Abstract
As adherence and entry of a pathogen into a host cell are key components to an infection, identifying the molecular mechanisms responsible for cellular association will provide a better understanding of a microbe's pathogenesis. We previously established an in vitro model for Borrelia burgdorferi infection of human neuroglial cells. To expand on our earlier study, we performed B. burgdorferi whole-genome expression analysis following a 20-hour infection of human neuroglial cells to identify borrelial genes that were differentially regulated during host-cell association compared with cultured Borrelia in cell-free medium. This study identifies several regulated genes, the products of which may be important mediators of cellular pathogenesis.
Lyme disease is a complex illness caused by infection with the tick-borne spirochetal bacterium Borrelia burgdorferi and can present with multiple manifestations, such as arthritis, carditis, and neurological syndromes.
B. burgdorferi adapts to disparate environments when transmitted between ticks and mammals. In naturally and experimentally infected murine hosts, B. burgdorferi has been found in a variety of tissues, including heart, bladder, joints, and ears (1, 2, 6, 58). In vitro, B. burgdorferi has been observed to adhere to endothelial cells, chondrocytes, synovial cells, peripheral blood fibrocytes, skin fibroblasts, tick cells, macrophages, and neuroglial cells of several species (12, 23, 27, 34, 35, 41, 42, 62). This ability to inhabit diverse environments implies that B. burgdorferi has the capability to adapt its physiology and membrane structure in response to its particular tissue location within arthropod and mammalian hosts. For example, investigators have demonstrated that B. burgdorferi inversely regulates the expression of outer surface proteins OspC and OspA as the spirochete migrates from the mid-gut of the tick to a mammal (45, 59). B. burgdorferi gene expression is orchestrated by many factors, including pH (7), temperature (46, 51, 59, 61), and host immune response (36-38). However, little is known about B. burgdorferi gene expression during spirochetal colonization of different tissues or cell types. Furthering our knowledge of B. burgdorferi differential gene expression during cellular infection would increase our understanding of the pathogen's adaptive mechanisms responsible for dissemination and colonization to host cells and identify novel targets for development of new treatments and diagnostics.
Subsequent to human infection with B. burgdorferi, neurological syndromes occur in roughly 15% of the untreated patients diagnosed with Lyme disease (60). The current case definition for diagnosis of acute neurological manifestations of Lyme disease includes meningitis, radiculopathy, cranial neuropathy, mononeuropathy complex and, rarely, encephalomyelitis (67). Mechanisms by which B. burgdorferi affects the nervous system are not known, but association with and/or invasion of neural cells by this bacterium could be a basis for the clinical manifestations seen in neuroborreliosis.
The nonhuman primate (NHP) is currently the most widely accepted model used to study late Lyme disease and neuroborreliosis (49). B. burgdorferi-infected NHPs show both central and peripheral nervous system involvement. In NHPs infected with B. burgdorferi, researchers have detected spirochetes in central nervous system (CNS) tissue by PCR and, moreover, spirochetes have also been visualized by histopathological staining (5, 17, 48, 49, 55, 56). Although studies of Lyme disease in NHPs have yielded valuable data, in vitro cellular association studies have the ability to provide insights into the molecular mechanisms utilized by B. burgdorferi that contribute to human neuroborreliosis.
We have previously shown that an infectious strain of B. burgdorferi invades human endothelial cells and human neural cells in vitro (40). To obtain a more comprehensive understanding of the borrelial mechanisms involved during cellular association, including attachment and invasion, we used microarray analysis to determine the expression profile of B. burgdorferi upon encountering human neuroglial cells. Identifying genes that are differentially regulated during association with human host cells may provide clues to understanding borrelial mechanisms of neuropathogenesis and present potential targets for development of novel diagnostics, prevention tools, and/or treatments for Lyme disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. burgdorferi strain B31 A3 is a clonal, low-passage infectious strain (16). Frozen stocks of all B. burgdorferi strains were maintained in 60% glycerol at −70°C. All bacterial strains were grown in liquid Barbour-Stoenner-Kelly (BSK-II) complete medium at 35°C with 5% CO2.
Mammalian cell culture.
The human neuroglioma cell line H4 was obtained from the American Type Culture Collection (Manassas, VA). H4 cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 4 mM l-glutamine containing 4.5 g/liter glucose, 1.5 g/liter sodium bicarbonate and supplemented with 10% fetal bovine serum. Cells were grown at 37°C with 5% CO2 in a humidified cell incubator. Cells were harvested from confluent monolayers using 0.05% trypsin with 10 mM EDTA and enumerated with a hemacytometer.
B. burgdorferi infection of mammalian cells.
Cell infections and calculations were performed as described in earlier publications, although they were scaled up (40). Briefly, mammalian cells were allowed to grow to confluence on a T-225 flask (Corning) and were trypsinized and enumerated the morning of the experiment. After trypsinization, 1.9 × 107 H4 cells were allowed to readhere to the flask for 4 h at 37°C. B. burgdorferi cultures were grown for 3 days until they reached mid-logarithmic phase and enumerated using a Petroff Hausser counting chamber. Bacteria were centrifuged at 500 × g for 20 min and resuspended in prewarmed DMEM. B. burgdorferi was added to the mammalian cells at a multiplicity of infection of 40 (7.8 × 108 bacteria) and was incubated at 35°C with 5% CO2 for approximately 20 h. Previous calculations of 2 bacteria associated per cell and an internalization efficiency of 0.013 per cell (40) approximated 3.8 × 107 and 2.4 × 105 Borrelia cells adherent to and internalized in the H4 cells, respectively. Each experimental sample was done in triplicate.
RNA purification, labeling, and microarray hybridization.
After incubation with B. burgdorferi for 20 h, monolayers were washed three times in phosphate-buffered saline (PBS) to remove unassociated spirochetes. Cells were then scraped from the flask and suspended in PBS. The cells were centrifuged at 125 × g for 5 min, and total RNA was isolated from infected monolayers using the RNAqueous-Midi kit (Ambion, Austin, TX). RNA was similarly extracted from uninfected H4 monolayers and from B. burgdorferi organisms incubated in cell-free DMEM for 20 h. Polyadenylated and ribosomal host RNA was removed from the total RNA extracted from the infected monolayers by using MicrobEnrich columns (Ambion). RNA samples were subjected to DNase treatment using Turbo DNA-free (Ambion). RNA samples were tested for contaminating DNA by PCR analysis for flaB (flagellin gene) prior to use in the cDNA synthesis, microarray, and quantitative reverse transcription-PCR (qRT-PCR) assays. Purified RNA was used to prepare fragmented and biotin-dUTP-labeled cDNA according to standard Affymetrix prokaryotic target preparation methods (www.affymetrix.com/support/downloads/manuals/expression_s3_manual.pdf). Briefly, cDNA was synthesized from ∼10 μg of total RNA in 1× first-strand buffer containing 25 ng/μl random primers (Invitrogen Life Technologies). Remaining RNA was removed by hydrolysis with 1 N NaOH, and the reaction mixture was neutralized with 1 N HCl. cDNA (∼3 to 6 μg/sample) was fragmented using 0.6 units/μg of DNase I (Amersham). The average size of the fragmented cDNA was 100 bp. The 3′ termini of the fragmented cDNA were labeled using Affymetrix GeneChip DNA labeling reagent. The reaction was completed as indicated by Affymetrix using the Midi format.
