SPS signaling due to the Q439R mutation further suggests that Ptr3 hyperphosphorylation is a key determination factor for pathway activation.
FIG. 4.
An activating mutant allele of PTR3, PTR3(Q439), leads to Ptr3 hyperphosphorylation. (A) A PTR3(Q439R) mutation, due to a change of residue 439 from glutamine to arginine, leads to constitutive activation of an AGP1-lacZ reporter gene. An integrative plasmid carrying PTR3(Q439)-myc (pZL2323) was targeted to the PTR3 genomic locus in ptr3Δ cells carrying an AGP1-lacZ reporter gene (ZLY1917). The resultant transformant and wild-type control cells were grown in SD medium with or without leucine as indicated, and AGP1-lacZ expression was analyzed as described previously. (B) Ptr3(Q439R)-myc is hyperphosphorylated. Total cellular proteins from cells expressing the Ptr3(Q439R)-myc mutant or wild-type (WT) Ptr3-myc were separated by SDS-PAGE and probed with anti-myc antibody. The numbers (average from three independent experiments) indicate the ratios of the levels of hyperphosphorylated (hyperphos) to hypophosphorylated (hypophos) forms of Ptr3-myc.
Activation of SPS signaling due to an rts1Δ mutation correlates with Ptr3 hyperphosphorylation.
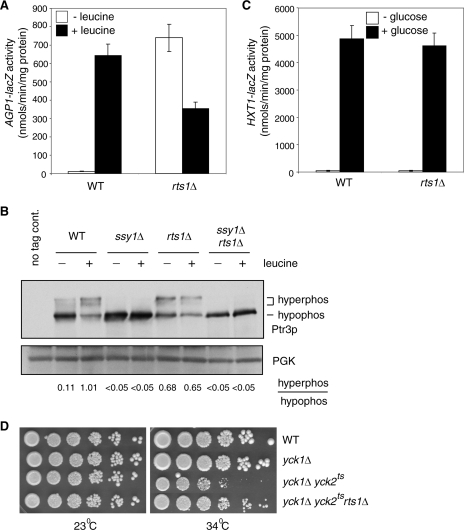
Our data so far demonstrate a positive regulatory role for the hyperphosphorylation of Ptr3 in the activation of SPS sensing. A phosphatase may be required to turn off the pathway when amino acids have been exhausted in the extracellular environment. A recent report demonstrated that mutations in Rts1, a regulatory subunit of the PP2A phosphatase complex, activate SPS sensing (9). However, the underlying mechanism is unknown. We hypothesize that an rts1 mutation, by reducing the activity of PP2A phosphatase toward its substrates, could result in hyperphosphorylation of Ptr3, leading to the activation of the SPS-sensing pathway. To test this possibility, we introduced an rts1Δ mutation into our strains and found, as reported previously (9), that it activates the expression of the AGP1-lacZ reporter gene in the absence of amino acids (Fig. 5A).
FIG. 5.
An rts1Δ mutation results in the hyperphosphorylation of Ptr3 and the activation of SPS sensing. (A) An rts1Δ mutation constitutively activates AGP1-lacZ reporter gene expression. Wild-type (WT; ZLY044) and rts1Δ (ZLY2507) cells with an integrated AGP1-lacZ reporter gene were grown in SD medium with or without 0.02% leucine as indicated, and β-galactosidase activities were determined. (B) An rts1Δ mutation results in increased levels of hyperphosphorylated Ptr3. Wild-type (ZLY2142), ssy1Δ (ZLY2848), rts1Δ (ZLY2512), and ssy1Δ rts1Δ (ZLY2820) cells expressing Ptr3-myc from the genomic PTR3 locus were grown in SD medium with or without 0.02% leucine as indicated. Total cellular proteins were separated by SDS-PAGE and Ptr3-myc was probed with anti-myc antibody. The numbers indicate the ratios of the levels of hyperphosphorylated (hyperphos) and hypophosphorylated (hypophos) forms of Ptr3-myc. 3-Phosphoglycerate kinase (PGK) was used as a loading control. (C) An rts1Δ mutation does not affect expression of an HXT1-lacZ reporter gene. Wild-type (ZLY652) and rts1Δ (ZLY2538) cells with an integrated HXT1-lacZ reporter gene were grown in YNBCasRaffGal (− glucose) or YNBcasD (+ glucose) medium to mid-log phase, and β-galactosidase activities were determined. (D) An rts1Δ mutation partially rescues the cell growth defect in yck1Δ yck2ts mutant cells grown at a semipermissive temperature (34°C). Wild-type (LRB341), yck1Δ (ZLY2021), yck1Δ yck2ts (ZLY2518), and yck1Δ yck2ts rts1Δ (ZLY2627) cells were serially diluted fivefold and spotted onto YPD medium at either 23°C (permissive temperature) or 34°C and grown for 3 to 4 days.
