Energy conservation in chemotrophic organisms is generally coupled to redox reactions in catabolic pathways. In the oxidative part or branch, “energy-rich” compounds are formed, from which ATP is generated via substrate-level phosphorylation (SLP). In the reductive branch the electron carriers are reoxidized by a terminal acceptor; in this way an electrochemical ion gradient (ΔμH+ or ΔμNa+) at the cytoplasmic membrane is established, which is used for ATP synthesis, transport across membranes, and motility. This second type of energy conservation is called respiration or electron transport phosphorylation (ETP). Bacterial fermentations are considered apparent exceptions to this generalization because they are thought to lack ETP (41). In these fermentations the substrate serves not only as an electron donor but also as a terminal acceptor, since oxygen, nitrate, fumarate, etc. are absent (44). An example is the fermentation of glutamate via 3-methylasparate by the closely related anaerobic bacteria Clostridium tetani, Clostridium tetanomorphum, and Clostridium pascui to ammonia, acetate, butyrate, and molecular hydrogen according to equation 1 (Fig. 1) (8, 16, 51):
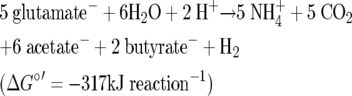 |
(1) |
In the first part of this pathway, glutamate is converted to ammonia, acetate, and pyruvate, which is oxidized by ferredoxin and coenzyme A (CoA) to acetyl-CoA and CO2. In order to regenerate the oxidant, acetyl-CoA and protons are reduced to butyryl-CoA and hydrogen, respectively. From 2 butyryl-CoA and 1 acetyl-CoA, 3 ATP are obtained via SLP. But the free enthalpy required to synthesize 1 ATP (−317/3 = −106 kJ mol−1) is still much higher than that needed in other systems; i.e., the efficiency is low (η = 42%). Usually, −75 ± 5 kJ mol−1 (η = 60%) is considered the minimum free enthalpy for ATP synthesis (44). It should be noted that without hydrogen formation the ATP yield would drop to 2.5 mol ATP (5 mol glutamate)−1, because in order to maintain the redox balance, acetyl-CoA has to be completely reduced to butyrate. Thus, hydrogen formation increases SLP. If all the reducing equivalents dissipated as hydrogen and no butyrate was formed, the yield would rise to 5 mol ATP (5 mol glutamate)−1. This, however, is thermodynamically not possible (−46 kJ mol glutamate−1).
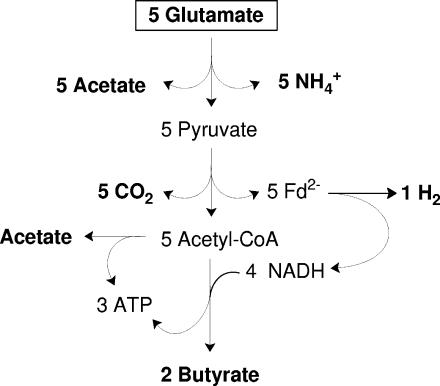
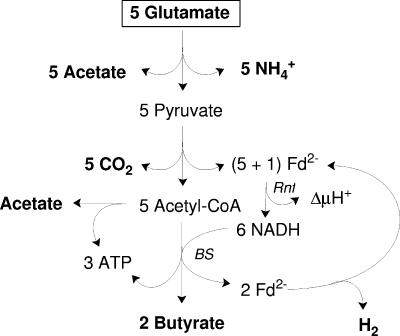
FIG. 1.
Traditional scheme of glutamate fermentation via 3-methylaspartate in C. tetanomorphum. Five pyruvates derived from 5 glutamates are oxidized by 5 ferredoxins to 5 CO2 and 5 acetyl-CoA. The 5 reduced ferredoxins (Fd2−) formed are reoxized by 2 H+ and 4 NAD+, yielding 1 H2 and 4 NADH, respectively. Four of the 5 acetyl-CoA formed are reduced by 4 NADH to 2 butyryl-CoA (see Fig. 2). Three ATP are generated from 2 butyryl-CoA and the remaining 1 acetyl-CoA (see Fig. 5).
The oxidation of pyruvate derived from glutamate yields reduced ferredoxin (E0′ ≤ −420 mV), whereas NADH (E0′ = −320 mV) is the reductant in butyrate synthesis. The difference (≥100 mV) could be used for additional energy conservation. Recently, we discovered the enzyme that catalyzes the reduction of NAD+ with reduced ferredoxin (Fig. 1) and characterized it as an Rnf-type NADH ferredoxin oxidoreductase localized in the membrane of C. tetanomorphum. Although it has not been demonstrated yet that this clostridial Rnf protein pumps protons or sodium ions, its cellular localization and the homology of four of its six subunits with four of the subunits of NADH-quinone reductase (Nqr) from Vibrio alginolyticus and Vibrio cholerae strongly suggest that this enzyme is involved in energy conservation via an electrochemical ion gradient (5). Furthermore, the designation Rnf is derived from “Rhodobacter capsulatus nitrogen fixation,” and in this species it most likely catalyzes the NADH-dependent reduction of ferredoxin driven by ΔμH+, also called reversed electron transfer (see below). Taking the Rnf-like NADH-ferredoxin oxidoreductase from C. tetanomorphum into consideration, the yield of conserved energy from the fermentation of glutamate would significantly improve (5, 19, 24; E. Jayamani, C. D. Boiangiu, and W. Buckel, unpublished results). A further improvement (up to 0.9 mol ATP mol glutamate−1) is proposed below. Taken together, these findings are an example of energy conservation by anaerobic respiration in fermentative bacteria. It should be noted that the clostridium-type ferredoxins, like the ferredoxin from Acidaminococcus fermentans (43), contain two [4Fe-4S]1+/2+ clusters with somewhat different redox potentials (E0′ = −405 mV and E0′ = −340 mV). Depending on the partial pressure of the hydrogen produced, either the more negative cluster or both clusters could be involved in electron transfer. For the sake of simplicity we assume that upon reduction oxidized ferredoxin (Fd) accepts two electrons, resulting in Fd2−.