Microarray.
The Rocky Mountain Lab custom chip 1 (RMLchipa510998 GEO, platform GPL2129, EMBL-EBI A-AFFY-48) is an Affymetrix platform, antisense oligonucleotide expression array containing genomes from six different genera. Each gene or probe set consists of 16 randomized, individual probe pairs containing a perfect match (PM) probe and a mismatch (MM) probe. The MM probe has a single substitution at base position 13 relative to the PM probe. The Borrelia component of the RML custom chip 1 consists of probe sets specific to 1,323 full-length genomic open reading frames (ORFs; 846 chromosomal ORFs and 477 plasmid ORFs) of the sequenced Borrelia burgdorferi B31 strain. Further information is available at http://www.ebi.ac.uk/microarrayas/aer/result?queryFor=PhysicalArrayDesign and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL2129.
Probe sets not unique for a specific ORF were not included in order to eliminate cross-hybridization. B. burgdorferi contains 21 plasmids; however, 9 of those are virtual duplicates. Because of this duplication, the full 869 plasmid ORFs could not be represented without cross-hybridization. Therefore, the chromosome has 97% coverage on the array and the plasmids are at a strict 55% coverage with arguably over 90% coverage once duplication is removed. Data analysis of all chips was performed as previously described with minor modifications (a detailed analysis protocol is included with the supplemental material). In brief, all chips were globally scaled to an arbitrary value of 500 using GCOS v1.4 and a scale filter for B. burgdorferi probe sets. Average signal intensities for each probe set were used to combine biological replicates with three separate statistical tests to produce P values. The t test, significance analysis of microarrays (SAM) (66), and analysis of variance P values were generated for each probe set, producing a final gene list consisting of only probe sets significant by all three measures. A complete set of microarray data are provided in Table S1 in the supplemental material and are posted on the Gene Expression Omnibus (GEO) database website (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=hrgvhouwkaoqsfs&acc=GSE8219).
qRT-PCR.
Reverse transcription was performed on 5 μg of total RNA isolated from infected cell monolayers used for the microarray and on 5 ng of RNA from cultured B. burgdorferi cells by use of Superscript II (Invitrogen). TaqMan PCR was performed using cDNA generated from the reverse transcription as follows. TaqMan PCR primers and probe sequences were designed using Integrated DNA Technologies software and Primer Express software and were synthesized with the probe containing 5′ -6-carboxyfluorescein and 3′-Black Hole Quencher (see Table 5, below). Real-time PCR was performed in a reaction volume of 50 μl with a final concentration of 1 μM of each primer, 0.15 μM probe, 1× TaqMan Universal PCR master mix (Applied Biosystems, Foster City, CA), and 1 μl cDNA (from the 20-μl RT reaction mixture). All test sample PCRs were performed in triplicate in 96-well PCR iCycler plates (Bio-Rad, Hercules, CA) under the parameters of 1 cycle at 95°C for 3 min and 50 cycles at 95°C for 30 s, 65°C for 30 s, and 72° for 30 s using a Bio-Rad iCycler. Crossing threshold (CT) values were determined by the iCycler software. Relative quantitation of gene expression in infected cells was calculated by the 2−ΔΔCT method described by Livak and Schmittgen, whereby transcript levels were normalized against the constitutively expressed borrelial flaB gene and analyzed relative to cell-free expression (39). Each plate running PCRs also contained samples with cDNA from B. burgdorferi incubated in DMEM for relative gene expression calculation purposes and included water as a no-template negative control.
TABLE 5.
Primer and probe sequences for qRT-PCR
| Gene | Forward and reverse primer (5′-3′) | Probe |
|---|---|---|
| flaB/BB0147 | TCTTTTCTCTGGTGAGGGAGCT | AAACTGCTCAGGCTGCACCGGTTC |
| TCCTTCCTGTTGAACACCCTCT | ||
| BBA52 | ATCCAAGCCAAGAATCAGACGAGC | GACGATGAGCTAGAAAGCGAAGATCCAG |
| CGATCATCTTGGTACTCTCTTTCCC | ||
| ospC/BBB19 | CGGATTCTAATGCGGTTTTACTTG | TGTGAAAGAGGTTGAAGCGTTGCTGTC |
| CAATAGCTTTAGCAGCAATTTCATCT | ||
| dbpA/BBA24 | AGACGCTGCTCTTAAGGGCGTAAA | AGTGCGAGCTACTACAGTAGCGGAAA |
| CACCACTACTTCCAGTTTCTTTGAG | ||
| dpbB/BBA25 | GGAAACAGTGGTCAATTCTTGGCT | AGTGCGAGCTACTACAGTAGCGGAAA |
| CCGCAAGCAATCTTTCAGCTGTGT | ||
| ospA/BBA15 | GCGTTTCAGTAGATTTGCCTGGTG | GACGGCAAGTACGATCTAATTGCAACAGT |
| ACGCCTTCAAGTACTCCAGATCCA | ||
| BBK32 | GTTTAAATTCCCTTAGCGGTGAAA | TGGTGAATTGGAGGAGCCTATTGAAAGTAATGA |
| TTTGGCCTTAAATCAGAATCTATAGTAAGA | ||
| cheW/BB0670 | TTGGTTGTGGTAGTGGAAAGGAG | ACTATGGCTTTAGCCAATGCTTTGTCTGAA |
| ACACCAACCTAGAAGTTTCAACAAC | ||
| cheX/BB0671 | TGGATGCTGCTTCTTCGGTT | ATAGAAATGGGTAAGCCCGGGCTT |
| ACAGACCCAGCAAGCCCTACTATT | ||
| BBA72 | ACTTCATGTGCTGTTAATCCAATTG | TCCAAAAGTAAAAAGCCGCACCGA |
| CCAGATTTTTGGTTGCTTTCTTTT | ||
| S1 antigen/BBA05 | GCGCAATTTAACAGACGAAGAA | AATGTTGCAGGGTTTTGTTGGGCG |
| AAGCCTTTTAAACAAATCGTCAAGA | ||
| BB0327 | GACCCTTTCTTTGGGATACAATGC | AGTGTATCCACCGCACGCATTTGTTG |
| ACACGCCTTTGCCTAGCATGTTCT | ||
| BBA64 | AGCCCGCAGCTGGAAAA | AATCCCAACGCTAATGCCAACAATGCT |
| AGCTTTGCAACTTCAGGATCTAATATT | ||
| BBA73 | GGATGCAAAGCTGATGGAAA | ATTCTCGGAGAAGTAATAAGGGTTGG |
| GCATACTTTTATTTGAGTCGAGTTCA | ||
| BBA71 | CAGCCCCGATTACGAAAATATAA | TATAGGACTACTTTATCACAAAGCATTGGGCATTCA |
| AATTCTGCGTGTATTAGTAGATCGTTTAA |
RESULTS
Analysis of global B. burgdorferi gene expression upon infection of human cells in vitro.