Next, we examined the phosphorylation state of Ptr3 in rts1Δ cells. Figure 5B shows that the hyperphosphorylated forms of Ptr3 are sixfold greater in rts1Δ cells grown in the absence of leucine than in wild-type cells grown under the same conditions, suggesting that Rts1/PP2A phosphatase is responsible for Ptr3 dephosphorylation. In both ssy1Δ single and ssy1Δ rts1Δ double deletion mutant cells with or without leucine treatment, Ptr3 fails to be hyperphosphorylated, indicating that the rts1Δ mutant effect on the hyperphosphorylation of Ptr3-myc also requires Ssy1 (Fig. 5B). Because both an ssy1Δ single mutation and a yck1Δ yck2ts double mutation result in a failure to hyperphosphorylate Ptr3, we propose that Ssy1 recruits Yck1/2 to phosphorylate Ptr3. Therefore, the phosphorylation state of Ptr3 is likely to be determined by opposing activities from Yck1/2 and Rts1/PP2A. We hypothesize that in cells without amino acid stimulus, a basal level of Ptr3 hyperphosphorylation is due to the Rts1/PP2A phosphatase activity and a low kinase activity from Yck1/2. An rts1Δ mutation would shift the balance toward a high level of Ptr3 hyperphosphorylation in cells grown in the absence of amino acids. We propose that upon stimulus with amino acids, Ssy1 potently activates Yck1/2 and the resultant robust kinase activity shifts the Ptr3 phosphorylation state toward hyperphosphorylation (Fig. 1D and 5B). Because Ptr3 is not hyperphosphorylated in ssy1Δ cells, an rts1Δ mutation in ssy1Δ cells will not change the balance and it will not result in the appearance of hyperphosphorylated forms of Ptr3. In response to leucine stimulation, the hyperphosphorylation of Ptr3 is greatly induced in wild-type cells, while there is little change in Ptr3 phosphorylation in rts1Δ cells (Fig. 5B). A block in Ptr3 hyperphosphorylation in ssy1Δ rts1Δ double mutant cells provides an explanation for the observation that an rts1Δ mutation fails to activate AGP1-lacZ reporter gene expression in ssy1Δ mutant cells (9). To exclude the possibility that the activation of SPS signaling in rts1Δ cells is due to an indirect effect from leakage or to the increased secretion of intracellular amino acids into the growth medium, we examined AGP1-lacZ expression in wild cells cocultured with an equal amount of rts1Δ cells without the AGP1-lacZ reporter gene. We found that the β-galactosidase activity from the AGP1-lacZ reporter gene in cocultured cells without leucine treatment is about half of what is observed for the single culture of wild cells carrying the AGP1-lacZ reporter gene grown in the absence of leucine (see Fig. S2 in the supplemental material), indicating that the activation of SPS signaling in rts1Δ cells is not due to increased secretion or leakage of amino acids into their extracellular environment. Taken together, these data suggest that the activation of SPS sensing in rts1Δ cells is due to the hyperphosphorylation of Ptr3 and that the hyperphosphorylated forms of Ptr3 are targets for the Rts1/PP2A phosphatase complex.
To determine whether the Rts1/PP2A phosphatase complex plays a more general role in antagonizing CKI activities, we examined whether an rts1Δ mutation affects the glucose signal transduction pathway. In response to extracellular glucose stimulation, Std1 and Mth1, two negative regulatory factors in this pathway, are proposed to be phosphorylated by Yck1 and Yck2, leading to their degradation and the activation of the glucose signal transduction pathway (16, 22, 32). If the Rts1/PP2A phosphatase complex plays a role in dephosphorylating Mth1 and Std1, increased phosphorylation of these two proteins in rts1Δ mutant cells would lead to their increased turnover, resulting in a constitutive activation of target gene expression, e.g., of HXT1 in the glucose signal transduction pathway. We found that an rts1Δ mutation does not affect HXT1-lacZ reporter gene expression under either noninduced or glucose-induced conditions (Fig. 5C), suggesting that the Rts1/PP2A phosphatase complex is unlikely to play an important role in the CKI-mediated glucose signal transduction pathway.
To examine whether other targets of CKI are subject to regulation by PP2A phosphatase as a possible general mode of regulation by these proteins, we analyzed the lethal phenotype observed for yck1Δ yck2ts double mutant cells (29, 35). The target(s) for this lethal phenotype is still unknown, so we determined whether an rts1Δ mutation might rescue the cell growth of yck1Δ yck2ts mutant cells. At 34°C, a semipermissive temperature for yck1Δ yck2ts mutant cells, an rts1Δ mutation partially suppresses the cell growth defect (Fig. 5D), suggesting that PP2A phosphatase is likely to antagonize the activities of CKI proteins on a hitherto-unidentified essential target protein(s). Together, the Rts1/PP2A phosphatase complex antagonizes CKI proteins in the regulation of the phosphorylation of Ptr3 and an unidentified essential target protein(s) of Yck1/2.
Interaction of Ptr3 with itself and with Ssy5 is independent of Ptr3 hyperphosphorylation.