Here we propose how one step in the synthesis of butyrate, the NADH-dependent reduction of crotonyl-CoA (2-butenoyl-CoA) to butyryl-CoA, could be used for additional energy conservation. Anaerobic bacteria produce crotonyl-CoA either by fermentation of glutamate, lysine, threonine, and methionine or by synthesis from two acetyl-CoA molecules (Fig. 2) derived from glucose or from glutamate via 3-methylasparate as indicated above (7). The reduction of crotonyl-CoA (E0′ = −10 mV) by NADH (E0′ = −320 mV) is highly exergonic and irreversible under physiological conditions (ΔG°′ = −60 kJ mol−1). Therefore, it has been suggested that this reaction could be involved in energy conservation (41), like the combined action of the membrane enzymes NADH-quinone oxidoreductase (complex I) and fumarate reductase, which generate ΔμH+ via the reduction of fumarate (E0′ = 30 mV) by NADH (ΔG°′ = −68 kJ mol−1) (26). Ongoing experiments in our laboratory have demonstrated, however, that the reduction of crotonyl-CoA is catalyzed by a single soluble enzyme closely related to the acryloyl-CoA reductase complex (18). The flavin adenine dinucleotide (FAD)-containing crotonyl-CoA reductases from C. tetanomorphum and C. pascui are composed of three different subunits with the probable stoichiometry α2βγ. N-terminal sequencing identified the α-subunits (40 kDa) as butyryl-CoA dehydrogenase (Bcd) and the β-subunit (36 kDa) and γ-subunit (28 kDa) as the subunits of an electron-transferring flavoprotein (Etf). As determined by gel filtration, the whole Etf-Bcd complex had a molecular mass of 360 kDa, suggesting that there was a dimer of the tetramer. Furthermore, polyclonal antibodies raised against the Etf-Bcd complex in combination with gold labeling and electron microscopy showed that the enzyme was evenly distributed in the whole cytoplasm and revealed no localization at the cytoplasmic membrane (G. Herrmann, E. Mörschel, and W. Buckel, unpublished data). These preliminary results indicated that thte Etf-Bcd complex is unable to conserve energy via ΔμH+ and prompted us to propose a novel but indirect type of energy conservation in clostridia, which also may be applied to other bacteria.
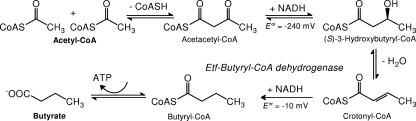
FIG. 2.
Butyrate synthesis from 2 acetyl-CoA in members of the genus Clostridium.
Acyl-CoA dehydrogenases have been characterized mainly from mammals. They are homotetrameric enzymes with one FAD per monomer as a prosthetic group (25). They catalyze the oxidation of acyl-CoA to E-enoyl-CoA with a separate Etf as an electron acceptor in vivo (Fig. 3). In vitro, the best acceptor is ferricenium hexafluorophosphate, which is reduced by one electron to ferrocene (27). The only well-studied and structurally characterized bacterial enzyme is butyryl-CoA dehydrogenase from Megasphaera elsdenii (in the family Acidaminococcaceae the order Clostridiales) (25). This enzyme catalyzes the NADH-dependent reduction of crotonyl-CoA mediated by Etf (Fig. 3), which also has been purified from this organism (33). Notably, in the related anaerobe A. fermentans (in the family Acidaminococcaceae) butyryl-CoA dehydrogenase and Etf are also separate enzymes (52). In members of the genus Clostridium, however, acyl-CoA dehydrogenase and Etf form a stable complex (18).
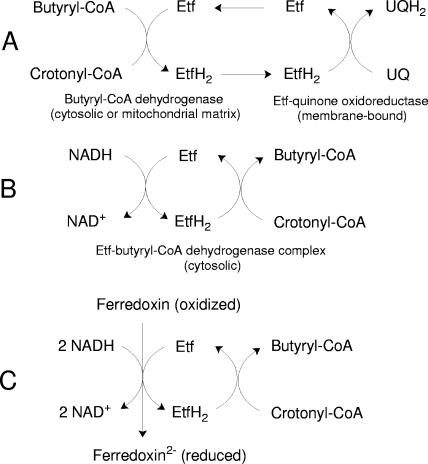
FIG. 3.
Different functions of Etf. (A) In aerobic organisms Etf transfers the electrons from butyryl-CoA dehydrogenase in the cytoplasm or mitochondrial matrix to ubiquinone (UQ), which is mediated by the integral membrane enzyme Etf-quinone oxidoreductase. (B) In anaerobic bacteria Etf transfers the electrons from NADH to the cytosolic butyryl-CoA dehydrogenase, which reduces crotonyl-CoA. (C) Additional reduction of ferredoxin by the Etf-butyryl-CoA dehydrogenase complex as proposed in this work.
The Etfs from anaerobic bacteria and from mammals both interact with acyl-CoA dehydrogenases, but they have different functions (Fig. 3). The mammalian protein transports electrons from the dehydrogenase to the mitochondrial membrane enzyme Etf-quinone oxidoreductase, which delivers them to ubiquinone in the respiratory chain (4). The bacterial protein is reduced by NADH and guides the electrons to the acyl-CoA dehydrogenase and to ferredoxin, as hypothesized below. The crystal structure of human Etf revealed that FAD and AMP are prosthetic groups (35). In contrast, M. elsdenii Etf contains 2 mol of FAD per heterodimer but no AMP; one FAD is tightly bound to the protein, whereas the other appears to readily dissociate from the enzyme and has unusual spectral properties (37). Similarly, reduction of crotonyl-CoA by NADH mediated by the clostridial complexes requires addition of FAD; this is not the case for the ferricenium-dependent acyl-CoA dehydrogenase activity of these enzymes, in which neither of the Etf subunits is involved.
HYPOTHESIS: FERREDOXIN REDUCTION COUPLED TO CROTONYL-CoA REDUCTION
We propose that Etf-Bcds from anaerobic bacteria are able to bifurcate the two electrons from NADH (E0′ = −320 mV); one electron proceeds to the more positive electron acceptor butyryl-CoA dehydrogenase and finally to crotonyl-CoA (E0′ = −10 mV), and the other electron is transported in the reverse direction to the more negative acceptor ferredoxin (Fd → Fd−) or flavodoxin (E0′ ≤ −420 mV). The next NADH delivers the second electron to complete the reduction of crotonyl-CoA to butyryl-CoA and the reduction of Fd− to Fd2−. Hence, the exergonic reduction of crotonyl-CoA drives the endergonic reduction of ferredoxin (equation 2) (Fig. 3). This could be achieved by the prosthetic group FAD, which is reduced by NADH to the hydroquinone form. One electron is then transferred to the special FAD and further to ferredoxin, whereas the other is captured by the dehydrogenase. The reduced ferredoxin obtained in this way can be reoxidized either by NAD+ (catalyzed by Rnf, with generation of ΔμH+ or ΔμNa+) or by protons to form molecular hydrogen, mediated by an [Fe-Fe]-hydrogenase. This mode of hydrogen generation saves crotonyl-CoA as an electron acceptor and leaves more of this intermediate for β-oxidation to two acetyl-CoA molecules, which increases the ATP yield via SLP, as mentioned above.