We examined B. burgdorferi differential gene expression during interaction with human neuroglial cells using an Affymetrix GeneChip microarray. H4 neuroglial cells were chosen for this analysis, as B. burgdorferi has previously been shown to associate extra- and intracellularly with these cells, which did not affect Borrelia or host cell viability (40, 64). A 20-hour incubation was performed, which allowed for optimum spirochete-cell interaction. This time point included B. burgdorferi cells that were both intra- and extracellularly localized to the H4 cells with extracellular adherent borrelial cells at approximately a 160-fold-higher ratio versus internalized Borrelia (40). Therefore, adherent spirochetes were represented at higher numbers than internalized organisms. After thorough washing with PBS, there were virtually no unassociated spirochetes detected by bacterial plating of the final wash (data not shown).
After B. burgdorferi infection of these cells, the total cellular RNA preparation (including H4 RNA and Borrelia RNA) was enriched for prokaryotic RNA, which concentrated the bacterial message but did not completely eliminate the eukaryotic RNA (data not shown). Therefore, as a control in our arrays, eukaryotic RNA from the human H4 cells was also purified, reverse transcribed, and hybridized to the array. Differential B. burgdorferi gene expression following H4 cell infection was measured relative to cell-free cultured B. burgdorferi gene expression. To control for the spirochetes' transfer into the H4 cell growth medium from BSK medium, we standardized (normalized) the gene expression data from the experimental conditions (B. burgdorferi-infected human cells) to borrelial expression after a 20-h incubation in DMEM tissue culture medium. All samples were assayed in triplicate.
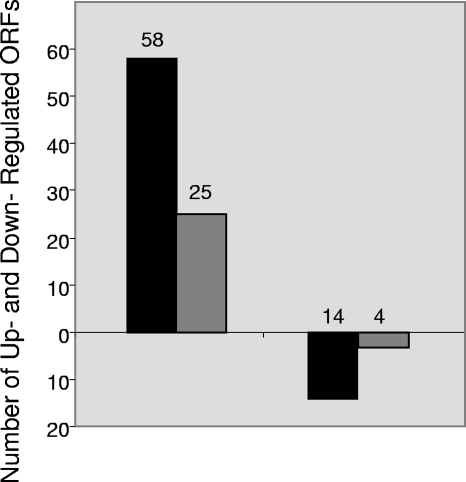
An analysis of gene expression of the 1,323 genes in the Affymetrix array under the experimental conditions was conducted. A threshold of twofold up- or down-regulation was statistically derived as a conservative foundation for all of the following interpretations. Expression levels below this threshold may have biological relevance, but the more stringent analysis was employed in this study. The complete data set is included in Table S1 in the supplemental material and is posted on the GEO database website. We performed statistical analyses of the microarray data consisting of SAM, analysis of variance, and a t test. We evaluated the B. burgdorferi genes which passed all three of these statistical analyses to minimize the false discovery rate. Using these criteria, 72 genes from the H4 infection were found to be differentially regulated (Fig. 1 and Tables 1 and 2). In referring to genes, we adopted the gene nomenclature and gene designation delineated by The Institute for Genomic Research (TIGR; http://cmr.tigr.org/). Genes that were differentially expressed during human cell association were divided into categories based on function, genomic location, and gene family. Hypothetical proteins comprised the largest category of differentially regulated genes and totaled 40% (29 genes) of the total differentially regulated genes (Fig. 1).
FIG. 1.
Overview of differentially expressed B. burgdorferi ORFs after infection of human neuroglial cells. ORFs (n = 72) from the microarray included in this graph passed three statistical tests, SAM, partek, and t tests, and were up- or down-regulated at least twofold. Black bars indicate the fraction of total ORFs up- or down-regulated. Gray bars represent the fraction of up- or down-regulated genes of the total ORFs which encode hypothetical proteins.
TABLE 1.
Mean up-regulation of B. burgdorferi genes on the array after infection of human H4 neuroglial cells
| Gene | Functional description | Fold change | Replicon | Paralogous gene family |
|---|---|---|---|---|
| BBA71 | Hypothetical protein | 25.11 | lp54 | 54 |
| BBA37 | Hypothetical protein | 16.42 | lp54 | |
| BBA72 | Hypothetical protein | 11.63 | lp54 | |
| BBA64 | p35 antigen | 10.97 | lp54 | 54 |
| BBA73 | p35 antigen, putative | 10.21 | lp54 | 54 |
| BBB19 | OspC, outer surface protein C | 9.82 | cp26 | |
| BB0641 | Spermadine/putrescineABC transporter, permease protein | 8.85 | Chromosome | 41 |
| BB0640 | Spermadine/putrescineABC transporter, permease protein | 7.11 | Chromosome | 41 |
| BBJ24 | Hypothetical protein | 5.99 | lp38 | 106 |
| BB0549 | Hypothetical protein | 5.94 | Chromosome | |
| BB0129 | Conserved hypothetical protein | 5.92 | Chromosome | |
| BB0670 | CheW3, purine-binding chemotaxis protein | 5.14 | Chromosome | 33 |
| BBJ26 | ABC transporter, ATP-binding protein | 4.90 | lp38 | 4 |
| BB0654 | Hypothetical protein | 4.85 | Chromosome | |
| BBJ28 | Hypothetical protein | 4.43 | lp38 | |
| BB0353 | Hypothetical protein | 4.41 | Chromosome | |
| BB0681 | Methyl-accepting chemotaxis protein | 4.27 | Chromosome | 13 |
| BB0270 | FlhF, flagellum-associated GTP-binding protein | 3.81 | Chromosome | 10 |
| BB0548 | PolA, DNA polymerase I | 3.81 | Chromosome | |
| BB0255 | Hypothetical protein | 3.81 | Chromosome | 2 |
| BB0768 | PdxK, pyridoxal kinase | 3.69 | Chromosome | |
| BB0541 | Hypothetical protein | 3.62 | Chromosome | |
| BB0276 | FliZ, flagellar biosynthesis protein | 3.49 | Chromosome | |
| BB0259 | Hypothetical protein | 3.45 | Chromosome | |
| BBA05 | S1 antigen | 3.39 | lp54 | |
| BB0063 | Hypothetical protein | 3.37 | Chromosome | |
| BB0790 | Hypothetical protein | 3.34 | Chromosome | |
| BBA07 | ChpAI protein, putative | 3.32 | lp54 | |
| BB0808 | Hypothetical protein | 3.31 | Chromosome | |
| BB0354 | Hypothetical protein | 3.27 | Chromosome | |
| BBJ25 | Hypothetical protein | 3.26 | lp38 | |
| BB0607 | Rep:Rep helicase, single-stranded DNA-dependent ATPase | 3.14 | Chromosome | 18 |
| BB0306 | Conserved hypothetical protein | 3.13 | Chromosome | |
| BB0589 | Pta, phosphate acetyltransferase | 3.13 | Chromosome | |
| BB0459 | Hypothetical protein | 3.04 | Chromosome | |
| BB0027 | Hypothetical protein | 2.97 | Chromosome | |
| BB0099 | RsgA, ribosome small subunit-dependent GTPase A | 2.92 | Chromosome | |
| BB0318 | MglA, methylgalactoside ABC transporter, ATP-binding protein | 2.86 | Chromosome | 4 |
| BB0803 | TruB, tRNA pseudouridine 55 synthase | 2.84 | Chromosome | |
| BB0172 | Hypothetical protein | 2.81 | Chromosome | 121 |
| BB0091 | V-type ATPase, subunit I, putative | 2.78 | Chromosome | |
| BB0018 | Conserved hypothetical protein | 2.66 | Chromosome | |
| BB0671 | CheX, chemotaxis operon protein | 2.51 | Chromosome | |
| BB0095 | Hypothetical protein | 2.51 | Chromosome | |
| BB0090 | V-type ATPase, subunit K, putative | 2.50 | Chromosome | |
| BB0128 | Cmk, cytidylate kinase | 2.44 | Chromosome | |
| BB0567 | CheA1, chemotaxis histidine kinase | 2.41 | Chromosome | 134 |
| BB0788 | TilS, tRNA(lle)-lysidine synthetase. | 2.26 | Chromosome | |
| BB0232 | tRNAIle-lysidine synthetase, HbbU protein | 2.20 | Chromosome | |
| BB0443 | Jag, SpoIIIJ-associated protein | 2.19 | Chromosome | |
| BB0442 | Inner membrane protein | 2.19 | Chromosome | |
| BB0014 | PriA, primosomal protein N | 2.18 | Chromosome | |
| BB0468 | Conserved hypothetical protein | 2.17 | Chromosome | |
| BB0581 | RecG, DNA recombinase | 2.14 | Chromosome | 20 |
| BB0257 | Cell division protein, putative | 2.11 | Chromosome | |
| BB0625 | N-Acetylmuramoyl-l-alanine amidase, putative | 2.07 | Chromosome | |
| BB0290 | FliG2, flagellar motor switch protein | 2.06 | Chromosome | 38 |
| BB0171 | Hypothetical protein | 2.01 | Chromosome |
TABLE 2.