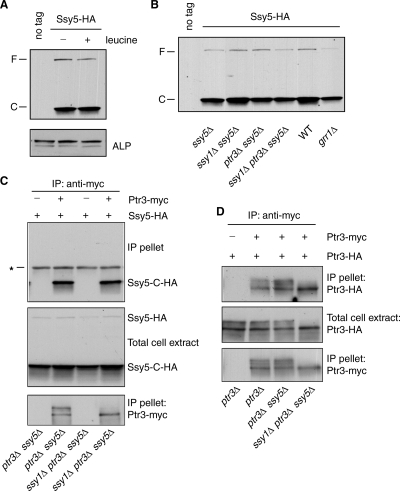
Ssy5 is known to be posttranslationally processed (1, 3, 26). Recent results suggest that Ssy5 contains a C-terminal chymotrypsin-like serine protease. We examined a functional Ssy5-HA, which contains a C-terminal 3×HA tag. As reported (1, 3, 26), Ssy5-HA is processed into a C-terminal fragment, Ssy5-C-HA (Fig. 6A). We found that the steady-state level of Ssy5-C-HA is constant, irrespective of the presence of leucine in the growth medium. Its processing into the C-terminal activity domain is also independent of other positive regulatory factors, namely, Ssy1, Ptr3, and Grr1 (Fig. 6B).
FIG. 6.
Interaction of Ptr3 with itself or Ssy5 is not affected by the hyperphosphorylation of Ptr3. (A) Ssy5 is constitutively processed. ssy5Δ cells (ZLY1939) expressing nontagged Ssy5 (no tag; pZL736) or Ssy5-HA (pZL1668) as indicated were grown in SD medium with or without 0.02% leucine. Total cellular proteins were separated by SDS-PAGE, and Ssy5-HA was probed with anti-HA antibody. F and C indicate full-length Ssy5 and the C-terminal fragment of Ssy5, respectively. Alkaline phosphatase (ALP) was used as the loading control. (B) Ssy5 proteolytic processing is independent of Ssy1, Ptr3, or Grr1. Wild-type (WT; PLY126), ssy5Δ (RBY909), ssy1Δ ssy5Δ (RBY873), ptr3Δ ssy5Δ (RBY875), ssy1Δ ptr3Δ ssy5Δ (RBY951), and grr1Δ (ZLY175) cells transformed with a centromeric plasmid expressing Ssy5-HA (pZL1668) were grown in SD medium with 0.02% leucine to mid-log phase. HA-tagged proteins were detected as described for panel A. (C) Interaction between Ptr3 and Ssy5 is independent of Ptr3 hyperphosphorylation and of Ssy1. Indicated strains carrying centromeric plasmids expressing Ptr3-myc (pZL1239) and Ssy5-HA (pZL1668) were grown in SD medium with 0.02% leucine. Ptr3-myc was immunoprecipitated with anti-myc antibody. Ptr3-myc and Ssy5-HA were probed with anti-myc and anti-HA antibody, respectively. Ssy5 and Ssy5-C-HA indicate full-length and the C-terminal fragment of Ssy5. *, the heavy chain of anti-myc antibody. (D) Self-interaction of Ptr3 is independent of its phosphorylation status, Ssy1, and Ssy5. Indicated strains carrying centromeric plasmids expressing Ptr3-myc (pZL1239) and Ptr3-HA (pZL835) were grown in SD medium with 0.02% leucine. Ptr3-myc was immunoprecipitated with anti-myc antibody. Ptr3-myc and Ptr3-HA were probed with anti-myc and anti-HA antibodies, respectively.
Ptr3 is known to interact with Ssy5 and itself in a two-hybrid assay (5). To confirm these interactions and to determine whether Ptr3 phosphorylation affects its interaction with Ssy5, we coexpressed Ptr3-myc with Ssy5-HA in otherwise wild-type or ssy1Δ mutant cells. Total cell extracts were obtained and subjected to immunoprecipitation with anti-myc antibody. The C-terminal Ssy5 fragment, Ssy5-C-HA, is recovered with tagged Ptr3-myc but not with nontagged Ptr3 (Fig. 6C). In the ssy1Δ mutant strain background, Ptr3 fails to be hyperphosphorylated, and the hypophosphorylated form of Ptr3 efficiently interacts with Ssy5-C-HA, suggesting that amino acid-induced Ptr3 hyperphosphorylation does not regulate its interaction with Ssy5. To exclude the possibility that the loss-of-function mutation of T525A in Ptr3 shown in Fig. 3 is due to gross misfolding of Ptr3, we examined its ability to interact with Ssy5-C-HA and found that the T525A mutation does not affect Ptr3's interaction with Ssy5-C-HA (see Fig. S3 in the supplemental material). Similarly, coimmunoprecipitation experiments revealed that the self-interaction of Ptr3 is also independent of its phosphorylation status, Ssy1, and Ssy5 (Fig. 6D). It has been proposed that Ssy1, Ptr3, and Ssy5 form a complex (11). Our data show that Ptr3 and Ssy5 can assemble into a complex independent of Ssy1 (Fig. 6C). Interaction between Ptr3 and Ssy5-C raises the possibility that Ptr3 may function as a positive regulator by protecting Ssy5-C from degradation. To this end, we analyzed the stability of Ssy5-C by use of the cycloheximide chase assay. Figure S4 in the supplemental material shows that Ssy5-C is relatively unstable and that neither an ssy1Δ nor a ptr3Δ mutation increases the instability of Ssy5-C, indicating that the Ssy1-dependent hyperphosphorylation of Ptr3 does not regulate the stability of Ssy5-C. Together, these data suggest that the hyperphosphorylation of Ptr3 does not affect its interaction with Ssy5-C. A possible activation mechanism for the SPS-sensing pathway may thus be due to the activation of Ssy5-C protease activity toward Stp1/2 induced by Ptr3 hyperphosphorylation.