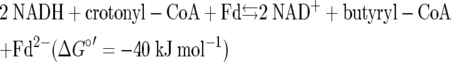 |
(2) |
This mechanism recalls the so-called “archerases,” which raise the energy of an electron to a more negative redox potential by hydrolysis of ATP. Two different types of these proteins are found in nature; the G-protein-related type is involved in nitrogen fixation or chlorophyllide reduction, whereas the other type, belonging to the ASKHA protein family, activates (R)-2-hydroxyacyl-CoA dehydratases or overcomes the resonance energy of benzoyl-CoA during reduction to cyclohexadiene carboxyl-CoA. These enzymes are homodimeric proteins with two ATP-binding sites and one [4Fe-4S]1+/2+ cluster in between (E0′, ca. −350 mV), which is readily reduced by ferredoxin. Hydrolysis of 2 ATP should decrease the redox potential of the cluster, probably to less than −800 mV (9).
Experimental evidence confirming the hypothesis that there is electron bifurcation in clostridial Etfs comes from work of R. K. Thauer and K. Jungermann. This research team discovered that cell extracts from Clostridium kluyveri or Clostridium pasteurianum catalyzed the formation of hydrogen from NADH provided that ferredoxin and acetyl-CoA were present (22, 23, 46). The concentrations of NADH and acetyl-CoA were kept constant using the NAD+/galactose/galactose dehydrogenase and acetylphosphate/reduced CoA/phosphate acetyltransferase regenerating systems, respectively. Thauer and Jungermann, who were aware that reduction of ferredoxin by NADH is a thermodynamically unfavorable reaction, concluded that acetyl-CoA acted as a highly specific activator of this process, because the equally “energy-rich” compounds propionyl-CoA and formyl-CoA were ineffective. Therefore, the possibility of exergonic synthesis of butyryl-CoA from two acetyl-CoA was not taken into consideration. These results can now be readily explained by our hypothesis. In the cell extract, which contained all of the soluble enzymes, 2 acetyl-CoA were converted by 2 NADH to butyryl-CoA (ΔG°′ = −57 kJ mol−1) (Fig. 2), and the free enthalpy change powered the reduction of ferredoxin. Obviously, the enzymes involved in butyrate synthesis do not use propionyl-CoA or formyl-CoA as a substrate. On the other hand, the intermediates of this pathway, acetoacetyl-CoA, (S)-3-hydroxybutyryl-CoA, and crotonyl-CoA, were not tested. In addition to hydrogenase and ferredoxin, the system is composed of at least four enzymes. Therefore, all attempts to purify this apparent NADH-ferredoxin reductase as a single protein were unsuccessful. The fact that the pure Etf-butyryl-CoA dehydrogenase complex from C. kluyveri uses NADH to reduce crotonyl-CoA together with ferredoxin is demonstrated in the accompanying paper (28).
ENHANCEMENT OF EITHER SLP OR ETP BY BUTYRATE SYNTHESIS
Ferredoxin reduction by NADH coupled to butyrate synthesis nicely explains catabolism in several anaerobic bacteria. C. pasteurianum ferments glucose to acetate, butyrate, CO2, and H2 according to equation 2, and 3.3 ATP/glucose is formed via SLP (equation 3) (23).
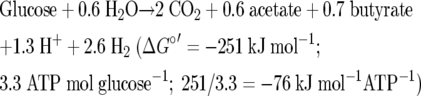 |
(3) |
In this fermentation glucose is converted via the Embden-Meyerhof-Parnas pathway to 2 pyruvates concomitant with the formation of 2 ATP and 2 NADH. Pyruvate is then oxidized by ferredoxin to CO2 and acetyl-CoA. For reoxidation of NADH butyrate is synthesized (equation 3), which results in formation of additional reduced ferredoxin. Thus, 2.0 H2 results from the oxidation of pyruvate and the remaining 0.6 H2 from the reduction of crotonyl-CoA. The 0.7 butyryl-CoA formed and the residual 0.6 acetyl-CoA give rise to 1.3 ATP via SLP. The ratio of butyrate to the remaining H2 (0.7/0.6) is close to 1.0, and this indicates that crotonyl-CoA reductase indeed diverts the two electrons from NADH, one to crotonyl-CoA and one to ferredoxin according to equation 2.
In contrast to C. pasteurianum, Clostridium aminobutyricum (15) and Fusobacterium nucleatum (21) produce butyrate without hydrogen formation. C. aminobutyricum ferments 4-aminobutyrate (γ-aminobutyrate) via dehydration of 4-hydroxybutyryl-CoA to crotonyl-CoA and finally to 1 acetate and 0.5 butyrate according to equation 4, and 1 ATP is formed by SLP (30).
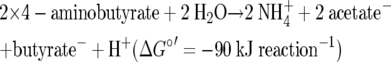 |
(4) |
(The ΔGf° of 4-aminobutyrate, which was not found in previous publications, was calculated to be −362 kJ mol−1 from the decarboxylation of glutamate, assuming that ΔG°′ is −20 kJ mol−1; ΔGf° is the free enthalpy of formation of a compound from elements.)