Mean down-regulation of B. burgdorferi genes on the array after infection of human H4 neuroglial cells
| Gene | Functional description | Fold change | Replicon | Paralogous gene family |
|---|---|---|---|---|
| BBH37 | Hypothetical protein | −7.14 | lp28-3 | |
| BBH37 | Predicted coding region | −7.14 | lp28-3 | 12 |
| BBD21 | Conserved hypothetical protein | −5.56 | lp17 | 32 |
| BB0658 | GmpA, phosphoglycerate mutase | −4.17 | Chromosome | |
| BBJ36 | Hypothetical protein | −3.70 | lp38 | 92 |
| BBQ41 | Conserved hypothetical protein | −3.70 | lp56 | 49 |
| BB0348 | Pyk, pyruvate kinase | −3.45 | Chromosome | 16 |
| BB0645 | PtsG, PTS system, glucose-specific IIBC component | −3.33 | Chromosome | |
| BB0215 | Psts, phosphate ABC transporter, periplasmic phosphate-binding protein | −2.94 | Chromosome | |
| BB0031 | LepB2, signal peptidase | −2.86 | Chromosome | |
| BB0241 | GlpK, glycerol kinase | −2.78 | Chromosome | |
| BB0518 | DnaK2, heat shock protein 70 | −2.78 | Chromosome | 9 |
| BBA52 | Outer membrane protein | −2.63 | lp54 | |
| BB0337 | Eno, enolase | −2.50 | Chromosome | |
| BB0693 | XylR1, xylose operon regulatory protein | −2.04 | Chromosome |
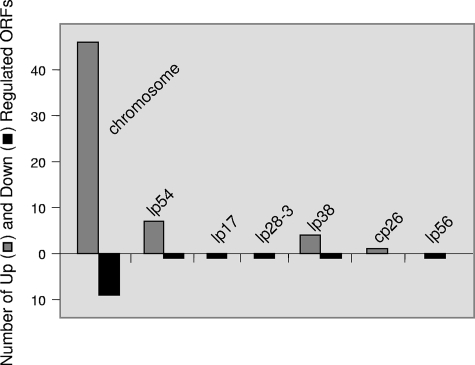
Distribution of differentially expressed ORFs among the B. burgdorferi genome.
B. burgdorferi is unique in that its genome consists of a chromosome and a number of smaller linear and circular plasmids (8, 22). Differential regulation on the chromosome and plasmids was observed, demonstrating the ability of B. burgdorferi to regulate genes in response to environmental stimuli, i.e., interactions with human cells (Fig. 2). The linear chromosome accounts for roughly two-thirds of the borrelial genome, and this fraction correlated with the proportion of differentially expressed genes that localized to the chromosome in our experiments. The majority of the differentially expressed genes lie on the chromosome (55 genes, 76% of total differentially expressed). Of the plasmid-encoded differentially expressed genes, eight were on the 54-kb linear plasmid (lp54), which represented the most on a single plasmid (Fig. 2). lp54 (plasmid A) encodes 76 genes, including genes expressed in tick or mammalian infections: the ospA/B, dbpA/B operons, and the paralogous gene family 54 (pgf54). The importance of this plasmid in nature is underscored in that no isolates have been discovered missing lp54 or an ortholog of lp54.
FIG. 2.
Distribution of differentially expressed ORFs among B. burgdorferi replicons after infection of human neuroglial cells in vitro. ORFs (n = 72) included in this graph passed three statistical tests, SAM, partek, and t tests, and were up- or down-regulated at least twofold. The graph displays the number and replicon location of differentially expressed B. burgdorferi genes after infection of H4 neuroglial cells. Gray bars represent the number of up-regulated genes, and the black bars represent the number of down-regulated genes.
Paralogous gene families of B. burgdorferi.
Upon sequencing of the B. burgdorferi genome, many genes and pseudogenes were grouped into paralogous gene families (8). In all, the genome harbors 161 paralogous gene families, of which 107 contain plasmid-borne members. These family members are largely based on sequence homology and suggest a survival advantage to the spirochete in carrying multiple paralogs on different plasmids. The microarray was designed to include 16 unique 25-bp probes randomly dispersed across the array for each B. burgdorferi gene (for potential ORFs of <400 bp probes can overlap, or if fewer than 16 probes can accommodate a region then fewer are used). Additionally, the identical 16 probes containing a single-base mismatch at bp 13 of 25 were also placed on the array to help control for cross-hybridization. The paralogous gene families of differentially expressed genes are included in Table 1. Notably, pgf54 had three members that were up-regulated in the H4 infection, which represents the gene family with the highest ratio of up-regulated genes (that passed the statistical criteria) in our study. The majority of genes in this family were also differentially regulated in independent microarray studies of B. burgdorferi gene expression in environments mimicking B. burgdorferi transmission from the tick into the mammalian host (4, 46, 54, 65).
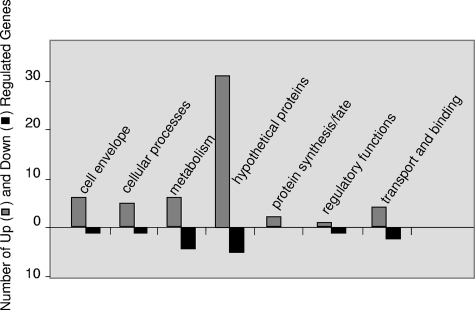
Distribution of differentially expressed B. burgdorferi ORFs by functional category.