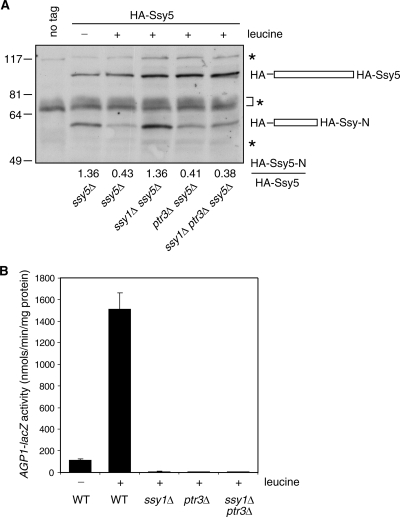
A reduced level of the N-terminal fragment of Ssy5 is not sufficient for activation of SPS signaling.
Ssy5 has been suggested to undergo autoproteolytic processing to generate an N-terminal prodomain and a C-terminal activity domain, which remain associated after processing (1, 3, 26). One report suggests that the N-terminal prodomain of Ssy5, Ssy5-N, may keep the C-terminal activity domain in an inactive state when the SPS-sensing pathway is not activated (3). That hypothesis was based on the observation that the activation of SPS sensing leads to a reduced level of Ssy5-N, whose reduction also requires Ssy1 and Ptr3, following a 30-min treatment with amino acids. We examined a functional N-terminal 6× HA-tagged Ssy5 and found that the steady-state level of HA-Ssy5-N is reduced ∼3-fold in cells grown in the presence of leucine, which requires Ssy1 (Fig. 7A). However, in contrast to a 30-min treatment with amino acids, under the steady-state condition, an amino acid-induced reduction of HA-Ssy5-N is still observed both for ptr3Δ mutant cells and for ssy1Δ ptr3Δ double mutant cells, suggesting that Ptr3 does not act by regulating the abundance of the N-terminal fragment of Ssy5. Under steady-state conditions, the activation of SPS sensing is blocked in ssy1Δ, ptr3Δ, and ssy1Δ ptr3Δ mutant cells (Fig. 7B). Clearly, the level of the N-terminal prodomain of Ssy5, Ssy5-N, does not always correlate with the activity of the SPS-sensing pathway. It has been reported that an N-terminal HA-tagged Ssy5 constitutively activates SPS signaling (3). However, our HA-SSY5 construct maintains leucine-induced activation of an AGP1-lacZ reporter gene and is not constitutively active (see Fig. S5 in the supplemental material). Together, our data indicate that the increased turnover of Ssy5-N is not sufficient to activate the SPS-sensing pathway. However, our data do not exclude the possibility that a decreased level of the N-terminal fragment of Ssy5 can contribute to the short-term activation of SPS signaling immediately following an amino acid stimulus. It is also likely that amino acid-induced Ptr3 hyperphosphorylation and a reduced level of the N-terminal fragment of Ssy5 work together to activate SPS signaling.
FIG. 7.
The level of the N-terminal fragment of Ssy5 does not correlate with the activity of the SPS-sensing pathway. (A) Under steady-state conditions, a leucine-induced reduction of the N-terminal fragment of Ssy5, HA-Ssy5-N, is abolished in ssy1Δ but not in ptr3Δ single or ssy1Δ ptr3Δ double deletion mutant cells. ssy5Δ (RBY909), ssy1Δ ssy5Δ (RBY873), ptr3Δ ssy5Δ (RBY875), and ssy1Δ ptr3Δ ssy5Δ (RBY951) cells expressing an N-terminal 6× HA-tagged Ssy5 from a centromeric plasmid (pZL840) were grown in SD medium for at least six generations with or without 0.02% leucine as indicated. Total cellular proteins were separated by SDS-PAGE and HA-tagged proteins were probed with anti-HA antibody. Numbers at the bottom of the figure indicate the ratios of the levels of the N-terminal fragment of Ssy5, HA-Ssy5-N, to full-length HA-Ssy5. *, cross-reacting species. (B) Leucine-induced expression of an AGP1-lacZ reporter gene is abolished in ssy1Δ, ptr3Δ, and ssy1Δ ptr3Δ mutant cells. Wild-type cells (WT; RBY721) and ssy1Δ (HKY20), ptr3Δ (HKY31), and ssy1Δ ptr3Δ (RBY923) cells carrying an AGP1-lacZ report gene on a centromeric plasmid (pZL465) were grown in SD medium with or without 0.02% leucine as indicated for at least six generations to reach an optical density at 600 nm of 0.5 to 0.8, and β-galactosidase activities were determined.