Growth experiments with C. aminobutyricum yielded 7.6 mg (dry weight) cells (mmol 4-aminobutyrate)−1, which is more than the expected yield, 5 mg mmol−1 calculated from a YATP of 10 (10 g [dry weight] cells mol ATP−1) (41). (Note that a YATP of 10 is an average value based on many fermentations.) When acetate and/or CO2 rather than C3 and C4 building blocks requiring less ATP are the carbon sources, the YATP is <10 (42), whereas a YATP of >10 is certainly due to additional energy-conserving processes, as discussed here. The conservation of more than 1.0 ATP equivalent from two 4-aminobutyrate is supported by thermodynamic data, because the −90 kJ mol butyrate−1 released in 4-aminobutyrate fermentation is sufficient for about 1.3 ATP. The hypothesis presented in this paper can explain this observation. Reduction of crotonyl-CoA to butyryl-CoA gives rise to reduced ferredoxin, which in C. aminobutyricum and F. nucleatum is not used for H2 formation but may be reoxidized by NAD+ catalyzed by Rnf. Thus, cycling of NADH conserves additional energy via ΔμH+ or ΔμNa+. The detection of high membrane-bound NADH dehydrogenase (Rnf) activities in C. aminobutyricum and F. nucleatum confirms this conclusion (unpublished results).
F. nucleatum has a more complex catabolism (equation 5 and Fig. 4 without H2 production) because it conserves additional energy from decarboxylation of glutaconyl-CoA via ΔμNa+ (3). Two glutamates give rise to 1 ATP via SLP, 0.25 ATP via ΔμH+ mediated by Rnf plus Etf-Bcd, and 2 × 0.25 ATP via ΔμNa+ mediated by glutaconyl-CoA decarboxylase. Thus, the energy conserved from 2 glutamates amounts to about 1.75 ATP, which according to equation 5 is thermodynamically possible: 2 × (−67) × 1.75−1 = −77 kJ mol ATP−1.
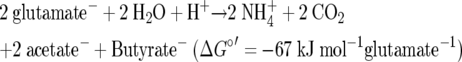 |
(5) |
The comparison of C. pasteurianum with C. aminobutyricum or F. nucleatum showed that there are two ways to increase the conserved energy; either SLP is increased by H2 production, or ΔμH+ is enhanced by recycling NADH. Therefore, the choice between hydrogenase and Rnf or both depends on the enzymes in the organism. The fermentation of glutamate by A. fermentans and Clostridium symbiosum (equation 1 and Fig. 4) is an example in which SLP is increased by H2 production and ΔμNa+ and ΔμH+ are formed.
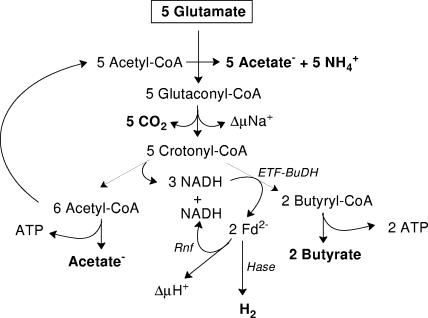
FIG. 4.
Hydroxyglutarate pathway of glutamate fermentation proposed here. Note that 3 of 5 crotonyl-CoA are oxidized to 6 acetyl-CoA by 3 NAD+. Two of the 3 NADH formed and the 1 NADH recycled are used to reduce 2 crotonyl-CoA to 2 butyryl-CoA, which drives the reduction of 2 ferredoxins by the other 2 NADH. One ferredoxin reduces protons to H2, and the other reduces NAD to NADH, which is recycled. Hase = hydrogenase (Hyd); ETF-BuDH, Etf-Bcd complex; Fd2−, reduced ferredoxin.
ENERGY CONSERVATION IN GLUTAMATE-FERMENTING HYDROGENIC BACTERIA
Glutamate is fermented by hydrogenic bacteria by two different pathways, but identical products are formed; one pathway occurs in closely related members of the genus Clostridium and involves (2S,3S)-3-methylasparate (Fig. 1 and 5), and the other involves (R)-2-hydroxyglutarate (Fig. 4) and is used by a more diverse group of anaerobes (8). The latter pathway is almost identical to that in F. nucleatum, but the additional hydrogen production changes the acetate/butyrate ratio from 2 to 3. In this pathway (R)-2-hydroxyglutaryl-CoA is dehydrated to glutaconyl-CoA and decarboxylated to crotonyl-CoA. NADH is formed by oxidation of (S)-3-hydroxybutyryl-CoA to acetoacetyl-CoA and is used for reduction of crotonyl-CoA to butyryl-CoA (Fig. 4). This leads to 2 acetates and 1 butyrate, and the additional thioester bond generated in the thiolase reaction gives rise to 0.5 ATP/glutamate (equation 5). The formation of hydrogen via reduced ferredoxin from the reduction of crotonyl-CoA changes the acetate/butyrate ratio from 2 to 3 and increases the ATP yield via SLP from 0.5 to 0.6 ATP/glutamate. In a previous paper we speculated that ferredoxin is reduced with NADH with mediation by the Rnf-related ferredoxin NAD+ reductase (5). However, this reduction would require energy via ΔμNa+ generated by the Na+ pump glutaconyl-CoA decarboxylase (50), which is available for membrane processes. If one assumes that Etf-butyryl-CoA dehydrogenase bifurcates the electrons from NADH to ferredoxin and crotonyl-CoA (equation 2), then only one half of the reduced ferredoxin would be required for hydrogen production, whereas the other half could be used to recycle NADH mediated by Rnf. The total energy conserved from 5 glutamates would amount to 3 ATP via SLP, 5 × 0.25 ATP via decarboxylation of glutaconyl-CoA catalyzed by the membrane-bound Na+ pump, and an additional 0.25 ATP via Rnf, which would lead to 0.9 mol ATP mol glutamate−1 and to a requirement for 63.4 kJ mol glutamate−1 (70 kJ mol ATP−1). Thus, respiration via Etf-Bcd and Rnf increases the total ATP yield by 50% to the highest value that is thermodynamically possible.
FIG. 5.
Methylasparate pathway of glutamate fermentation proposed here (see Fig. 1). Note that 4 acetyl-CoA are reduced by 4 NADH to 2 butyryl-CoA; the fifth NADH and the 1 recycled NADH are used to reduce 2 ferredoxins, driven by the reduction of crotonyl-CoA to butyryl-CoA. BS, butyrate-synthesizing enzymes, including the Etf-Bcd complex; Fd2−, reduced ferredoxin.