TIGR has delineated functional categories based on sequence homology to known genes across different bacterial species (clusters of orthologous genes [COG]) and grouped the borrelial genes into these functional categories. Although the genome has been sequenced, functions for many of the borrelial genes have not been identified. Therefore, it is not surprising that the majority of the differentially expressed genes encode hypothetical proteins (Fig. 3). Further investigation into the functions of these genes is required, but their differential expression suggests that some of the genes may be important in the biology of dissemination and cell attachment of B. burgdorferi within the host.
FIG. 3.
Number of differentially expressed B. burgdorferi ORFs grouped by putative protein functions. ORFs (n = 72) included in this graph passed three statistical tests, SAM, partek, and t tests, and were up- or down-regulated at least twofold. The graph displays the number of differentially expressed B. burgdorferi genes, grouped by putative protein function as assigned by TIGR as COGs, after infection of H4 neuroglial cells. Gray bars represent the number of up-regulated genes, and the black bars represent the number of down-regulated genes.
Lipoproteins fell into the gene category of “cell envelope.” The B. burgdorferi proteome is predicted to have 136 putative lipoproteins (8), and 8 of these were differentially expressed upon borrelial interaction with neuroglial cells (Table 3), suggesting that the spirochete may alter its membrane structure upon infection of human neuroglial cells. The human neuroglial cell infection also yielded a number of up-regulated genes associated with metabolism (BB014, BB0091, BB0232, BB0548, BB0581, and BB0607; see Table 1 for functional characterizations), suggesting different energy requirements and growth properties among cell-free Borrelia and inter- and intracellular interactions. Additionally, during interaction with these cells in vitro, we saw a reduction in the expression of the gene products which catalyze the final three steps of glycolysis (gmpA for phosphoglyceromutase, eno for enolase, and puk for pyruvate kinase).
TABLE 3.
Differential expression of B. burgdorferi lipoprotein genes upon infection of H4 neuroglial cells
| Effect on B. burgdorferi lipoprotein gene expression
| |
|---|---|
| Up-regulated genes | Down-regulated genes |
| BBA05 | BBH37 |
| BBA07 | BBJ36 |
| BBA64 (P35) | |
| BBA73 | |
| BBA72 | |
| BBB19 (ospC) | |
Chemotaxis and flagellar synthesis.
One gene category that was differentially regulated between Borrelia cells grown in culture and Borrelia cells associating with neuroglial cells contained genes related to chemotaxis and motility. In the H4 infection, four chemotaxis-associated genes were up-regulated (BB0670/cheW3, BB0681/methyl-accepting chemotaxis protein, BB0671/cheX, and BB0567/cheA1). There are two well-conserved operons among spirochetes related to chemotaxis and motility: one operon includes flaA, cheA2, cheW3, cheX, and cheY3 (BB0668 to BB0670), and another includes cheW2, BB0566, cheA1, cheB2, BB0569, and cheY2 (BB0566 to BB05700) (reviewed in reference 9). Two chemotaxis-related genes that were up-regulated in our study (BB0670/cheW3 and BB0671/cheX) reside on one of these operons, suggesting an increase in expression of the entire operon. Although only BB0670 and BB0671 passed all three statistical tests in the microarray, two of the other five genes in this operon were also slightly up-regulated (although they did not pass all three statistical tests [see Table S1 in the supplemental material]). Interestingly, other investigators have also reported an up-regulation in chemotaxis genes under experimental conditions that mimic the mammalian environment (54, 65). Revel et al. reported an increase in three of the five genes in the one of these operons (flaA, cheA2, and cheX) and two of the six genes in another operon (cheW2 and cheA1) (54) within B. burgdorferi cells grown in dialysis membrane chambers implanted in rats. Tokarz et al. similarly saw an increase in cheX and cheW2 expression while reporting cheA1 and cheA2 as being up-regulated in Borrelia with blood added to the culture medium (65).
Additionally, three flagellar synthesis genes were up-regulated in our study (BB0270/flhF, BB0276/fliZ, and BB0290/fliG2). The B. burgdorferi genome contains a large operon consisting of 26 genes associated with flagellar synthesis (24). Of the 26 genes in this operon, only 3 passed all three statistical tests used in our microarray analysis. However, after analysis of the raw microarray data, we observed that 21 of 26 genes were slightly up-regulated (i.e., >1.0), and 3 were unchanged when less-stringent statistical tests were employed (see Table S1 in the supplemental material). Since the operon is under the transcriptional control of one promoter, this suggests that the genes within the operon were transcriptionally up-regulated as B. burgdorferi associates with human neuroglial cells. Because microarray studies give a semiquantitative measurement of the gene expression trends observed under certain circumstances, more precise data regarding regulation of genes in the flgB operon would need to be obtained and validated by qRT-PCR. A previous study of mutant B. burgdorferi cells lacking flagella demonstrated a decreased ability to penetrate endothelial cells in vitro (57), thereby leading some investigators to characterize motility and chemotaxis in spirochetes as major virulence factors. Roughly 5 to 6% of the genomes of Treponema pallidum and B. burgdorferi encode putative chemotaxis and motility genes (9). Motility and chemotaxis gene regulation may be a critical factor(s) to foster B. burgdorferi dissemination into or out of neuroglial cells.
Validation of microarray data by real-time PCR.
As the microarray data provided a “snapshot” of the transcriptome under experimental conditions and gave a general view of the expression of the borrelial genes, it was important to validate the trends observed in the microarray by qRT-PCR. A subset of genes was chosen for further analysis from several functional categories and plasmid locations. Additionally, we analyzed four genes (ospA, dbpA, dbpB, and BBK32) that did not pass all three statistical tests in the microarray but were of interest because of their functions as adhesins and importance during borrelial transmission (29, 30, 50, 59). The same B. burgdorferi-infected H4 cell total RNA preparation analyzed in the microarray analysis was utilized for the qRT-PCR studies. Relative quantitation of gene expression was calculated by the 2−ΔΔCT method (39), whereby transcript levels were normalized against the constitutively expressed B. burgdorferi flaB gene and analyzed relative to expression in organisms incubated in DMEM. The changes measured by qRT-PCR were compared to the microarray data (Table 4). Primer and probe sequences for the gene targets are listed in Table 5. The microarray was successful in predicting the amount of up- or down-regulation seen with qRT-PCR analysis 70% of the time (7 of the 10 genes analyzed by qPCR corroborated the microarray data) (Table 4). The microarray field has adopted a wide variety of methods for use in data analysis. Differences in technology and approaches employed for data analysis produce a wide range of acceptable methods for data confirmation. We chose a simplified approach to analyze our qRT-PCR validation of the microarray, as we felt this was a more accurate measure of comparison. By qRT-PCR, we observed significant up-regulation of genes related to chemotaxis, cheX and cheW. Additionally, the adhesins dbpA and dbpB were strongly up-regulated 21- and 19-fold, respectively. This illustrates the importance of these proteins as mediators in the attachment of B. burgdorferi, especially in human tissue (20, 21). We saw roughly a threefold down-regulation in the gene encoding fibronectin-binding protein BBK32, suggesting this adhesin is not important to neuroglial cell association after 20 h of Borrelia infection. Interestingly, ospA was strongly up-regulated while ospC was slightly down-regulated. This is in contrast to the reciprocal ospA/ospC expression observed as B. burgdorferi migrates from the tick mid-gut to a mammalian host (45, 59), although not inconsistent with previous findings regarding differential gene expression during the progression of infection. For example, down-regulation of the OspC gene during infection has been demonstrated as a mechanism to counter the host's specific humoral response to this antigen (36).