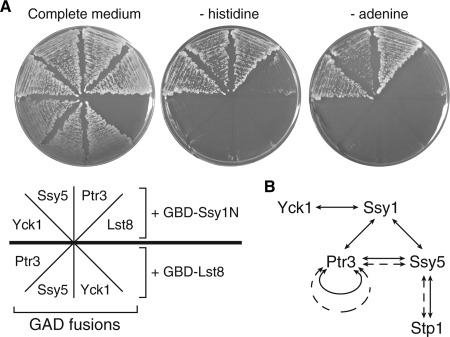
The signal transduction domain of Ssy1 interacts with both Ptr3 and Yck1.
Unlike bona fide amino acid transporters, Ssy1 has an N-terminal cytoplasmic extension, which has been proposed to function as a signal transduction domain (14, 17). To gain understanding of how Ssy1 transmits amino acid signals to downstream factors, we examined whether there exist interactions between Ssy1 and Ptr3, Ssy5, or Yck1, three factors localized close to the top of the signal transduction cascade. Our attempts to detect an interaction between Ssy1 and these three factors via immunoprecipitation were not successful (data not shown). Since Ssy1 is a multitransmembrane protein, this may have complicated our detection of interactions between Ssy1 and other factors, and we therefore employed a yeast two-hybrid assay to detect whether the signal transduction domain of Ssy1, Ssy1N, interacts with Ptr3, Yck1, and Ssy5. To this end, a yeast two-hybrid strain expressing a Gal4 DNA binding domain-Ssy1N fusion (GBD-Ssy1N) was crossed to another yeast two-hybrid strain expression Gal4 transcriptional activation domain (GAD) fused to Ptr3, Ssy5, or Yck1 or a control protein, Lst8, an essential WD repeat protein (28). The resultant diploid strains also carry GAL2-ADE2 and GAL1-HIS3 reporter genes. In the presence of a positive interaction, these diploid cells would be able to grow in medium without adenine or histidine. The coexpression of GBD-Ssy1N and GAD-Lst8 does not allow cells to grow in the absence of adenine or histidine (Fig. 8A). In contrast, the coexpression of GBD-Ssy1N, but not GBD-Lst8, with GAD-Ptr3, GAD-Ssy5, or GAD-Yck1 enables cells to grow in the absence of either adenine or histidine, suggesting interactions between Ssy1N and these three factors. We failed to detect a positive interaction between Ptr3 and Yck1 by two-hybrid analysis (data not shown), suggesting that Ssy1's N-terminal signal transduction domain may function by bringing Ptr3 and Yck1 together to allow Ptr3 to be hyperphosphorylated by Yck1. Figure 8B summarizes protein interactions among factors in the SPS signaling pathway from this study and others (3, 5). It is conceivable that the N-terminal signal transduction domain of Ssy1 functions by recruiting or activating Yck1 toward Ptr3, thus resulting in the hyperphosphorylation of Ptr3. This protein interaction network allows the transmission of amino acid signals from the amino acid sensor Ssy1 to the transcription factor Stp1.
FIG. 8.
Interactions between the N-terminal signal transduction domain of Ssy1, Ssy1N, and Ptr3, Yck1, and Ssy5. (A) A yeast two-hybrid analysis of protein interactions between Ssy1N and Ptr3, Ssy5, and Yck1. Yeast two-hybrid strains coexpressing GBD-Ssy1N or GBD-Lst8 and GAD-Lst8, Ptr3, Ssy5 or Yck1 were grown on medium as indicated. (B) A schematic representation of protein interactions in the amino acid-sensing pathway. Solid and dotted lines indicate protein interactions revealed by yeast two-hybrid analysis and coimmunoprecipitation, respectively.