In the fermentation of glutamate via 3-methylaspartate by C. tetanomorphum, C. pascui, and C. tetani, no Na+-pumping decarboxylase is present (Fig. 1), but in this pathway Rnf is much more important (Fig. 5). Here reduced ferredoxin, obtained by oxidation of pyruvate, is funneled into Rnf, forming NADH for butyrate synthesis, which produces additional reduced ferredoxin. One half of this extra reduced ferredoxin is used for H2 production, whereas the other half recycles NADH. The efficiency is identical to that of the 2-hydroxyglutarate pathway: 5 glutamates yield 3 ATP via SLP and 1.5 ATP equivalents (6 × 0.25) by the formation of 6 NADH via Rnf (0.9 mol ATP mol−1 glutamate).
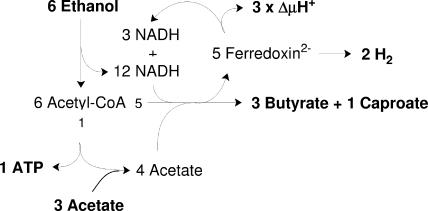
ENERGY CONSERVATION IN C. KLUYVERI
C. kluyveri thrives on the synthesis of butyrate and caproate from acetate and ethanol, approximately according to equation 6 (Fig. 6):
 |
(6) |
Previously, the energy metabolism of C. kluyveri has been explained by the exergonic synthesis of 3 butyrates and 1 caproate from 3 + 1 acetates and 5 ethanols, which drives the endergonic oxidation of the sixth ethanol to 2 H2, 1 acetate, and 1 ATP. It has been assumed that the two processes are coupled by ΔμH+ (34), but all the enzymes thought to be involved in this process are soluble. The new hypothesis presented here readily clarifies this 40-year-old enigma. The two processes are not separated. Reduced ferredoxin is generated during reduction of crotonyl-CoA and caprenoyl-CoA (hex-2-enoyl-CoA) by NADH (Fig. 3). Part of the ferredoxin (40%) reduces protons to hydrogen, and 60% recycles NADH. In this way 1.0 ATP via SLP and 3 × 0.25 ATP via Rnf are conserved, which amounts to 1.75 ATP per reaction or 105 kJ mol ATP−1. Growth experiments revealed the formation of 9.2 mg (dry weight) of cells per 2 mmol H2, from which 1.0 ATP per reaction (equation 6) was calculated, using a YATP of 9 (45). Since one-third of the cell carbon of C. kluyveri comes from CO2 and two-thirds comes from acetate, YATP is certainly less than 9 (42) (see above). A value of 5.3 would be in perfect agreement with the calculated 1.75 ATP per reaction.
FIG. 6.
Proposed catabolism of C. kluyveri. Substrates and products are indicated by bold type.
REDUCTION OF ACRYLOYL-CoA TO PROPIONYL-CoA IN C. PROPIONICUM
Clostridium propionicum thrives on the fermentation of alanine to ammonia, CO2, acetate, and propionate (2) (equation 7).
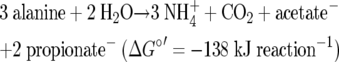 |
(7) |
The free enthalpy of reaction 7 conserves up to 2 ATP. However, SLP gives rise to only 1 ATP via acetyl-CoA, which is derived via pyruvate in the oxidative branch. In the reductive branch acryloyl-CoA generated from the dehydration of lactyl-CoA is reduced to propionyl-CoA. The enzyme, which, similar to Etf-Bcd, has been characterized as the Etf/propionyl-CoA dehydrogenase complex (18), should catalyze the reduction of acryloyl-CoA and the reduction of ferredoxin by NADH. Together with the reduced ferredoxin from pyruvate ferredoxin oxidoreductase, up to 3 × 0.25 ATP (i.e., 0.75 ATP) could be conserved via Rnf, since no hydrogen is produced. However, the presence of Rnf in this organism has yet to be established. Hence, up to 1.75 ATP could be conserved, leading to −79 kJ mol ATP−1. Growth yield studies with Clostridium homopropionicum, which uses the same pathway, suggested, however, that the organism sacrifices extra ATP in order to have a higher growth rate (40).
CAFFEATE REDUCTION IN A. WOODII
Acetobacterium woodii thrives on respiration using H2 as the donor and caffeate [3-(3,4-dihydroxyphenyl)acrylate] as the acceptor. The sodium ion has been established as the coupling factor between respiration and ATP synthesis, but how ΔμNa+ is generated has remained an enigma (20). Recently, an Rnf-related ferredoxin-NAD+ reductase was detected in this organism, and the enzyme catalyzing the reduction of caffeate was characterized as an NADH-dependent Etf-Bcd-related caffeoyl-CoA reductase (19). Hence, energy can be conserved from reduced ferredoxin generated directly from hydrogen and by reduction of caffeoyl-CoA to 3-(3,4-dihydroxyphenyl)propionyl-CoA, assuming that the mechanism for the reductase is identical to that of Etf-Bcd.
SYNTROPHIC OXIDATION OF BUTYRATE TO ACETATE
The genome of Syntrophus aciditrophicus contains genes encoding all the enzymes necessary for activation of butyrate and β-oxidation of butyryl-CoA to acetate, including acyl-CoA dehydrogenase and both subunits of Etf (31). The oxidant, however, must be NAD+ rather than a quinone, because NADH is the precursor of hydrogen, although the pressure is very low (ca. 1 Pa). Thus, the problem of how butyryl-CoA is oxidized with NAD+ arises. An aerobic pathway via quinone followed by reverse electron transport appears to be very unlikely because only menaquinone (E0′ = −74 mV) is present in S. aciditrophicus and NADH-ubiquinone oxidoreductase (complex I) is missing. A solution to this problem could be the reverse of equation 2, the NAD+-dependent oxidation of butyryl-CoA driven by reduced ferredoxin. Although this is still an endergonic process, the concentrations of the substrates and products could deviate far from standard conditions, yielding a negative ΔG′, since syntrophism requires only 1 Pa H2 (E0′ = −265 mV). The reduced ferredoxin involved in the oxidation of butyryl-CoA could be provided by NADH mediated by Rnf and driven by ΔμH+/Na+ in the reverse direction.