TABLE 4.
Comparison of qRT-PCR and microarray dataa
| ORF | Fold up- or down-regulation
|
|
|---|---|---|
| Microarray | qRT-PCRb | |
| S1 antigen BBA05 | 3.39 | 1.2 |
| BBA64 | 10.97 | 2.46 |
| BBA71 | 25.1 | 1.48 |
| BBA72 | 11.6 | 3.5 |
| BBA73 | 10.2 | 1.8 |
| BB0337 | −2.5 | 3.4 |
| cheX BB0671 | 2.51 | 9.8 |
| cheW BB0670 | 5.14 | 22.6 |
| BBA52 | −2.63 | 1.5 |
| OspC BBB19 | 9.82 | −1.43 |
| ospA BBA15 | 3.41 | 8.9* |
| dbpA BBA24 | 2.60 | 21* |
| dbpB BBA25 | 2.01 | 19* |
| BBK32 | 1.35 | −3.33* |
The agreement between the two methods was 70%.
ORFs marked with an asterisk did not pass all three statistical tests in the microarray analysis.
DISCUSSION
Since the B. burgdorferi genome has been sequenced (8, 22), investigators have used microarray technology to study B. burgdorferi gene expression under a variety of conditions. Microarray studies have been published examining the differential gene expression of B. burgdorferi grown under conditions mimicking unfed ticks compared to conditions similar to ticks feeding on mammals and by simulation of mammalian infection (4, 46, 54, 65). Additionally, Narasimhan et al. studied in vivo B. burgdorferi gene expression in the heart and medulla of infected NHPs by using microarray technology in conjunction with DECAL (differential expression using customized amplification libraries), which selectively amplifies specific prokaryotic message (43). Alternatively, some investigators interested in the host response to B. burgdorferi infection have used gene arrays to monitor the expression of human genes, specifically matrix metalloproteinases, following B. burgdorferi infection of human chondrocytes, murine cartilage, and synovial fluid from Lyme disease patients (3).
These gene expression studies have provided important information to elucidate B. burgdorferi responses to environmental changes associated with infection. However, with the exception of the study reported by Narasimhan et al., the above studies using B. burgdorferi genomic microarrays were indirect depictions of the host environment, as they were all extrapolations of cultured Borrelia grown under different medium conditions in vitro without host-cell interactions. Our study is the first comprehensive analysis of changes in B. burgdorferi gene expression as it infects human cells in vitro. Study of borrelial gene expression in this model can help in understanding the transcriptional responses when spirochetes encounter human cells.
In vitro cell culture models have been used by other investigators to examine differential gene expression of bacteria as they invade mammalian cells in vitro. Microarray analysis studying the change in gene expression of Mycobacterium tuberculosis as it crossed the blood-brain barrier yielded insights into the virulence factors associated with CNS invasion (32). The intracellular gene expression profile of Listeria monocytogenes as it invades mouse macrophages and human epithelial cells has also been described (10, 33). Similarly, the expression profiles of other bacteria, including Salmonella enterica, Streptococcus pneumonia, Neisseria gonorrhoeae, and Escherichia coli, as they invade mammalian cells in vitro have been examined by using microarrays to gain insights into the pathogenesis of these bacteria (13-15, 18, 19, 47, 63).
Currently, there is little knowledge regarding B. burgdorferi pathogenesis in the CNS, although previous studies have provided interesting findings. Grab et al. used an in vitro model system of the blood-brain barrier to investigate the ability of B. burgdorferi to invade human brain microvascular endothelial cells (28). In cultured rhesus monkey astrocytes, lipidated OspA caused astrogliosis and stimulated interleukin-6 and tumor necrosis factor alpha production (52). Additionally, borrelial lipoproteins activated the p38 and Erk1/2 mitogen-activated protein kinase pathway in B. burgdorferi-infected astrocytes, demonstrating a dysregulation in the inflammatory response (53).
The goal of this study was to investigate B. burgdorferi differential gene expression during in vitro infection of human H4 glial cells using microarray to identify putative borrelial mediators of cellular infection. After conservative statistical analysis, our data set revealed that B. burgdorferi infecting H4 cells had 72 genes that were differentially regulated compared to cell-free Borrelia grown in culture. This implies that B. burgdorferi differentially expresses a wide variety of genes to adapt and survive in host tissues. The majority of the differentially expressed genes in this data set encode hypothetical proteins, suggesting that these proteins may be novel virulence factors and important markers of cellular association. Further study of these genes could provide insights to the molecular mechanisms involved in cellular infection. In addition to a large number of hypothetical proteins, the Borrelia proteome is predicted to have 136 putative lipoproteins (8), and 8 of these were differentially expressed upon borrelial association with neuroglial cells. As lipoproteins are often surface exposed and immunogenic, they could be important in mediating the attachment of B. burgdorferi to different cell types.
Among the differentially regulated genes, a group that was up-regulated in the neuroglial cells is related to chemotaxis and motility, including flagellar synthesis genes. B. burgdorferi has two characterized operons related to chemotaxis. Gene products of both operons were “up” in our study, a finding that corroborates with other microarray studies of Borrelia grown under conditions mimicking the mammalian environment (54, 65). The B. burgdorferi genome encodes a large motility operon, flgB, which includes 26 genes relevant to motility and flagellar synthesis. Since B. burgdorferi lacks σ28 (a sigma factor which typically regulates flagellar gene transcription in bacteria), transcription from this promoter is regulated by σ70 (31). It has been shown that B. burgdorferi synthesizes its periplasmic flagella throughout its life cycle; therefore, up-regulation of this operon could signal the bacterium to disseminate upon encountering neuroglial cells (24). Bacterial motility has been implicated as an important virulence factor in bacteria, and transcriptional control of genes coordinating bacterial motility and chemotaxis may enhance the capacity of B. burgdorferi to disseminate within the host.
Of the 72 differentially regulated genes in our data set, 55 of them (including genes related to motility) lie on the chromosome. However, eight of them lie on lp54, and three belong to pgf54. lp54 has 8 of the 12 members of pgf54 clustered toward the end of this linear plasmid. Other investigators have observed that lp54 has the highest ratio of differentially expressed genes in response to changes in temperature and pH of the medium (4, 46, 54). Serological data from Lyme disease patients have shown antibody responses against BBA64 and BBA66, indicating that these proteins are immunogenic and are expressed during host infection (11, 26, 44). Our data highlight the potential importance of this paralogous gene family in the infectious process of B. burgdorferi. Interestingly, in a long-term murine infection, BBA64 gene expression was down-regulated as measured by B. burgdorferi cells localized to ear tissues, despite the host eliciting a strong immune response against this antigen (25). The up-regulation of the BBA64 gene observed after neural cell infection suggests that this gene is expressed during interaction with specific host tissues.