DISCUSSION
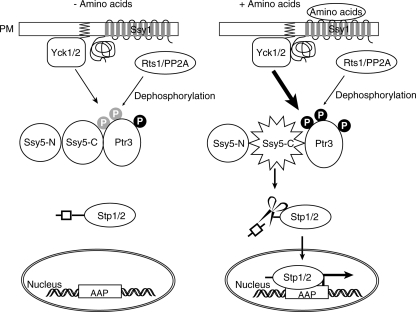
Our data establish a strong correlation between Ptr3 hyperphosphorylation and the activation of the SPS-sensing pathway. We propose that the phosphorylation of Ptr3 is a key regulatory point of SPS sensing. We base our proposal on the following observations. (i) An ssy1Δ mutation and a yck1 yck2 double mutation block both the hyperphosphorylation of Ptr3 and the activation of the SPS-sensing pathway. (ii) An rts1Δ mutation, which reduces PP2A phosphatase activity, increases Ptr3 hyperphosphorylation and results in the constitutive activation of SPS sensing. (iii) Mutations in threonine 525 of Ptr3 result in an inability to hyperphosphorylate Ptr3 and in a concomitant inactivation of SPS sensing by blocking the processing of Stp1. (iv) An activating mutant allele of PTR3, PTR3(Q439R), results in Ptr3 hyperphosphorylation. (v) A grr1Δ mutation blocks both pathway activation and the amino acid-induced hyperphosphorylation of Ptr3. Our results are summarized in a model diagrammed in Fig. 9. The binding of amino acids to Ssy1 leads to the recruitment of Yck1/2 through its N-terminal putative signal transduction domain to hyperphosphorylate Ptr3 in the Ptr3-Ssy5-C complex, and this results in the activation of the protease activity of Ssy5-C toward Stp1/2. The Rts1/PP2A phosphatase complex modulates the SPS-sensing pathway by dephosphorylating Ptr3.
FIG. 9.
A model for the regulation of the SPS-sensing pathway in yeast. Amino acid sensing is initiated by amino acid binding to Ssy1, which transmits signals through Ptr3 and Ssy5 and activates two transcription factors, Stp1 and Stp2, by enabling them to enter the nucleus. Once in the nucleus, Stp1/2 activate the expression of target genes encoding amino acid permeases (AAP). Ssy5, a protease, is constitutively processed into an N-terminal prodomain, Ssy5-N, and a C-terminal activity domain, Ssy5-C, which removes an N-terminal sequence from Stp1/2, allowing Stp1/2 to enter the nucleus. These two fragments of Ssy5 interact with each other after processing. Ptr3 constitutively interacts with Ssy5-C. In the absence of amino acids, Ptr3 is only partially phosphorylated (indicated by “P” in gray circles; “P” in black circles indicates robust phosphorylation). That form of Ptr3 is unable to activate the protease activity of Ssy5-C toward Stp1/2. Upon binding to amino acids, Ssy1 is likely to recruit Yck1/2, which is associated with the plasma membrane (PM) through prenylation (indicated by zigzag lines in PM), to hyperphosphorylate Ptr3. Hyperphosphorylated Ptr3 increases the Ssy5-C protease activity toward Stp1/2. The Rts1/PP2A phosphatase complex mediates SPS sensing by dephosphorylating Ptr3.
We propose that the hyperphosphorylation of Ptr3 is mediated by opposing activities from two CKI proteins, Yck1 and Yck2, and from the Rts1/PP2A phosphatase complex. Increased hyperphosphorylation of Ptr3 in cells grown in the presence of amino acids can be due to an increased activity of Yck1 and Yck2 toward Ptr3, a decreased activity of PP2A phosphatase, or some combination of these activities. Many transmembrane signaling pathways rely on receptor-mediated phosphorylation by recruiting pathway-specific kinases. CKI proteins Yck1 and Yck2 are known to be prenylated and associated with the plasma membrane (34). We propose that amino acid-bound Ssy1 increases its interaction with Yck1/2, which leads to the hyperphosphorylation of Ptr3. We have obtained a mutant form of Yck1 with mutations in its last six residues, including the last two cysteine residues, which serve as a prenylation motif, which in turn has been shown to be required for the plasma membrane localization of CKI proteins (34). This mutant is unable to complement a yck1 yck2 double mutation to activate SPS sensing (data not shown), suggesting that the plasma membrane localization of Yck1 is required for the phosphorylation of Ptr3.
The hyperphosphorylation of Ptr3 is dependent on Ssy1, but not on Ssy5, consistent with the current view that signals are transmitted in the order Ssy1-Ptr3-Ssy5 (5). Ptr3, independent of its phosphorylation status and of Ssy1, can form a complex with Ssy5. These findings suggest that activated Ssy1 can signal to the preexisting Ptr3/Ssy5 complex. How does Ptr3 hyperphosphorylation lead to pathway activation? We have demonstrated that Ptr3 hyperphosphorylation correlates with processing of Stp1 to its active form (Fig. 3C and D), which is believed to be due to the protease activity of Ssy5. Because Ptr3 hyperphosphorylation does not affect its interaction with Ssy5, we propose that a conformational change in the Ptr3-Ssy5 complex due to Ptr3 hyperphosphorylation results in Ssy5 activation. The activation of a protease toward its substrate by the phosphorylation of its interaction partner represents a mechanism for regulating protease activity that to our knowledge is novel. Among eight CKI consensus sites in Ptr3 we examined, only threonine 525 is essential for Ptr3 functionality. There are two distinct hyperphosphorylated forms of Ssy5, suggesting that the amino acid-induced hyperphosphorylation of Ptr3 involves at least two phosphorylation sites. The abolishment of both hyperphosphorylated forms of Ptr3 in threonine 525 mutants suggests that threonine 525 either serves as one of the Ptr3 phosphorylation sites, whose phosphorylation is a prerequisite for the phosphorylation of the second site, or is a critical residue required for phosphorylation of both sites. The correlation between failures to hyperphosphorylate Ptr3 due to mutations in threonine 525 or addition of a 3× myc tag to the C-terminal end of Ptr3 and Ptr3 inactivation strongly suggests that Ptr3 hyperphosphorylation plays an important role in pathway activation.