These possibilities agree well with previous observations made with whole-cell suspensions of Syntrophomonas wolfei grown on either butyrate or crotonate (48). Upon incubation with 20 mM butyrate, butyrate-grown cells produced up to 7 Pa H2, which could be completely inhibited by addition of the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) or preincubation with the FoF1 ATPase inhibitor N,N′-dicyclohexylcarbodiimide (DCCD). In contrast, no inhibition of H2 formation by CCCP or DCCD was observed with crotonate-grown cells when they were incubated with crotonate. As proposed above, the oxidation of butyryl-CoA to crotonyl-CoA requires ferredoxin reduced by NADH, which is catalyzed by the ΔμH+/Na+-driven Rnf. Thus, CCCP uncouples Rnf, and DCCD inhibits the formation of ΔμH+/Na+ from ATP, which is obtained by SLP from acetyl-CoA. On the other hand, the disproportionation of crotonate via crotonyl-CoA to butyrate and acetate generates reduced ferredoxin without involvement of ΔμH+/Na+ and ATPase.
NITROGEN FIXATION
Flavoproteins related to Etf with NADH as a reductant not only are present in anaerobes but also have been detected in aerobic or phototrophic nitrogen-fixing bacteria. Nitrogen fixation requires a more powerful reductant than NADH, either reduced ferredoxin or flavodoxin. As shown above, clostridia and other anaerobes have no problem with providing these agents, but nitrogen-fixing aerobes and phototrophs do have difficulties. One solution is Rnf from R. capsulatus, which is thought to catalyze the NADH-dependent reduction of ferredoxin driven by ΔμH+ (36, 38). Rhodospirillum rubrum, however, lacks the rnfABCDFG genes, but it contains the fixABCX cluster that is also present in several other diazotrophic bacteria (11-13). FixAB is related to Etf, and FixCX is related to Etf-quinone reductase. The function of the FixABCX proteins is unknown, but it has been proposed that they form a complex attached to the cytoplasmic membrane and catalyze the electron transfer from NADH to nitrogenase concomitant with respiration. Furthermore, it has been shown that deletion of fixC abolishes nitrogen fixation (13). In view of the main proposal in this work, we hypothesized that the electrons from NADH are again bifurcated by FixAB; one electron goes to ferredoxin and then to nitrogen fixation, whereas the other is captured via FixCX by ubiquinone in the respiratory chain. Hence, the endergonic reduction of ferredoxin by NADH is driven by the exergonic oxidation of NADH via the respiratory chain. The nitrogenases of all bacteria and archaea, including both the iron and molybdenum-iron proteins, appear to have a common origin, but the sources of the electrons are very diverse. Nature developed at least four methods to reduce ferredoxin, all of which are employed by nitrogen-fixing organisms: the Etfs described in this work, the Rnf proteins, the 2-oxoacid-ferredoxin oxidoreductases, and molecular hydrogen together with hydrogenase.
HISTORICAL CONSIDERATIONS
Until the mid-1960s, the ethanol-acetate fermentation of C. kluyveri challenged Lipmann's ingenious concept of “energy-rich” compounds (29), because workers had no idea how this organism might conserve its energy (41). At this time the relatively small amount of hydrogen produced was neglected, which resulted in a change in the stoichiometry of equation 6 that did not lead to net ATP synthesis via acetyl-CoA (equation 8):
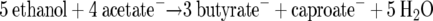 |
(8) |
Barker assumed, however, that the very exergonic reduction of crotonyl-CoA might contribute to energy conservation by “oxidative phosphorylation” (1), but no mechanism was proposed. A preliminary report of ATP production coupled to crotonyl-CoA reduction by NADH was later dismissed (14). Thauer et al. (45) and Schoberth and Gottschalk (39) demonstrated that the growth yield of C. kluyveri was proportional to the amount of hydrogen produced. Furthermore, it was shown that ATP was formed by SLP via oxidation of ethanol to acetyl-CoA and 2 H2 (45). Thus, the concept of SLP via an “energy-rich” compound was rehabilitated, but now the question concerned the coupling between the exergonic butyrate synthesis and the endergonic ethanol oxidation. Following the triumph of Mitchell and Moyle's chemiosmotic hypothesis in the late 1960s (32), ΔμH+ was assigned to be the coupling factor, but conclusive evidence was never obtained (34).
CONCLUSION
We propose that Etf bifurcates the two electrons from NADH (E0′ = −320 mV) via FAD, with one electron going to the more negative compound ferredoxin or flavodoxin (E0′ ≤ −420 mV) and the other electron going to the more positive compound crotonyl-CoA (E0′= −10 mV). Thus, the exergonic reduction of crotonyl-CoA drives the endergonic reduction of ferredoxin, which has been experimentally verified, as described in the accompanying paper. This process might be considered “reversed electron transport,” but this term is used mainly for reductions driven by the proton motive force, including the reduction of NAD+ by ubiquinol catalyzed by complex I in phototrophic proteobacteria and certain lithotrophs (17) or, most likely, the reduction of ferredoxin by NADH catalyzed by Rnf in R. capsulatus (38). In biology there are two types of energy conservation, direct ATP synthesis by SLP and indirect conservation via an electrochemical ion gradient, the so-called ETP or, more generally, ion gradient phosphorylation. In the last two decades the term ETP has been extended to all exergonic processes which produce ΔμH+ or ΔμNa+, not only electron transport itself but also decarboxylation (6, 10), methyl transfer (49), and electrogenic end product efflux (30). Electron bifurcation may be better described as a third type of energy conservation, because reduced ferredoxin can function like ATP plus NAD(P)H in various respects. (i) It increases SLP in the oxidative part of fermentations by dissipating reducing equivalents as molecular hydrogen (Fig. 1); (ii) it most likely generates an electrochemical ion gradient via Rnf; and (iii) it propels anabolism by synthesis of acetyl-CoA from 2 CO2, reductive carboxylation of acetyl-CoA to pyruvate, and direct reduction of 3-phosphoglycerate to 3-phosphoglyceraldehyde without 1,3-bisphosphoglycerate as an intermediate. Hence, in primordial biochemical pathways, ferredoxin or other iron-sulfur clusters might have preceded ATP plus NADPH as a driving force for anabolism (47). One mechanism to reduce these clusters could have been electron bifurcation, which may have been prevalent in ancient times.
Acknowledgments
We thank Rudolf K. Thauer (Max-Planck-Institut für terrestrische Mikrobiologie, Marburg, Germany) for many very helpful discussions. The valuable suggestions by two anonymous reviewers are gratefully acknowledged.
This work was supported by funds from the Deutsche Forschungsgemeinschaft (DFG) and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Barker, H. A. 1956. Bacterial fermentations. Wiley, New York, NY.