Using this in vitro model of human cell infection, we have provided the first global gene expression analysis of B. burgdorferi during the active process of cellular infection. Preliminary studies in our laboratory utilizing the microarray to analyze B. burgdorferi infection of human endothelial cells demonstrated a different pattern of gene expression than that observed with the H4 cells (data not shown), suggesting that a unique set of borrelial genes are expressed when interacting with specific cell types. Regulation of gene expression in response to dissemination and host tissue tropism is a complex process researchers in this field are only beginning to understand. Elucidation of the mechanisms of cell association is an important step to understanding how B. burgdorferi responds to tissue colonization. These data and analyses are a starting point for further research into the events and mechanisms leading to the pathogenesis of Lyme borreliosis.
Supplementary Material
Acknowledgments
J.A.L. was supported by a postdoctoral fellowship from the American Society for Microbiology and Centers for Disease Control and Prevention.
We thank Philip Stewart for critical reading and intellectual contributions to the project, and we gratefully acknowledge the contribution of Daniel Sturdevant and the Research Technologies Branch of the Rocky Mountain Laboratories for providing the expertise for the microarray data generation and interpretation. We also thank Patti Rosa for the generous donation of the Affymetrix chips.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Editor: A. Camilli
Footnotes
Published ahead of print on 5 November 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anderson, J. F., R. C. Johnson, L. A. Magnarelli, and F. W. Hyde. 1985. Identification of endemic foci of Lyme disease: isolation of Borrelia burgdorferi from feral rodents and ticks (Dermacentor variabilis). J. Clin. Microbiol. 2236-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthold, S. W., D. H. Persing, A. L. Armstrong, and R. A. Peeples. 1991. Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am. J. Pathol. 139263-273. [PMC free article] [PubMed] [Google Scholar]
- 3.Behera, A. K., E. Hildebrand, J. Scagliotti, A. C. Steere, and L. T. Hu. 2005. Induction of host matrix metalloproteinases by Borrelia burgdorferi differs in human and murine Lyme arthritis. Infect. Immun. 73126-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 713371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadavid, D., T. O'Neill, H. Schaefer, and A. R. Pachner. 2000. Localization of Borrelia burgdorferi in the nervous system and other organs in a nonhuman primate model of Lyme disease. Lab. Investig. 801043-1054. [DOI] [PubMed] [Google Scholar]
- 6.Callister, S. M., W. A. Agger, R. F. Schell, and K. M. Brand. 1989. Efficacy of the urinary bladder for isolation of Borrelia burgdorferi from naturally infected, wild Peromyscus leucopus. J. Clin. Microbiol. 27773-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infect. Immun. 686677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35490-516. [DOI] [PubMed] [Google Scholar]
- 9.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 3647-73. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 741323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clifton, D. R., C. L. Nolder, J. L. Hughes, A. J. Nowalk, and J. A. Carroll. 2006. Regulation and expression of bba66 encoding an immunogenic infection-associated lipoprotein in Borrelia burgdorferi. Mol. Microbiol. 61243-258. [DOI] [PubMed] [Google Scholar]
- 12.Comstock, L. E., and D. D. Thomas. 1989. Penetration of endothelial cell monolayers by Borrelia burgdorferi. Infect. Immun. 571626-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahan, S., S. Knutton, R. K. Shaw, V. F. Crepin, G. Dougan, and G. Frankel. 2004. Transcriptome of enterohemorrhagic Escherichia coli O157 adhering to eukaryotic plasma membranes. Infect. Immun. 725452-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Cello, F., Y. Xie, M. Paul-Satyaseela, and K. S. Kim. 2005. Approaches to bacterial RNA isolation and purification for microarray analysis of Escherichia coli K1 interaction with human brain microvascular endothelial cells. J. Clin. Microbiol. 434197-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du, Y., J. Lenz, and C. G. Arvidson. 2005. Global gene expression and the role of sigma factors in Neisseria gonorrhoeae in interactions with epithelial cells. Infect. Immun. 734834-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 702139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.England, J. D., R. P. Bohm, Jr., E. D. Roberts, and M. T. Philipp. 1997. Mononeuropathy multiplex in rhesus monkeys with chronic Lyme disease. Ann. Neurol. 41375-384. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47103-118. [DOI] [PubMed] [Google Scholar]
- 19.Faucher, S. P., S. Porwollik, C. M. Dozois, M. McClelland, and F. Daigle. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. USA 1031906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fikrig, E., P. K. Coyle, S. E. Schutzer, M. Chen, Z. Deng, and R. A. Flavell. 2004. Preferential presence of decorin-binding protein B (BBA25) and BBA50 antibodies in cerebrospinal fluid of patients with neurologic Lyme disease. J. Clin. Microbiol. 421243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, J. R., N. Parveen, L. Magoun, and J. M. Leong. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 1007307-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fuji, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390580-586. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Monco, J. C., B. Fernandez-Villar, and J. L. Benach. 1989. Adherence of the Lyme disease spirochete to glial cells and cells of glial origin. J. Infect. Dis. 160497-506. [DOI] [PubMed] [Google Scholar]
- 24.Ge, Y., I. G. Old, I. Saint Girons, and N. W. Charon. 1997. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus σ70 promoter. J. Bacteriol. 1792289-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilmore, R. D., Jr., R. R. Howison, V. L. Schmit, A. J. Nowalk, D. R. Clifton, C. Nolder, J. L. Hughes, and J. A. Carroll. 2007. Temporal expression analysis of the Borrelia burgdorferi paralogous gene family 54 genes BBA64, BBA65, and BBA66 during persistent infection in mice. Infect. Immun. 752753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmore, R. D., Jr., K. J. Kappel, and B. J. Johnson. 1997. Molecular characterization of a 35-kilodalton protein of Borrelia burgdorferi, an antigen of diagnostic importance in early Lyme disease. J. Clin. Microbiol. 3586-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girschick, H. J., H. I. Huppertz, H. Russmann, V. Krenn, and H. Karch. 1996. Intracellular persistence of Borrelia burgdorferi in human synovial cells. Rheumatol. Int. 16125-132. [DOI] [PubMed] [Google Scholar]
- 28.Grab, D. J., G. Perides, J. S. Dumler, K. J. Kim, J. Park, Y. V. Kim, O. Nikolskaia, K. S. Choi, M. F. Stins, and K. S. Kim. 2005. Borrelia burgdorferi, host-derived proteases, and the blood-brain barrier. Infect. Immun. 731014-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, B. P., E. L. Brown, D. W. Dorward, L. C. Rosenberg, and M. Hook. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30711-723. [DOI] [PubMed] [Google Scholar]