Yck1 and Yck2 play a redundant, essential role in yeast. This is not due to a block in the SPS-sensing pathway. Mutations in ssy1, ptr3, or ssy5 result in synthetic lethality with a leu2 mutation in cells grown on amino acid-rich medium. This synthetic lethal phenotype is not observed on minimal medium. However, on minimal medium, the yck1Δ yck2ts double mutant cells are still nonviable at a restrictive temperature (data not shown). Thus, Yck1/2 must have other target proteins whose phosphorylation is essential for cell viability. Our observation that an rts1Δ mutation can partially suppress cell growth defects observed for the yck1 yck2 double mutant suggests that PP2A phosphatase is likely to be involved in an essential cellular process that is disrupted in yck1 yck2 mutant cells. Std1 and Mth1, two negative regulators of the glucose signal transduction pathway, are phosphorylated by Yck1/2, but they are unlikely to be targets of the Rts1/PP2A phosphatase complex, because an rts1Δ mutation does not affect the expression of a reporter gene expression in the glucose signal transduction pathway.
The amino acid-sensing pathway and the glucose signal transduction pathway both employ sensors, which are homologous to functional nutrient transporters. Unlike bona fide amino acid or hexose transporters, these sensors have an extra cytoplasmic extension, functioning as a putative signal transduction domain. The glucose sensors Rgt2 and Snf3 have an extra C-terminal cytoplasmic domain (23), whereas Ssy1 has an N-terminal cytoplasmic extension (14). These domains may function similarly by recruiting Yck1/2 to phosphorylate downstream effectors, Std1 and Mth1, in the glucose signal transduction pathway and Ptr3 in the amino acid-sensing pathway. Although Yck1/2-dependent phosphorylation events lead to pathway activation of both glucose and SPS sensing, the underlying mechanisms differ: in the glucose signal transduction pathway, there is an inactivation of two negative regulators, whereas in the SPS-sensing pathway, a positive regulator is activated. Despite a common requirement of Yck1/2 for the activation of these two pathways, there seems to be no cross talk between them: amino acids are not sufficient to activate the glucose signal transduction pathway and glucose is not sufficient to activate SPS sensing, suggesting that the pathway-specific phosphorylation of downstream effectors by Yck1/2 lies with the glucose or amino acid sensors. Besides Yck1/2, Grr1 is another common factor shared by both pathways. For the glucose signal transduction pathway, it has been proposed that the SCFGrr1 E3 ubiquitin ligase is required to degrade Std1 and Mth1, thus activating the pathway. In the SPS-sensing pathway, Grr1 is required for amino acid-stimulated but not basal-level hyperphosphorylation of Ptr3. Based on multiple lines of evidence between Ptr3 hyperphosphorylation and pathway activation, a Grr1-dependent increase of Ptr3 hyperphosphorylation provides a good explanation as to how Grr1 functions as a positive regulator in this pathway. Skp1, a component of the SCFGrr1 E3 ubiquitin ligase, and Cdc34, a ubiquitin-conjugating enzyme, two factors generally required for the Grr1-dependent degradation of substrate proteins, are also required for the activation of SPS sensing (6). However, the leucine-rich repeat domain of Grr1, which is generally believed to be the substrate binding domain, and the proteasome function are dispensable for SPS sensing (1, 4, 32). Although there are two proteolytic events in the SPS-sensing pathway, the processing of Ssy5 is independent of Grr1, and the processing of Stp1/2 has been attributed to activated Ssy5. The lack of proteasome function in the activation of SPS signaling raises an interesting possibility that a hitherto unknown Grr1-mediated ubiquitination event plays a regulatory role rather than by mediating some target protein for degradation. Protein ubiquitination has been implicated in activating IκB kinase activity independent of proteasome function (33), raising the possibility that Grr1 may be required for activating Yck1/2 kinases toward Ptr3. Further studies will be required to determine the mechanism by which Grr1 functions as a positive regulator in the SPS-sensing pathway.
Supplementary Material
Acknowledgments
This work was supported by grants GM22525 from the NIH (to R.A.B.) and I-0642 from The Robert A. Welch Foundation (to R.A.B.).
We thank Per O. Ljungdahl, Lucy C. Robinson, and Michael Snyder for yeast strains and members of the Butow laboratory for helpful discussions.