- 2.Barker, H. A. 1961. Fermentations of nitrogenous organic compounds, p. 151-207. In I. C. Gunsalus and R. Y. Stanier (ed.), The bacteria, vol. 2. Academic Press Inc., New York, NY. [Google Scholar]
- 3.Beatrix, B., K. Bendrat, S. Rospert, and W. Buckel. 1990. The biotin-dependent sodium ion pump glutaconyl-CoA decarboxylase from Fusobacterium nucleatum (subsp. nucleatum). Comparison with the glutaconyl-CoA decarboxylases from gram-positive bacteria. Arch. Microbiol. 154362-369. [DOI] [PubMed] [Google Scholar]
- 4.Beckmann, J. D., and F. E. Frerman. 1985. Electron-transfer flavoprotein-ubiquinone oxidoreductase from pig liver: purification and molecular, redox, and catalytic properties. Biochemistry 243913-3921. [DOI] [PubMed] [Google Scholar]
- 5.Boiangiu, C. D., E. Jayamani, D. Brügel, G. Herrmann, J. Kim, L. Forzi, R. Hedderich, I. Vgenopoulou, A. J. Pierik, J. Steuber, and W. Buckel. 2005. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J. Mol. Microbiol. Biotechnol. 10105-119. [DOI] [PubMed] [Google Scholar]
- 6.Buckel, W. 2001. Sodium ion-translocating decarboxylases. Biochim. Biophys. Acta 150515-27. [DOI] [PubMed] [Google Scholar]
- 7.Buckel, W. 2001. Unusual enzymes involved in five pathways of glutamate fermentation. Appl. Microbiol. Biotechnol. 57263-273. [DOI] [PubMed] [Google Scholar]
- 8.Buckel, W., and H. A. Barker. 1974. Two pathways of glutamate fermentation by anaerobic bacteria. J. Bacteriol. 1171248-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckel, W., and B. T. Golding. 2006. Radical enzymes in anaerobes. Annu. Rev. Microbiol. 6027-49. [DOI] [PubMed] [Google Scholar]
- 10.Dimroth, P., and B. Schink. 1998. Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch. Microbiol. 17069-77. [DOI] [PubMed] [Google Scholar]
- 11.Edgren, T., and S. Nordlund. 2005. Electron transport to nitrogenase in Rhodospirillum rubrum: identification of a new fdxN gene encoding the primary electron donor to nitrogenase. FEMS Microbiol. Lett. 245345-351. [DOI] [PubMed] [Google Scholar]
- 12.Edgren, T., and S. Nordlund. 2004. The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J. Bacteriol. 1862052-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgren, T., and S. Nordlund. 2006. Two pathways of electron transport to nitrogenase in Rhodospirillum rubrum: the major pathway is dependent on the fix gene products. FEMS Microbiol. Lett. 26030-35. [DOI] [PubMed] [Google Scholar]
- 14.Gunsalus, I. C., and C. W. Shuster. 1961. Energy-yielding metabolism in bacteria, p. 1-58. In I. C. Gunsalus and R. Y. Stanier (ed.), The bacteria, vol. 2. Academic Press Inc., New York, NY. [Google Scholar]
- 15.Hardman, J. K., and T. C. Stadtman. 1963. Metabolism of omega-amino acids. V. Energetics of the gamma-aminobutyrate fermentation by Clostridium aminobutyricum. J. Bacteriol. 851326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Härtel, U., and W. Buckel. 1996. Sodium ion-dependent hydrogen production in Acidaminococcus fermentans. Arch. Microbiol. 166350-356. [DOI] [PubMed] [Google Scholar]
- 17.Herter, S. M., C. M. Kortluke, and G. Drews. 1998. Complex I of Rhodobacter capsulatus and its role in reverted electron transport. Arch. Microbiol. 16998-105. [DOI] [PubMed] [Google Scholar]
- 18.Hetzel, M., M. Brock, T. Selmer, A. J. Pierik, B. T. Golding, and W. Buckel. 2003. Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur. J. Biochem. 270902-910. [DOI] [PubMed] [Google Scholar]
- 19.Imkamp, F., E. Biegel, E. Jayamani, W. Buckel, and V. Müller. 2007. Dissection of the caffeate respiratory chain in the acetogen Acetobacterium woodii: identification of an Rnf-type NADH dehydrogenase as a potential coupling site. J. Bacteriol. 1898145-8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imkamp, F., and V. Müller. 2002. Chemiosmotic energy conservation with Na+ as the coupling ion during hydrogen-dependent caffeate reduction by Acetobacterium woodii. J. Bacteriol. 1841947-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackins, H. C., and H. A. Barker. 1951. Fermentative processes of the fusiform bacteria. J. Bacteriol. 61101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jungermann, K., E. Rupprecht, C. Ohrloff, R. Thauer, and K. Decker. 1971. Regulation of the reduced nicotinamide adenine dinucleotide-ferredoxin reductase system in Clostridium kluyveri. J. Biol. Chem. 246960-963. [PubMed] [Google Scholar]
- 23.Jungermann, K., R. K. Thauer, G. Leimenstoll, and K. Decker. 1973. Function of reduced pyridine nucleotide-ferredoxin oxidoreductases in saccharolytic clostridia. Biochim. Biophys. Acta 305268-280. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J., M. Hetzel, C. D. Boiangiu, and W. Buckel. 2004. Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of alpha-amino acids by anaerobic bacteria. FEMS Microbiol. Rev. 28455-468. [DOI] [PubMed] [Google Scholar]
- 25.Kim, J. J., and R. Miura. 2004. Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences. Eur. J. Biochem. 271483-493. [DOI] [PubMed] [Google Scholar]
- 26.Kröger, A., S. Biel, J. Simon, R. Gross, G. Unden, and C. R. Lancaster. 2002. Fumarate respiration of Wolinella succinogenes: enzymology, energetics and coupling mechanism. Biochim. Biophys. Acta 155323-38. [DOI] [PubMed] [Google Scholar]
- 27.Lehman, T. C., and C. Thorpe. 1990. Alternate electron acceptors for medium-chain acyl-CoA dehydrogenase: use of ferricenium salts. Biochemistry 2910594-10602. [DOI] [PubMed] [Google Scholar]
- 28.Li, F., J. Hinderberger, H. Seedorf, J. Zhang, W. Buckel, and R. K. Thauer. 2008. Coupled ferredoxin coenzyme A (CoA) and crotonyl-CoA reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190•••-•••. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipmann, F. 1941. Metabolic generation and utilization of phosphate bond energy. Adv. Enzymol. Relat. Subj. Biochem. 199-162. [Google Scholar]
- 30.Lolkema, J. S., B. Poolman, and W. N. Konings. 1995. Role of scalar protons in metabolic energy generation in lactic acid bacteria. J. Bioenerg. Biomembr. 27467-473. [DOI] [PubMed] [Google Scholar]
- 31.McInerney, M. J., L. Rohlin, H. Mouttaki, U. Kim, R. S. Krupp, L. Rios-Hernandez, J. Sieber, C. G. Struchtemeyer, A. Bhattacharyya, J. W. Campbell, and R. P. Gunsalus. 2007. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc. Natl. Acad. Sci. USA 1047600-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, P., and J. Moyle. 1967. Chemiosmotic hypothesis of oxidative phosphorylation. Nature 213137-139. [DOI] [PubMed] [Google Scholar]
- 33.O'Neill, H., S. G. Mayhew, and G. Butler. 1998. Cloning and analysis of the genes for a novel electron-transferring flavoprotein from Megasphaera elsdenii. Expression and characterization of the recombinant protein. J. Biol. Chem. 27321015-21024. [DOI] [PubMed] [Google Scholar]
- 34.Pfeiff, B. 1991. Untersuchungen zur Kopplung von H2-Bildung und Fettsäuresynthese in Clostridium kluyveri. (Investigations of the coupling of H2 formation and butyric acit/caproic acid synthesis in Clostridium kluyveri.) Diploma thesis. Philipps-University, Marburg, Germany.