- 30.Guo, B. P., S. J. Norris, L. C. Rosenberg, and M. Hook. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 633467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Indest, K. J., R. Ramamoorthy, and M. T. Philipp. 2000. Transcriptional regulation in spirochetes. J. Mol. Microbiol. Biotechnol. 2473-481. [PubMed] [Google Scholar]
- 32.Jain, S. K., M. Paul-Satyaseela, G. Lamichhane, K. S. Kim, and W. R. Bishai. 2006. Mycobacterium tuberculosis invasion and traversal across an in vitro human blood-brain barrier as a pathogenic mechanism for central nervous system tuberculosis. J. Infect. Dis. 1931287-1295. [DOI] [PubMed] [Google Scholar]
- 33.Joseph, B., K. Przybilla, C. Stuhler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klempner, M. S., R. Noring, and R. A. Rogers. 1993. Invasion of human skin fibroblasts by the Lyme disease spirochete, Borrelia burgdorferi. J. Infect. Dis. 1671074-1081. [DOI] [PubMed] [Google Scholar]
- 35.Kurtti, T. J., U. G. Munderloh, D. E. Krueger, R. C. Johnson, and T. G. Schwan. 1993. Adhesion to and invasion of cultured tick (Acarina: Ixodidae) cells by Borrelia burgdorferi (Spirochaetales: Spirochaetaceae) and maintenance of infectivity. J. Med. Entomol. 30586-596. [DOI] [PubMed] [Google Scholar]
- 36.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang, F. T., J. Yan, M. L. Mbow, S. L. Sviat, R. D. Gilmore, M. Mamula, and E. Fikrig. 2004. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 725759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 40.Livengood, J. A., and R. D. Gilmore, Jr. 2006. Invasion of human neuronal and glial cells by an infectious strain of Borrelia burgdorferi. Microbes Infect. 82832-2840. [DOI] [PubMed] [Google Scholar]
- 41.Ma, Y., A. Sturrock, and J. J. Weis. 1991. Intracellular localization of Borrelia burgdorferi within human endothelial cells. Infect. Immun. 59671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montgomery, R. R., M. H. Nathanson, and S. E. Malawista. 1993. The fate of Borrelia burgdorferi, the agent for Lyme disease, in mouse macrophages. Destruction, survival, recovery. J. Immunol. 150909-915. [PubMed] [Google Scholar]
- 43.Narasimhan, S., M. J. Caimano, F. T. Liang, F. Santiago, M. Laskowski, M. T. Philipp, A. R. Pachner, J. D. Radolf, and E. Fikrig. 2003. Borrelia burgdorferi transcriptome in the central nervous system of non-human primates. Proc. Natl. Acad. Sci. USA 10015953-15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowalk, A. J., R. D. Gilmore, Jr., and J. A. Carroll. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect. Immun. 743864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohnishi, J., J. Piesman, and A. M. de Silva. 2001. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl. Acad. Sci. USA 98670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ojaimi, C., C. Brooks, D. Akins, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katonah, J. Radolf, M. Caimano, J. Skare, K. Swingle, S. Sims, and I. Schwartz. 2002. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 358165-177. [DOI] [PubMed] [Google Scholar]
- 47.Orihuela, C. J., J. N. Radin, J. E. Sublett, G. Gao, D. Kaushal, and E. I. Tuomanen. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 725582-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pachner, A. R., D. Cadavid, G. Shu, D. Dail, S. Pachner, E. Hodzic, and S. W. Barthold. 2001. Central and peripheral nervous system infection, immunity, and inflammation in the NHP model of Lyme borreliosis. Ann. Neurol. 50330-338. [PubMed] [Google Scholar]
- 49.Philipp, M. T., M. K. Aydintug, R. P. Bohm, Jr., F. B. Cogswell, V. A. Dennis, H. N. Lanners, R. C. Lowrie, Jr., E. D. Roberts, M. D. Conway, M. Karacorlu, G. A. Peyman, D. J. Gubler, B. J. Johnson, J. Piesman, and Y. Gu. 1993. Early and early disseminated phases of Lyme disease in the rhesus monkey: a model for infection in humans. Infect. Immun. 613047-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Probert, W. S., and B. J. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 301003-1015. [DOI] [PubMed] [Google Scholar]
- 51.Ramamoorthy, R., and D. Scholl-Meeker. 2001. Borrelia burgdorferi proteins whose expression is similarly affected by culture temperature and pH. Infect. Immun. 692739-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramesh, G., A. L. Alvarez, E. D. Roberts, V. A. Dennis, B. L. Lasater, X. Alvarez, and M. T. Philipp. 2003. Pathogenesis of Lyme neuroborreliosis: Borrelia burgdorferi lipoproteins induce both proliferation and apoptosis in rhesus monkey astrocytes. Eur. J. Immunol. 332539-2550. [DOI] [PubMed] [Google Scholar]
- 53.Ramesh, G., and M. T. Philipp. 2005. Pathogenesis of Lyme neuroborreliosis: mitogen-activated protein kinases Erk1, Erk2, and p38 in the response of astrocytes to Borrelia burgdorferi lipoproteins. Neurosci. Lett. 384112-116. [DOI] [PubMed] [Google Scholar]
- 54.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 991562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts, E. D., R. P. Bohm, Jr., F. B. Cogswell, H. N. Lanners, R. C. Lowrie, Jr., L. Povinelli, J. Piesman, and M. T. Philipp. 1995. Chronic Lyme disease in the rhesus monkey. Lab Investig. 72146-160. [PubMed] [Google Scholar]
- 56.Roberts, E. D., R. P. Bohm, Jr., R. C. Lowrie, Jr., G. Habicht, L. Katona, J. Piesman, and M. T. Philipp. 1998. Pathogenesis of Lyme neuroborreliosis in the rhesus monkey: the early disseminated and chronic phases of disease in the peripheral nervous system. J. Infect. Dis. 178722-732. [DOI] [PubMed] [Google Scholar]
- 57.Sadziene, A., D. D. Thomas, V. G. Bundoc, S. C. Holt, and A. G. Barbour. 1991. A flagella-less mutant of Borrelia burgdorferi. Structural, molecular, and in vitro functional characterization. J. Clin. Investig. 8882-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwan, T. G., W. Burgdorfer, M. E. Schrumpf, and R. H. Karstens. 1988. The urinary bladder, a consistent source of Borrelia burgdorferi in experimentally infected white-footed mice (Peromyscus leucopus). J. Clin. Microbiol. 26893-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 922909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345115-125. [DOI] [PubMed] [Google Scholar]
- 61.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 634535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szczepanski, A., M. B. Furie, J. L. Benach, B. P. Lane, and H. B. Fleit. 1990. Interaction between Borrelia burgdorferi and endothelium in vitro. J. Clin. Investig. 851637-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teng, C. H., M. Cai, S. Shin, Y. Xie, K. J. Kim, N. A. Khan, F. Di Cello, and K. S. Kim. 2005. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect. Immun. 732923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas, D. D., D. Cadavid, and A. G. Barbour. 1994. Differential association of Borrelia species with cultured neural cells. J. Infect. Dis. 169445-448. [DOI] [PubMed] [Google Scholar]
- 65.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 725419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wormser, G. P., R. J. Dattwyler, E. D. Shapiro, J. J. Halperin, A. C. Steere, M. S. Klempner, P. J. Krause, J. S. Bakken, F. Strle, G. Stanek, L. Bockenstedt, D. Fish, J. S. Dumler, and R. B. Nadelman. 2006. The clinical assessment, treatment, and prevention of Lyme disease, human granulocytic anaplasmosis, and babesiosis: clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 431089-1134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.