Footnotes
Published ahead of print on 5 November 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
We dedicate this paper to the memory of Ronald A. Butow (1936-2007).
REFERENCES
- 1.Abdel-Sater, F., M. El Bakkoury, A. Urrestarazu, S. Vissers, and B. Andre. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 249771-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Andreasson, C., S. Heessen, and P. O. Ljungdahl. 2006. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 201563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 163158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, F., and B. Andre. 2001. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 41489-502. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, F., and B. Andre. 2001. Ubiquitin and the SCF(Grr1) ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 49681-85. [DOI] [PubMed] [Google Scholar]
- 7.Capasso, H., C. Palermo, S. Wan, H. Rao, U. P. John, M. J. O'Connell, and N. C. Walworth. 2002. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J. Cell Sci. 1154555-4564. [DOI] [PubMed] [Google Scholar]
- 8.Didion, T., B. Regenberg, M. U. Jorgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27643-650. [DOI] [PubMed] [Google Scholar]
- 9.Eckert-Boulet, N., K. Larsson, B. Wu, P. Poulsen, B. Regenberg, J. Nielsen, and M. C. Kielland-Brandt. 2006. Deletion of RTS1, encoding a regulatory subunit of protein phosphatase 2A, results in constitutive amino acid signaling via increased Stp1p processing. Eukaryot. Cell 5174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flotow, H., and P. J. Roach. 1991. Role of acidic residues as substrate determinants for casein kinase I. J. Biol. Chem. 2663724-3727. [PubMed] [Google Scholar]
- 11.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 4091-109. [DOI] [PubMed] [Google Scholar]
- 13.Holsbeeks, I., O. Lagatie, A. Van Nuland, S. Van de Velde, and J. M. Thevelein. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29556-564. [DOI] [PubMed] [Google Scholar]
- 14.Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles, A. Urrestarazu, and B. Andre. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karin, M., and T. Hunter. 1995. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr. Biol. 5747-757. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J. H., V. Brachet, H. Moriya, and M. Johnston. 2006. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot. Cell 5167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 195405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine, A. J., Z. Feng, T. W. Mak, H. You, and S. Jin. 2006. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 20267-275. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Z., T. Sekito, M. Spirek, J. Thornton, and R. A. Butow. 2003. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol. Cell 12401-411. [DOI] [PubMed] [Google Scholar]
- 20.MacDonald, P. E., J. W. Joseph, and P. Rorsman. 2005. Glucose-sensing mechanisms in pancreatic beta-cells. Philos. Trans. R. Soc. Lond. B 3602211-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 2981912-1934. [DOI] [PubMed] [Google Scholar]
- 22.Moriya, H., and M. Johnston. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 1011572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozcan, S., J. Dover, and M. Johnston. 1998. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 172566-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozcan, S., J. Dover, A. G. Rosenwald, S. Wolfl, and M. Johnston. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 9312428-12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panek, H. R., J. D. Stepp, H. M. Engle, K. M. Marks, P. K. Tan, S. K. Lemmon, and L. C. Robinson. 1997. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 164194-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poulsen, P., L. Lo Leggio, and M. C. Kielland-Brandt. 2006. Mapping of an internal protease cleavage site in the Ssy5p component of the amino acid sensor of Saccharomyces cerevisiae and functional characterization of the resulting pro- and protease domains by gain-of-function genetics. Eukaryot. Cell 5601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulsen, P., B. Wu, R. F. Gaber, and M. C. Kielland-Brandt. 2005. Constitutive signal transduction by mutant Ssy5p and Ptr3p components of the SPS amino acid sensor system in Saccharomyces cerevisiae. Eukaryot. Cell 41116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberg, K. J., S. Bickel, N. Rowley, and C. A. Kaiser. 1997. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics 1471569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson, L. C., E. J. Hubbard, P. R. Graves, A. A. DePaoli-Roach, P. J. Roach, C. Kung, D. W. Haas, C. H. Hagedorn, M. Goebl, M. R. Culbertson, et al. 1992. Yeast casein kinase I homologues: an essential gene pair. Proc. Natl. Acad. Sci. USA 8928-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 2769583-9586. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, M. C., R. R. McCartney, X. Zhang, T. S. Tillman, H. Solimeo, S. Wolfl, C. Almonte, and S. C. Watkins. 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 194561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spielewoy, N., K. Flick, T. I. Kalashnikova, J. R. Walker, and C. Wittenberg. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 248994-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, L., and Z. J. Chen. 2004. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16119-126. [DOI] [PubMed] [Google Scholar]
- 34.Vancura, A., A. Sessler, B. Leichus, and J. Kuret. 1994. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J. Biol. Chem. 26919271-19278. [PubMed] [Google Scholar]
- 35.Wang, P. C., A. Vancura, T. G. Mitcheson, and J. Kuret. 1992. Two genes in Saccharomyces cerevisiae encode a membrane-bound form of casein kinase-1. Mol. Biol. Cell 3275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26283-289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.