- 35.Roberts, D. L., F. E. Frerman, and J. J. Kim. 1996. Three-dimensional structure of human electron transfer flavoprotein to 2.1-A resolution. Proc. Natl. Acad. Sci. USA 9314355-14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeki, K., and H. Kumagai. 1998. The rnf gene products in Rhodobacter capsulatus play an essential role in nitrogen fixation during anaerobic DMSO-dependent growth in the dark. Arch. Microbiol. 169464-467. [DOI] [PubMed] [Google Scholar]
- 37.Sato, K., Y. Nishina, and K. Shiga. 2003. Purification of electron-transferring flavoprotein from Megasphaera elsdenii and binding of additional FAD with an unusual absorption spectrum. J. Biochem. (Tokyo) 134719-729. [DOI] [PubMed] [Google Scholar]
- 38.Schmehl, M., A. Jahn, A. Meyer zu Vilsendorf, S. Hennecke, B. Masepohl, M. Schuppler, M. Marxer, J. Oelze, and W. Klipp. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241602-615. [DOI] [PubMed] [Google Scholar]
- 39.Schoberth, S., and G. Gottschalk. 1969. Considerations on the energy metabolism of Clostridium kluyveri. Arch. Mikrobiol. 65318-328. [DOI] [PubMed] [Google Scholar]
- 40.Seeliger, S., P. H. Janssen, and B. Schink. 2002. Energetics and kinetics of lactate fermentation to acetate and propionate via methylmalonyl-CoA or acrylyl-CoA. FEMS Microbiol. Lett. 21165-70. [DOI] [PubMed] [Google Scholar]
- 41.Stadtman, E. R. 1966. Some considerations of the energy metabolism of anaerobic bacteria, p. 39-62. In N. O. Kaplan and E. P. Kennedy (ed.), Current aspects of biochemical energetics. Academic Press, New York, NY.
- 42.Stouthamer, A. H. 1979. The search for correlation between theoretical and experimental growth yields. Int. Rev. Biochem. 211-47. [Google Scholar]
- 43.Thamer, W., I. Cirpus, M. Hans, A. J. Pierik, T. Selmer, E. Bill, D. Linder, and W. Buckel. 2003. A two [4Fe-4S]-cluster-containing ferredoxin as an alternative electron donor for 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. Arch. Microbiol. 179197-204. [DOI] [PubMed] [Google Scholar]
- 44.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thauer, R. K., K. Jungermann, H. Henninger, J. Wenning, and K. Decker. 1968. The energy metabolism of Clostridium kluyveri. Eur. J. Biochem. 4173-180. [DOI] [PubMed] [Google Scholar]
- 46.Thauer, R. K., K. Jungermann, E. Rupprecht, and K. Decker. 1969. Hydrogen formation from NADH in cell-free extracts of Clostridium kluyveri. Acetyl coenzyme A requirement and ferredoxin dependence. FEBS Lett. 4108-112. [DOI] [PubMed] [Google Scholar]
- 47.Wächtershäuser, G. 1992. Groundworks for an evolutionary biochemistry: the iron-sulphur world. Prog. Biophys. Mol. Biol. 5885-201. [DOI] [PubMed] [Google Scholar]
- 48.Wallrabenstein, C., and B. Schink. 1994. Evidence of reversed electron transport in syntrophic butyrate or benzoate oxidation by Syntrophomonas wolfei and Syntrophus buswellii. Arch. Microbiol. 162136-142. [Google Scholar]
- 49.Weiss, D. S., P. Gärtner, and R. K. Thauer. 1994. The energetics and sodium-ion dependence of N5-methyltetrahydromethanopterin:coenzyme M methyltransferase studied with cob(I)alamin as methyl acceptor and methylcob(III)alamin as methyl donor. Eur. J. Biochem. 226799-809. [DOI] [PubMed] [Google Scholar]
- 50.Wendt, K. S., I. Schall, R. Huber, W. Buckel, and U. Jacob. 2003. Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase. EMBO J. 223493-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilde, E., M. D. Collins, and H. Hippe. 1997. Clostridium pascui sp. nov., a new glutamate-fermenting sporeformer from a pasture in Pakistan. Int. J. Syst. Bacteriol. 47164-170. [DOI] [PubMed] [Google Scholar]
- 52.Wohlfarth, G., and W. Buckel. 1985. A sodium ion gradient as energy source for Peptostreptococcus asaccharolyticus. Arch. Microbiol. 142128-135. [DOI] [PubMed] [Google Scholar]